Abstract
The beet cyst nematode Heterodera schachtii induces a feeding site, called syncytium, in roots of host plants. In Arabidopsis, one of the genes whose expression is strongly induced in these structures is Pdf2.1 which codes for an antimicrobial plant defensin. Arabidopsis has 13 plant defensin genes. Besides Pdf2.1, the Pdf2.2 and Pdf2.3 genes were strongly expressed in syncytia and therefore the expression of all three Pdf genes was studied in detail. The promoter of the Pdf2.1 gene turned out to be an interesting candidate to drive a syncytium-specific expression of foreign genes as RT-PCR showed that apart from the feeding site it was only expressed in siliques (seeds). The Pdf2.2 and Pdf2.3 genes were in addition expressed in seedlings, roots, leaves, stems, and flowers. These results were supported by the analysis of promoter::GUS lines. After infection with H. schachtii all GUS lines showed a strong staining in syncytia at 5 and 15 dpi. This expression pattern was confirmed by in situ RT-PCR.
Keywords: Plant defensin, Antimicrobial peptide, Syncytium, Heterodera schachtii, Arabidopsis, Roots
Abbreviations: Dpi, days post infection
Highlights
► 3 plant defensin genes are strongly expressed in syncytia. ► Pdf2.1 is expressed in syncytia and seeds. ► The Pdf2.1 promoter is useful for the expression of foreign genes in syncytia.
1. Introduction
Nematodes are a group of animals which include free-living bacterial feeders such as the intensively studied worm Caenorhabditis elegans as well as many pathogens of animals and plants. Obligate biotrophic plant-parasitic nematodes attack mainly the roots of many plant species, often causing severe damage to crop plants either directly or as virus vectors. Some of the economically most important species are the cyst and root-knot nematodes within the family Heteroderidae which enter the plant roots as second stage juveniles (J2) and establish specialized feeding structures. The worldwide crop losses due to nematode damage have been estimated at over $100 billion per year [1]. Root-knot nematodes (genus Meloidogyne) induce a feeding structure which is composed of several giant cells [2]. The feeding structure of cyst nematodes (genera Heterodera and Globodera) is a syncytium [3], which is initiated from a single root cell and then expands by incorporating up to several hundred neighbouring cells by local cell wall dissolution. The nematodes feed only from the syncytium which is thus a severe nutrient sink for the plant. Adult male cyst nematodes leave the root to mate with females. The fertilized female cyst nematode continues to feed but dies once egg development is completed, leaving several hundred eggs contained within its enlarged body. It subsequently hardens to form a cyst, which protects the eggs until infective J2 hatch in favourable conditions.
The development of the syncytium from the initial syncytial cell inside the central cylinder is probably initiated through secretions from the nematode and a coordinated expression of plant genes. Such plant genes are, for instance, expansins and cellulases that are important for the degradation of cell walls leading to incorporation of new cells into the growing syncytium [4–6]. But syncytial cell walls also undergo modifications which require the synthesis of new cell wall polysaccharides. This might involve the myo-inositol oxygenase pathway as all 4 Arabidopsis genes that code for myo-inositol oxygenase are strongly expressed in syncytia [7]. Protuberances are produced at the interface between syncytia and xylem vessels and these are thought to be important for the transport of water and solutes [3]. The cells that are incorporated into the syncytium undergo drastic changes in structure and activity. This includes fragmentation of the central vacuole into many small ones, accumulation of mitochondria and ribosomes in a dense granular cytoplasm and a proliferation of the endoplasmic reticulum [8,9]. To cope with this high metabolic activity, nuclei and nucleoli are enlarged and contain endoreduplicated DNA [10].
The sugar beet cyst nematode Heterodera schachtii completes its life cycle on Arabidopsis roots in vitro within six weeks [11] and this interaction has been established as a model system. The translucent Arabidopsis roots growing on artificial media facilitate the study of the development of this and other nematode species inside the root [12]. Using this system, we have recently analyzed the transcriptome of syncytia induced by H. schachtii at 5 and 15 days post infection (dpi) [13]. Our results revealed that the Pdf2.1 gene, coding for a plant defensin, was one of the genes that were strongly upregulated in syncytia as compared to control root sections from uninfected plants. This was in contrast to the majority of plant defense-related genes which were not expressed or even downregulated in syncytia [13].
Plant defensins are a group of antimicrobial peptides with a molecular weight in the range of 5 kDa that have been found in virtually all plants [14,15]. Peptides with a similar structure have also been found in a variety of animals, including humans [16] as well as in fungi [17]. All defensins are relatively small, basic peptides with a three-dimensional structure comprised of a triple-stranded β-sheet with a parallel α-helix [18–20]. In plant defensins this structure is usually stabilized by 4 disulfide bridges. All defensins have repeatedly been shown to have antimicrobial activity in vitro [21,22] and anti-insect activities [23]. In addition, especially those plant defensins that were originally called γ-thionins, inhibit α-amylase activity [24] and protein synthesis [25].
Arabidopsis has 13 Pdf genes that can be divided into two groups [14] and 10 of these genes are represented on the Arabidopsis GeneChip. Three genes of group 1 (Pdf1.2a, Pdf1.2b, and Pdf1.2c) are closely related and encode the same defensin peptide. Pdf1.2 is generally regarded as a marker gene for the pathogen specific induction through the ethylene and jasmonic acid pathways [26]. The other Pdf genes are constitutively expressed in certain plant tissues (Fig. S1) [27,28]. Group 1 Pdf genes, (Pdf1.1, Pdf1.2a, Pdf1.2b, Pdf1.2c, Pdf1.3 and to some extent Pdf1.4) are induced in the non-host response of Arabidopsis to the barley powdery mildew fungus [29]. Overexpression of PDF1.1 resulted in enhanced resistance of Arabidopsis plants against Cercospora beticola [30]. In addition to a role in plant resistance, plant defensins have also been shown to be involved in conferring zinc resistance in the zinc hyper-accumulating plant Arabidopsis halleri [31]. Furthermore, defensin-like peptides were identified as the male determinant of self-incompatibility in Brassica [32].
Cyst nematodes are a serious problem for a range of important crops and researchers are therefore testing several transgenic approaches to enhance their resistance [33] by targeting the nematode feeding site. This includes for instance the expression of protease inhibitors or RNAi for the downregulation of genes whose expression is vital for the development of syncytia. In many cases the CaMV 35S promoter is used which is active in most tissues of many plant species and might therefore lead to unwanted side effects. There is thus a need for specific promoters that are as specific as possible for nematode feeding sites. The strong upregulation of Pdf2.1 in syncytia according to the GeneChip data indicated that the Pdf2.1 promoter might be useful to drive the expression of transgenes in syncytia. Using different techniques we have therefore studied in detail the expression of Pdf2.1 and the closely related genes Pdf2.2 and Pdf2.3, which are expressed in syncytia and control root sections.
2. Results
A transcriptome analysis of syncytia induced by H. schachtii in Arabidopsis roots [13] revealed several Pdf genes that were strongly expressed in syncytia. Analysis of these data (Table 1) showed that from the 10 Pdf genes that were represented on the GeneChip (from a total of 13 Pdf genes in the Arabidopsis genome), three were strongly expressed in syncytia while the other Pdf genes were expressed at a very low level in both syncytia and in control root sections. The three genes with a strong expression in syncytia were Pdf2.1, Pdf2.2, and Pdf2.3. Pdf2.2 and Pdf2.3 were also strongly expressed in control root sections. Pdf2.2 had a significant fourfold induction in syncytia while Pdf2.3 was not significantly induced in syncytia (1.15fold). Pdf2.1, on the other hand, had a very low level of expression in control root sections and was very strongly induced in syncytia (200 fold). The strong induction of Pdf2.1 in syncytia made its promoter an interesting candidate to drive specific expression of foreign genes in nematode feeding sites. Therefore the expression of all three Pdf genes with strong expression in syncytia was studied in detail using RT-PCR, promoter::GUS lines and in situ RT-PCR.
Table 1.
Expression of defensin genes in syncytia and control root segments.
| ID | Gene | Control | Syncytium (5 + 15 dpi) | Control vs syncytium | q-value |
|---|---|---|---|---|---|
| At1G75830 | Pdf1.1 | 2.7 | 2.8 | 0.2 | 0.33 |
| At5G44420 | Pdf1.2a | 2.5 | 3.0 | 0.5a | 0.00 |
| At2G26020 | Pdf1.2b | 3.2 | 3.1 | −0.1 | 0.49 |
| At1G19610 | Pdf1.4 | 3.2 | 3.2 | 0.0 | 0.89 |
| At2G02120 | Pdf2.1 | 3.3 | 11.0 | 7.7a | 8.99E + 04 |
| At2G02100 | Pdf2.2 | 11.1 | 13.2 | 2.0a | 0.00 |
| At2G02130 | Pdf2.3 | 12.2 | 12.4 | 0.2 | 0.33 |
| At1G61070 | Pdf2.4 | 3.9 | 3.9 | 0.0 | 0.89 |
| At5G63660 | Pdf2.5 | 3.2 | 2.3 | −0.9 | 0.00 |
| At2G02140 | Pdf2.6 | 3.4 | 3.4 | 0.0 | 0.89 |
Data for microaspirated syncytia at 5 dpi and 15 dpi were combined and compared with control roots (elongation zone without root tip was used as control). All expression values have been normalized and are on a log2 scale (third and fourth column) and the differences (fold changes) between the pairwise samples displayed (fifth column) are accordingly normalized log2 ratios (see Methods section for details). q-values indicate significance after correction for multiple testing controlling the False Discovery Rate.
Indicates significant up- or downregulation (false discovery rate < 5%).
2.1. RT-PCR
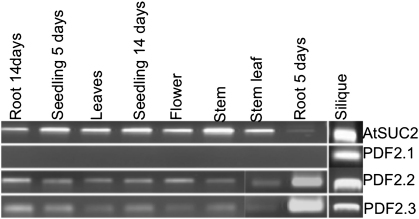
The expression of Pdf2.1, Pdf2.2, and Pdf2.3 in the plant was studied by RT-PCR. RNA was isolated from flowers, siliques, stems, roots and leaves and RT-PCR was performed with specific primers for all three genes as described in the Methods section. Expression of Pdf2.1 was only detected in siliques while expression of Pdf2.2 and Pdf2.3 genes was detected in all analysed plant organs with the strongest expression in 5 day old roots (Fig. 1). Primers for the Suc2 gene were used as a control. GeneChip expression data available in Genevestigator [34] also show that the Pdf genes are expressed in different organs and tissues (Table S1). According to those data Pdf2.1 is especially expressed in seeds while Pdf2.2 and Pdf2.3 are also expressed in roots and leaves which is in agreement with our results.
Fig. 1.
RT-PCR analysis of Pdf2.1, Pdf2.2 and Pdf2.3. Semi quantitative RT-PCR analysis of Pdf2.1, Pdf2.2, and Pdf2.3 in different Arabidopsis tissues. Pdf2.1 was only detected in siliques, whereas transcripts of Pdf2.2 and Pdf2.3 were detected in all plant tissues. Expression of both Pdf2.2 and Pdf2.3 was most intense in 5 days old root tissues. The Suc2 gene was used as a control.
2.2. Promoter::GUS analysis
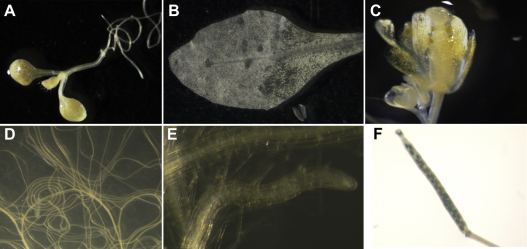
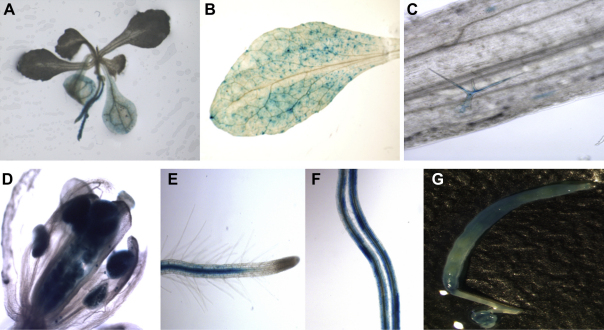
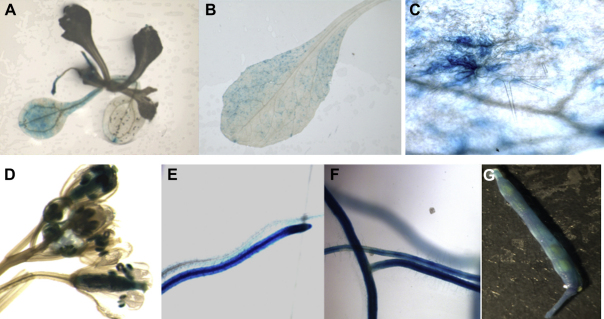
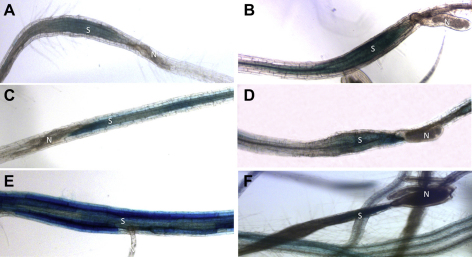
We produced promoter::GUS lines for Pdf2.1, Pdf2.2, and Pdf2.3. We selected representative homozygous lines and general GUS expression for Pdf2.1, Pdf2.2 and Pdf2.3 was assessed in different plant tissues of non-infected plants e.g. seedlings, leaves, roots, flowers and siliques. For Pdf2.1 (Fig. 2), expression in non-infected plants was restricted to siliques, where expression was observed in seeds and at the base (Fig. 2F). Only occasionally we observed a faint GUS expression in leaves and roots (data not shown). For Pdf2.2, (Fig. 3) there was GUS expression in tissues of non-infected plants which was especially strong in roots (Fig. 3E–F). In seedlings, GUS expression was observed in radicles and in cotyledons (Fig. 3A). Similarly, GUS expression driven by the Pdf2.2 promoter was observed in leaves, trichomes, flowers, roots, and siliques. Within root tissues, expression was mostly restricted to the central cylinder and there was no GUS expression observed in the root tip (Fig. 3E). The expression pattern of Pdf2.3 (Fig. 4) in non-infected tissues was similar to that of Pdf2.2, except that there was almost no expression in trichomes (Fig. 4C) but intense GUS staining in the whole root including root tips (Fig. 4E–F). Within root tissues, GUS expression was more intense in the central cylinder and in root tips (Fig. 4F). Twelve day old promoter::GUS lines of Pdf2.1, Pdf2.2, and Pdf2.3 were also infected with H. schachtii larvae. Roots of the infected plants were stained at 5 and 15 dpi (Fig. 5). Syncytia showed a strong staining at 5 dpi for all three genes (Fig. 5A, C, E). In the case of Pdf2.2 and Pdf2.3 staining was also detected in root tissues outside syncytia (Fig. 5C and E). This staining was very strong in the case of Pdf2.3 (Fig. 5E). At 15 dpi syncytia for all three genes also showed a GUS staining, and again, staining for Pdf2.2 and Pdf2.3 was also detected in root tissues outside syncytia (Fig. 5B, D, F).
Fig. 2.
GUS expression analysis pattern of the Pdf 2.1 promoter. No expression was observed for Pdf2.1 in seedlings (A), leaves (B), flowers (C) and roots (D, E). However, intense GUS expression for Pdf2.1 was observed in siliques of these plants (F).
Fig. 3.
GUS expression analysis pattern of the Pdf 2.2 promoter. GUS expression driven by the promoter of Pdf2.2 in seedlings (cotyledons and roots) (A), leaves (B), trichomes (C), flowers (D), roots (E, F) and siliques (G) of transgenic Arabidopsis plants was observed. Within root tissues, there was no expression in the elongation zone and root tip.
Fig. 4.
GUS expression analysis pattern of the Pdf 2.3 promoter. GUS expression driven by the promoter of Pdf2.3 in seedlings (cotyledons and roots) (A), leaves (B), flowers (D), roots (E, F), and siliques (G) of transgenic Arabidopsis plants was observed. In contrast to Pdf2.2, there was intense GUS expression in the elongation zone and the root tips. Trichomes did not show any staining for Pdf2.3 (C).
Fig. 5.
Expression of Pdf2.1, Pdf2.2, and Pdf2.3 promoter::GUS fusions in syncytia induced by H. schachtii. GUS expression driven by the Pdf2.1 promoter in nematode feeding sites at 5 dpi and 15 dpi (A, B). Root tissues surrounding the syncytium did not show any GUS expression. For Pdf2.2, there was also GUS expression in syncytia at 5 dpi and 15 dpi (C, D). However, this expression was not limited to the syncytium but was also found in surrounding tissues. For Pdf2.3, there was also GUS expression in syncytia as well as in surrounding tissues at 5 dpi and 15 dpi (E, F). This expression, especially at 5 dpi, was more intense in surrounding tissues than in syncytia. N, nematode; S, syncytium
2.3. Localization of Pdf gene expression by in situ RT-PCR
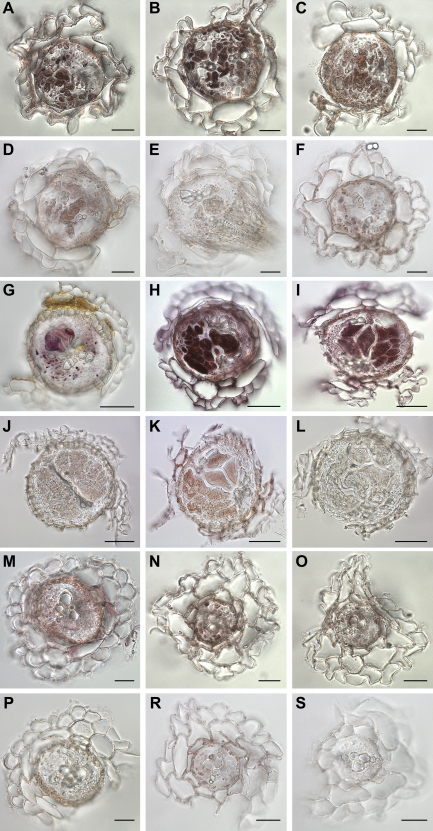
In situ RT-PCR analysis was performed with syncytia at 5 and 15 dpi and uninfected control roots with specific primers for Pdf2.1, Pdf2.2, and Pdf2.3 (Fig. 6). Expression of all three genes was clearly detected in syncytia at 5 dpi as well as 15 dpi. Transcripts of Pdf2.1 were restricted to the syncytium (Fig. 6A and G), while transcripts for Pdf2.2 (Fig. 6B and H) and Pdf2.3 (Fig. 6C and I) were also detected outside the feeding site within the central cylinder which is in agreement to the results obtained with promoter::GUS lines. In uninfected roots, transcripts for Pdf2.2 (Fig. 6N) and Pdf2.3 (Fig. 6O) accumulated in cells of the central cylinder, while expression of Pdf2.1 in these root sections was only very faint inside the central cylinder (Fig. 6M). Control reactions that were performed without polymerase always lacked any specific staining (Fig. 6D–F, J–L, P–S).
Fig. 6.
In situ RT-PCR. In situ RT-PCR analysis of Pdf2.1, Pdf2.2, and Pdf2.3 on sections of syncytia induced by Heterodera schachtii in roots of Arabidopsis as well as uninfected control roots. A, B and C – transcripts of Pdf2.1, Pdf2.2, and Pdf2.3, respectively, accumulate in the 5 dpi syncytia. D, E and F – control sections of 5 dpi syncytia show lack of the specific signal of Pdf2.1, Pdf2.2, and Pdf2.3, respectively. G, H and I – specific staining of Pdf2.1, Pdf2.2, and Pdf2.3, respectively, in 15 dpi syncytia. J, K and L – control sections of 15 dpi syncytia show lack of the specific staining of Pdf2.1, Pdf2.2, and Pdf2.3, respectively. M – very weak staining of Pdf2.1 within the central cylinder of the uninfected root. N – transcripts of Pdf2.2 accumulate in cells of the central cylinder of the uninfected root. O – transcripts of Pdf2.3 accumulate in cells of the central cylinder of the uninfected root. P, R and S – control sections of uninfected control roots show lack of the specific signal of Pdf2.1, Pdf2.2 and Pdf2.3, respectively. A–F and M–S scale bar = 20 μm; G–L scale bar = 50 μm.
For gene expression in roots data are also available from a transcriptome analysis of FACS purified root cells (Fig. S1) [35]. These data reveal a very weak expression of Pdf2.1 especially in older root tissues and a strong expression of Pdf2.2 and Pdf2.3 genes. Expression of Pdf2.3 is stronger in root tips and the root elongation zone as compared to Pdf2.2. These data are in agreement with our studies.
3. Discussion
Arabidopsis contains 13 plant defensin genes, some of which are induced by infection with fungi [27,29,30,36]. Pdf1.2, actually 3 very closely related genes that code for the same peptide, is frequently used as a marker gene for the ethylene and jasmonic acid pathway [26]. We have recently shown that this gene and other defense-related genes which are regulated through jasmonic acid, ethylene, or salicylic acid, are not expressed in syncytia [13]. It has, however, been reported that PR-3 and PR-4 genes, which are regulated through jasmonic acid, are induced in leaves of Arabidopsis plants infected with H. schachtii [37]. It is not known if this induction extends to other jasmonic acid regulated genes including Pdf1.2. In addition to Pdf1.2 also Pdf1.1, Pdf1.3, and to a lesser extent Pdf1.4, were induced during non-host interaction of Arabidopsis plants with the powdery mildew fungus Blumeria graminis f. sp. hordei. This induction is probably mediated by jasmonic acid for Pdf1.2, Pdf1.3, and Pdf1.4, while Pdf1.1 is only slightly inducible by methyle jasmonate as shown by experiments using treatment of the plants with methyle jasmonate [29]. The response of Pdf1.2 to jasmonic acid and ethylene is mediated through the transcription factor ORA59 which binds to GCC boxes in the Pdf1.2 promoter [38].
We found that expression of the Pdf2.1 gene was strongly upregulated in syncytia induced by the beet cyst nematode H. schachtii in Arabidopsis roots [13]. In addition, Pdf2.2 and Pdf2.3 were also strongly expressed in syncytia but contrary to the Pdf2.1 gene, both genes have a strong expression in uninfected roots. The ethylene/jasmonic acid marker gene Pdf1.2, however, was not induced in syncytia as were other genes that respond to ethylene, jasmonic acid, and salicylic acid [13]. The Pdf2.1, Pdf2.2, and Pdf2.3 genes, which are the focus of this publication, as well as Pdf2.4, Pdf2.5, and Pdf2.6 are not inducible by ethylene, jasmonic acid, and salicylic acid according to Genevestigator (data not shown).
Since Pdf2.1 was among the most strongly upregulated genes in syncytia, we were interested to use the promoter of this gene for the expression of foreign genes in feeding sites. We therefore studied its expression as well as the expression of Pdf2.2 and Pdf2.3 in detail. RT-PCR and GUS analysis showed that the Pdf2.2 and Pdf2.3 genes were expressed throughout the plant with the strongest expression in roots and siliques while expression of Pdf2.1 was restricted to siliques (especially seeds). GUS lines and in situ RT-PCR also demonstrated the expression of all three genes in syncytia. These results confirmed that the promoter of the Pdf2.1 gene can be used for a strong and specific expression in syncytia. Besides feeding sites, the Pfd2.1 gene was only expressed in seeds, however, this disadvantage has to be accepted as no gene is known to be exclusively expressed in syncytia but nowhere else in the plant. Thus, the promoter of the Pfd2.1 gene is a valuable tool for engineering plants with resistance against cyst nematodes which would involve a strong and specific expression in syncytia. One might envision that such resistance could be achieved by overexpression of genes that are downregulated in syncytia. We have identified a large number of such genes as compared to control root sections [13] which could be tested in such an approach. A strong and specific expression in syncytia is also needed if miRNAs [39] would be used to downregulate genes that are essential for the function and development of syncytia. Also a large number of candidate genes in this category have been identified in our previous transcriptome analysis of syncytia. We have already used this promoter for a specific downregulation of some of these genes in the syncytium using artificial miRNAs (Siddique and Bohlmann, unpublished results; Ali and Bohlmann, unpublished results). Furthermore, resistance against nematodes might also be achieved by expressing proteins or other compounds that lead to the death of the nematodes if taken up from the feeding sites [33]. Examples for the latter include proteinase inhibitors and dsRNA or miRNAs which are targeted against essential nematode genes. For all these approaches promoters that direct a strong and specific gene expression in feeding sites would be useful.
In contrast to cyst nematodes, root-knot nematodes, such as Meloidogyne incognita, induce several giant cells embedded in the gall tissue. The transcriptome of giant cells, induced by M. incognita in Arabidopsis roots, which were cut out from infected roots has been studied using CATMA microarrays [40]. The available data show that Pdf2.1 is, in contrast to syncytia, not induced in giant cells. No data are available for Pdf2.2 and Pdf2.3 but also the other Pdf genes that can be studied using CATMA microarrays did not show any induction in giant cells.
Cyst nematodes and root-knot nematodes live only from nutrients that are taken up from syncytia or giant cells, respectively, with the help of the stylet and specialized structures, called feeding tubes, that the nematodes produce within the feeding sites. Root-knot nematodes seem to have a higher size exclusion limit than the cyst nematodes. According to Urwin et al. [41], H. schachtii was able to take up the cystatin Oc-IDD86 (11.2 kDa) but not GFP (28 kDa). M. incognita, however, was able to take up GFP. Böckenhoff et al. [42] used microinjection of fluorescent probes to determine the size exclusion limit of H. schachtii that was estimated to be between 20 and 40 kDa. Taking these data together, this means that the plant defensins PDF2.1, PDF2.2, and PDF2.3 (approximately 5 kDa), which seem to be produced in large amounts in syncytia, could be easily taken up by the cyst nematodes. This would also indicate that these defensins have no toxic effect on the nematode and that they might rather be a rich source of nitrogen and sulfur for them. However, it cannot be excluded that the plant defensins could be retained in subcellular compartments that are not accessed by the feeding nematode or could be secreted to the apoplast outside the syncytium. To resolve these different possibilities would require the use of specific antibodies.
4. Conclusion
Our expression analysis of the Arabidopsis plant defensin genes Pdf2.1, Pdf2.2, and Pdf2.3 showed that they are strongly expressed in syncytia with Pdf2.1 being one of the most strongly induced genes. Our results also demonstrated that the Pdf2.1 promoter could be used for syncytium-specific expression of nematicidal products or gene products that would inhibit the development of the syncytium.
5. Methods
5.1. Plant cultivation
Arabidopsis seeds were surface-sterilized for 20 min in 6% (w/v) sodium hypochlorite and subsequently washed three times with sterile water. Seeds were placed into sterile Petri dishes (9 cm) on a modified Knop medium with 2% sucrose [11]. Seeds were grown in a growth chamber at 25 °C in a 16 h light and 8 h dark cycle.
5.2. Statistical analysis of microarray data
Affymetrix CEL files were analyzed using packages of the Bioconductor suite (www.bioconductor.org). Details are provided in Szakasits et al. [13]. For the statistical tests, individual gene variances have been moderated using an Empirical Bayes approach as described in Siddique et al. [7]. Tests were restricted to the subset of 10 Pdf genes (from a total of 13) that are included on the GeneChip. This considerably increases the statistical power of the testing procedure as it reduces the necessary correction for massive multiple testing.
5.3. RT-PCR
Total RNA of different plant tissues was extracted with the RNeasy Plant Mini Kit (Qiagen). Superscript III (Invitrogen) was used to transcribe total RNA into cDNA. Afterwards, 1 μl of cDNA was used to perform RT-PCR with forward and reverse primers as given in Table 2. AtSUC2 was used as control. Primers were designed by aligning all PDF genes using ClustalW and choosing the regions where these genes show maximum dissimilarity (see Fig S2).
Table 2.
Primer pairs used for PCR.
| Forward primer | Reverse primer | |
|---|---|---|
| Pdf2.1 | AAATGCGTGAGCGATACA | ACACACTAAACACGCATAC |
| Pdf2.2 | ACATGCGTGAGTGCATCA | TAGCTTTGTTATCAGAACATAGATTTT |
| Pdf2.3 | CCATGTGTGAGCACACAC | GACTCCGGTTATTAAAAACTTT |
| AtSuc2 | GCCTCTAAGAAGCTTTACAACGAC | CCCATAGTAGCTTTGAAGGCA |
| PromPdf2.1 | ATTAGGGTACCTTTGGAGTGACAGATTC | AGAGAACTCCATGGTTGGAGAAAGAGAA |
| PromPdf2.2 | GAGTGGTACCAGACACAAATCTCACTAGAT | AGAGAGCTCCATGGCAAGAGAGATAAAGA |
| PromPdf2.3 | TCAAGGTACCAATATGAAGTATAAAAACGTTT | AGAGAGCTCCATGGTTGAGAGGGATAGA |
5.4. GUS reporter analysis
Promoter regions (approx. 1000 bp) upstream the start codon of Pdf2.1, Pdf2.2, and Pdf2.3 were amplified using Arabidopsis genomic DNA as template. Primer pairs are given in Table 2. Forward and reverse primers included restriction sites for Kpn1 and Nco1, respectively. These restriction sites were subsequently used for cloning into pPZP3425 [43]. Promoter::GUS constructs were introduced into Agrobacterium tumefaciens (GV3101), which was then used for transformation of Arabidopsis Col-0 using the floral dip method [44]. Transformed plants were selected on MS medium containing 50 μg ml−1 kanamycin.
For analysis of GUS expression, Arabidopsis seeds were surface-sterilized for 5 min in 75% ethanol followed by 5 min in 10% (v:v) commercial bleach and subsequently washed three times in sterile water. Plants were grown on Knop medium [11] and infected with nematodes as described below (see nematode infection). The GUS expression was analysed at 5 and 15 dpi. For GUS expression analysis of aerial plant parts, plants were grown in soil in a climate chamber under long day light conditions. Plant tissues were separated and submerged in 100 mmol NaPO4 buffer (pH 7.0) containing 10 mmol EDTA, 0.01% Triton X-100, 0.5 mmol K3(Fe(CN)6), 0.5 mmol K4(Fe(CN)6) and 1 mg ml−1 5-bromo-4-chloro-3-indolyl glucuronide. Tissues were vacuum infiltrated for 5 min and then incubated in the dark at 37 °C for 5–6 h for Pdf2.2 and Pdf2.3 and overnight in case of Pdf2.1. For analysis of GUS expression in syncytia, roots were always incubated for 8 h.
5.5. In situ RT-PCR
This analysis was carried out according to the protocols described in Koltai and Bird [45] and Urbanczyk-Wlochniak et al. [46]. 5 and 15 dpi syncytia as well as 5 dpi uninfected control roots were dissected from the plates and immediately put into cold fixative (63% ethanol, v/v; 2% formalin, v/v). After a short incubation in vacuum they were placed on a horizontal shaker at 4 °C for 2 days. Root samples were washed and embedded in 4% low-melting agarose and 20 μm thick sections were prepared using a vibratom (VT100, Leica, http://www.leica.com/). Subsequently, DNase digestion and RT-PCR with digoxigenin-labelled dUTP were carried out. For the staining reaction nitro blue tetrazolium substrate (NBT/BCIP) was used and sections were photographed using an inverted microscope (Axiovert 200M, Zeiss, http://www.zeiss.com/) with an integrated camera (AxioCam MRc5, Zeiss, http://www.zeiss.com/). For a detailed description see [4].
5.6. Nematode infection
H. schachtii cysts were harvested from in vitro stock cultures on mustard (Sinapis alba cv. Albatros) roots growing on Knop medium supplemented with 2% sucrose [11]. Hatching of J2 was stimulated by adding 3 mM ZnCl2. The J2 were resuspended in 0.5% (w/v) gelrite (Duchefa, Haarlem The Netherlands) and 12 day old Arabidopsis roots were inoculated under sterile conditions with approximately 80–90 J2 per plant.
Acknowledgements
We appreciate the excellent technical assistance of Sabine Daxböck-Horvath. This research was supported by grants P16296-B06, P21067-B12, and P20471-B11 of the Austrian Science Fund (FWF). Shahid Masood Siddique was supported by Higher Education Commission (HEC) of Pakistan. DPK gratefully acknowledges support by the Vienna Science and Technology Fund (WWTF), Baxter AG, Austrian Research Centres (ARC) Seibersdorf, and the Austrian Centre of Biopharmaceutical Technology (ACBT).
Footnotes
Supplementary data associated with this article can be found in the online version, at doi:10.1016/j.plaphy.2011.07.005.
Contributor Information
Shahid Siddique, Email: shahid.siddique@boku.ac.at.
Krzysztof Wieczorek, Email: krzysztof.wieczorek@boku.ac.at.
Dagmar Szakasits, Email: dagmar.szakasits@boku.ac.at.
David P. Kreil, Email: david.kreil@boku.ac.at.
Holger Bohlmann, Email: holger.bohlmann@boku.ac.at.
Appendix. Supplementary data
References
- 1.Sasser J.N., Freckman D.W., Veech J.A., Dickson D.W. Society of Nematologists; Hyatssville: 1987. A World Perspective on Nematology: the Role of the Society, Vistas on Nematology. pp. 7–14. [Google Scholar]
- 2.Jones M.G.K., Payne H.L. Early stages of nematode-induced giant-cell formation in roots of Impatiens balsamina. Journal of Nematology. 1978;10:71–84. [PMC free article] [PubMed] [Google Scholar]
- 3.Jones M.G.K., Northcote D.H. Nematode-induced syncytium-A multinucleate transfer cell. Journal of Cell Science. 1972;10:789–809. doi: 10.1242/jcs.10.3.789. [DOI] [PubMed] [Google Scholar]
- 4.Wieczorek K., Golecki B., Gerdes L., Heinen P., Szakasits D., Durachko D.M., Cosgrove D.J., Kreil D.P., Puzio P.S., Bohlmann H., Grundler F.M. Expansins are involved in the formation of nematode-induced syncytia in roots of Arabidopsis thaliana. Plant Journal. 2006;48:98–112. doi: 10.1111/j.1365-313X.2006.02856.x. [DOI] [PubMed] [Google Scholar]
- 5.Wieczorek K., Hofmann J., Blochl A., Szakasits D., Bohlmann H., Grundler F.M. Arabidopsis endo-1,4-beta-glucanases are involved in the formation of root syncytia induced by Heterodera schachtii. Plant Journal. 2008;53:336–351. doi: 10.1111/j.1365-313X.2007.03340.x. [DOI] [PubMed] [Google Scholar]
- 6.Goellner M., Wang X., Davis E.L. Endo-beta-1,4-glucanase expression in compatible plant–nematode interactions. Plant Cell. 2001;13:2241–2255. doi: 10.1105/tpc.010219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siddique S., Endres S., Atkins J.M., Szakasits D., Wieczorek K., Hofmann J., Blaukopf C., Urwin P.E., Tenhaken R., Grundler F.M., Kreil D.P., Bohlmann H. Myo-inositol oxygenase genes are involved in the development of syncytia induced by Heterodera schachtii in Arabidopsis roots. New Phytologist. 2009;184:457–472. doi: 10.1111/j.1469-8137.2009.02981.x. [DOI] [PubMed] [Google Scholar]
- 8.Golinowski W., Grundler F.M.W., Sobczak M. Changes in the structure of Arabidopsis thaliana induced during development of females of the plant parasitic nematode Heterodera schachtii. Protoplasma. 1996;194:103–116. [Google Scholar]
- 9.Sobczak M., Golinowski W., Grundler F.M.W. Changes in the structure of Arabidopsis thaliana roots induced during development of males of the plant parasitic nematode Heterodera schachtii. European Journal of Plant Pathology. 1997;103:113–124. [Google Scholar]
- 10.Niebel A., De Almeida Engler J., Hemerly A., Ferreira P., Inz D., Van Montagu M., Gheysen G. Induction of cdc2a and cyc1At expression in Arabidopsis thaliana during early phases of nematode-induced feeding cell formation. Plant Journal. 1996;10:1037–1043. doi: 10.1046/j.1365-313x.1996.10061037.x. [DOI] [PubMed] [Google Scholar]
- 11.Sijmons P.C., Grundler F.M.W., von Mende N., Burrows P.R., Wyss U. Arabidopsis thaliana as a new model host for plant-parasitic nematodes. Plant Journal. 1991;1:245–254. [Google Scholar]
- 12.Wyss U., Grundler F.M.W. Heterodera schachtii and Arabidopsis thaliana, a model host-parasite interaction. Nematologica. 1992;38:488–493. [Google Scholar]
- 13.Szakasits D., Heinen P., Wieczorek K., Hofmann J., Wagner F., Kreil D.P., Sykacek P., Grundler F.M., Bohlmann H. The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant Journal. 2009;57:771–784. doi: 10.1111/j.1365-313X.2008.03727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomma B.P., Cammue B.P., Thevissen K. Plant defensins. Planta. 2002;216:193–202. doi: 10.1007/s00425-002-0902-6. [DOI] [PubMed] [Google Scholar]
- 15.Lay F.T., Anderson M.A. Defensins–components of the innate immune system in plants. Current Protein and Peptide Science. 2005;6:85–101. doi: 10.2174/1389203053027575. [DOI] [PubMed] [Google Scholar]
- 16.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nature Reviews Immunology. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 17.Mygind P.H., Fischer R.L., Schnorr K.M., Hansen M.T., Sonksen C.P., Ludvigsen S., Raventos D., Buskov S., Christensen B., De Maria L., Taboureau O., Yaver D., Elvig-Jorgensen S.G., Sorensen M.V., Christensen B.E., Kjaerulff S., Frimodt-Moller N., Lehrer R.I., Zasloff M., Kristensen H.H. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437:975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- 18.Bloch C., Jr., Patel S.U., Baud F., Zvelebil M.J., Carr M.D., Sadler P.J., Thornton J.M. 1H NMR structure of an antifungal gamma-thionin protein SIalpha1: similarity to scorpion toxins. Proteins. 1998;32:334–349. doi: 10.1002/(sici)1097-0134(19980815)32:3<334::aid-prot9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 19.Fant F., Vranken W., Broekaert W., Borremans F. Determination of the three-dimensional solution structure of Raphanus sativus antifungal protein 1 by 1H NMR. Journal of Molecular Biology. 1998;279:257–270. doi: 10.1006/jmbi.1998.1767. [DOI] [PubMed] [Google Scholar]
- 20.Bruix M., Jimenez M.A., Santoro J., Gonzalez C., Colilla F.J., Mendez E., Rico M. Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H NMR: a structural motif common to toxic arthropod proteins. Biochemistry. 1993;32:715–724. doi: 10.1021/bi00053a041. [DOI] [PubMed] [Google Scholar]
- 21.Segura A., Moreno M., Molina A., Garcia-Olmedo F. Novel defensin subfamily from spinach (Spinacia oleracea) FEBS Letters. 1998;435:159–162. doi: 10.1016/s0014-5793(98)01060-6. [DOI] [PubMed] [Google Scholar]
- 22.Moreno M., Segura A., Garcia-Olmedo F. Pseudothionin-St1, a potato peptide active against potato pathogens. European Journal of Biochemistry. 1994;223:135–139. doi: 10.1111/j.1432-1033.1994.tb18974.x. [DOI] [PubMed] [Google Scholar]
- 23.Chen K.C., Lin C.Y., Kuan C.C., Sung H.Y., Chen C.S. A novel defensin encoded by a mungbean cDNA exhibits insecticidal activity against bruchid. Journal of Agricultural and Food Chemistry. 2002;50:7258–7263. doi: 10.1021/jf020527q. [DOI] [PubMed] [Google Scholar]
- 24.Pelegrini P.B., Lay F.T., Murad A.M., Anderson M.A., Franco O.L. Novel insights on the mechanism of action of alpha-amylase inhibitors from the plant defensin family. Proteins. 2008;73:719–729. doi: 10.1002/prot.22086. [DOI] [PubMed] [Google Scholar]
- 25.Mendez E., Moreno A., Colilla F., Pelaez F., Limas G.G., Mendez R., Soriano F., Salinas M., de Haro C. Primary structure and inhibition of protein synthesis in eukaryotic cell-free system of a novel thionin, gamma-hordothionin, from barley endosperm. European Journal of Biochemistry. 1990;194:533–539. doi: 10.1111/j.1432-1033.1990.tb15649.x. [DOI] [PubMed] [Google Scholar]
- 26.Penninckx I.A., Thomma B.P., Buchala A., Metraux J.P., Broekaert W.F. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10:2103–2113. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epple P., Apel K., Bohlmann H. ESTs reveal a multigene family for plant defensins in Arabidopsis thaliana. FEBS Letters. 1997;400:168–172. doi: 10.1016/s0014-5793(96)01378-6. [DOI] [PubMed] [Google Scholar]
- 28.Thomma B.P.H.J., Broekaert W.F. Tissue-specific expression of plant defensin genes PDF2.1 and PDF2.2 in Arabidopsis thaliana. Plant Physiology and Biochemistry. 1998;36:533–537. [Google Scholar]
- 29.Zimmerli L., Stein M., Lipka V., Schulze-Lefert P., Somerville S. Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant Journal. 2004;40:633–646. doi: 10.1111/j.1365-313X.2004.02236.x. [DOI] [PubMed] [Google Scholar]
- 30.De Coninck B.M., Sels J., Venmans E., Thys W., Goderis I.J., Carron D., Delaure S.L., Cammue B.P., De Bolle M.F., Mathys J. Arabidopsis thaliana plant defensin AtPDF1.1 is involved in the plant response to biotic stress. New Phytologist. 2010;187:1075–1088. doi: 10.1111/j.1469-8137.2010.03326.x. [DOI] [PubMed] [Google Scholar]
- 31.Mirouze M., Sels J., Richard O., Czernic P., Loubet S., Jacquier A., Francois I.E., Cammue B.P., Lebrun M., Berthomieu P., Marques L. A putative novel role for plant defensins: a defensin from the zinc hyper-accumulating plant, Arabidopsis halleri, confers zinc tolerance. Plant Journal. 2006;47:329–342. doi: 10.1111/j.1365-313X.2006.02788.x. [DOI] [PubMed] [Google Scholar]
- 32.Schopfer C.R., Nasrallah M.E., Nasrallah J.B. The male determinant of self-incompatibility in Brassica. Science. 1999;286:1697–1700. doi: 10.1126/science.286.5445.1697. [DOI] [PubMed] [Google Scholar]
- 33.Fuller V.L., Lilley C.J., Urwin P.E. Nematode resistance. New Phytologist. 2008;180:27–44. doi: 10.1111/j.1469-8137.2008.02508.x. [DOI] [PubMed] [Google Scholar]
- 34.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 36.Thomma B.P., Eggermont K., Penninckx I.A., Mauch-Mani B., Vogelsang R., Cammue B.P., Broekaert W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in arabidopsis are essential for resistance to distinct microbial pathogens. Proceedings of the National Academy of Sciences U S A. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamamouch N., Li C., Seo P.J., Park C.M., Davis E.L. Expression of Arabidopsis pathogenesis-related genes during nematode infection. Molecular Plant Pathology. 2011;12:355–364. doi: 10.1111/j.1364-3703.2010.00675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pre M., Atallah M., Champion A., De Vos M., Pieterse C.M., Memelink J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiology. 2008;147:1347–1357. doi: 10.1104/pp.108.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jammes F., Lecomte P., de Almeida-Engler J., Bitton F., Martin-Magniette M.L., Renou J.P., Abad P., Favery B. Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant Journal. 2005;44:447–458. doi: 10.1111/j.1365-313X.2005.02532.x. [DOI] [PubMed] [Google Scholar]
- 41.Urwin P.E., Moller S.G., Lilley C.J., McPherson M.J., Atkinson H.J. Continual green-fluorescent protein monitoring of cauliflower mosaic virus 35S promoter activity in nematode-induced feeding cells in Arabidopsis thaliana. Molecular Plant Microbe Interactions. 1997;10:394–400. doi: 10.1094/MPMI.1997.10.3.394. [DOI] [PubMed] [Google Scholar]
- 42.Böckenhoff A., Grundler F.M.W. Studies on the nutrient uptake by the beet cyst nematode Heterodera schachtii by in situ microinjection of fluorescent probes into the feeding structures in Arabidopsis thaliana. Parasitology. 1994;109:249–254. [Google Scholar]
- 43.Szakasits D., Siddique S., Bohlmann H. An improved pPZP vector for Agrobacterium-mediated plant transformation. Plant Molecular Biology Reporter. 2007;25:115–120. [Google Scholar]
- 44.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 45.Koltai H., Bird D.M. High throughput cellular localization of specific plant mRNAs by liquid-phase in situ reverse transcription-polymerase chain reaction of tissue sections. Plant Physiology. 2000;123:1203–1212. doi: 10.1104/pp.123.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urbanczyk-Wochniak E., Filipecki M., Przybecki Z. A useful protocol for in situ RT-PCR on plant tissues. Cellular and Molecular Biology Letters. 2002;7:7–18. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.