Abstract
The nuclear export of large ribonucleoparticles is complex and requires specific transport factors. Messenger RNAs are exported through the RNA-binding protein Npl3 and the interacting export receptor Mex67. Export of large ribosomal subunits also requires Mex67; however, in this case, Mex67 binds directly to the 5S ribosomal RNA (rRNA) and does not require the Npl3 adaptor. Here, we have discovered a new function of Npl3 in mediating the export of pre-60S ribosomal subunit independently of Mex67. Npl3 interacts with the 25S rRNA, ribosomal and ribosome-associated proteins, as well as with the nuclear pore complex. Mutations in NPL3 lead to export defects of the large subunit and genetic interactions with other pre-60S export factors.
Keywords: mRNA export, ribosome transport, pre-60S export, export receptor, RNA-binding protein
Introduction
The compartmentalization between the nucleus and the cytoplasm requires an organized, selective and efficient transport of macromolecules. Transport occurs through nuclear pore complexes (NPCs), massive structures spanning the two membranes of the nuclear envelope that separates the nucleus from the cytoplasm, and is mediated by transport receptors, designated as karyopherins (Fried & Kutay, 2003; Tran & Wente, 2006). A different type of transport receptor is the heterodimeric shuttling transporter Mex67–Mtr2 (TAP–p15 or NXF1–NXT1 in higher eukaryotes) that mediates the export of messenger RNAs (mRNAs), or more precisely the messenger ribonucleoparticles (mRNPs; Kohler & Hurt, 2007). Mex67–Mtr2 requires the shuttling serine/arginine (SR)-type mRNA-binding proteins as adaptor proteins because Mex67–Mtr2 does not directly form contacts with the mRNAs (Erkmann & Kutay, 2004; Huang & Steitz, 2005). Npl3 is a well-known SR protein in yeast that functions as an adaptor protein between the mRNA and the export receptor Mex67 to mediate the nuclear export of mRNAs (Gilbert & Guthrie, 2004). Consistent with its adaptor function in mRNA export, Npl3 is essential in most yeast strain backgrounds and temperature-sensitive mutations in its gene lead to mRNA export defects (Lee et al, 1996).
Other large transport cargoes besides mRNPs are the small (40S) and the large (60S) ribosomal subunits. On maturation from a pre-90S particle in the nucleolus, the pre-60S and the pre-40S ribosomal subunit are released into the nucleus, where nuclear export is initiated. Transport of both macromolecules depends on the karyopherin Xpo1 (CRM1 in higher eukaryotes; Tschochner & Hurt, 2003; Zemp & Kutay, 2007). Xpo1 contacts the large subunit through the export-signal-containing adaptor protein Nmd3 (Gadal et al, 2001; Ho et al, 2000; Thomas & Kutay, 2003).
Interestingly, the mRNA export receptor heterodimer Mex67–Mtr2 also functions in the export of the large ribosomal subunit, by directly contacting the 5S ribosomal RNA (rRNA; Yao et al, 2007). Auxiliary transport proteins are Arx1 and Ecm1, but in contrast to the other pre-60S export receptors, deletion of either gene alone does not lead to pre-60S export defects that could be monitored by an Rpl25–GFP (green fluorescent protein) reporter (Bradatsch et al, 2007; Hung et al, 2008; Yao et al, 2010). Nevertheless, several export factors contribute to the efficient export of this large transport substrate.
Here, we show that Npl3 functions as a new nuclear export factor for the pre-60S ribosomal subunit. However, unlike the mode of action in mRNA export, Npl3 does not operate through Mex67 but is an independent mediator of the pre-60S ribosomal export that interacts with the 25S rRNA and Rpl25 on one side and the NPC protein Nup60 on the other side.
Results And Discussion
Npl3 is needed for the export of large ribosomal subunits
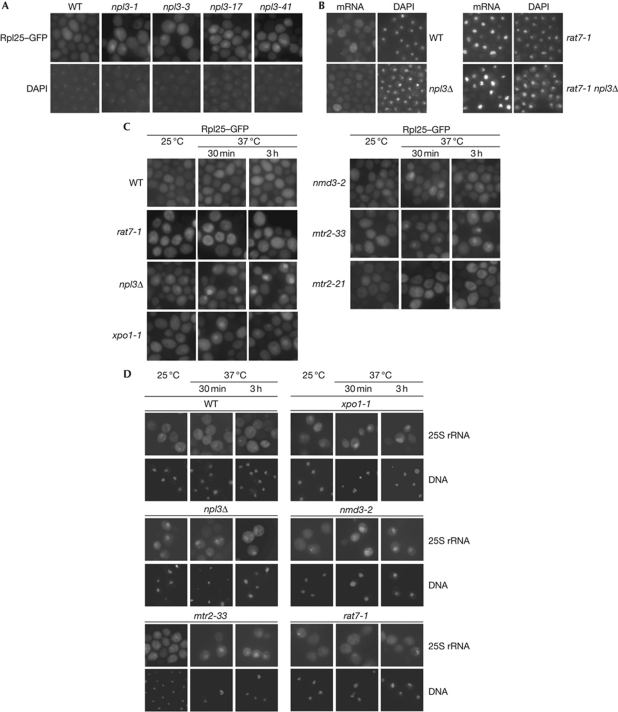
Npl3 is a shuttling mRNA-binding protein that has a known function in the nuclear export of mRNAs (Lee et al, 1996). Earlier studies showed that Npl3 also interacts with ribosomes and a specific mutation in NPL3 shows pre-60S localization defects (Stage-Zimmermann et al, 2000; Windgassen et al, 2004). Thus, we systematically tested whether Npl3 might have an extra function in transporting pre-60S ribosomal subunits from the nucleus to the cytoplasm. Several mRNA export mutants have been identified for Npl3 earlier (npl3-1, npl3-3, npl3-17 and npl3-41), all of which have amino-acid exchanges in the RNA recognition motifs, indicating that RNA binding is crucial for its transport function (Lee et al, 1996). We analysed the cellular localization of the 60S ribosomal subunit in these strains and found in all cases nuclear localization defects of the reporter protein Rpl25–GFP on a shift to the non-permissive temperature (Fig 1A). This suggests that functional Npl3 is important for the export of large ribosomal subunits, either directly or due to an indirect effect caused by the mRNA export block. Therefore, we were looking for a situation in which mRNA export is not impaired but ribosomal subunit export defects might still be visible. All Npl3 mutants were constructed in the Saccharomyces cerevisiae S288C-strain background where NPL3 is essential (Lee et al, 1996; supplementary Fig S1A online). Interestingly, the knockout of NPL3 is viable in the BY-strain background (supplementary Fig S1B online) used in the Saccharomyces genome deletion project (Winzeler et al, 1999) and shows no mRNA export defects (Fig 1B). By contrast, the nuclear porin mutant rat7-1, known for its mRNA export defects (Gorsch et al, 1995), shows a nuclear mRNA accumulation in BY cells, as does the double mutant rat7-1 npl3Δ in this background, suggesting that no assay limitations, for example, due to potential defects in generation of the poly(A) tail on the mRNAs in npl3Δ, prevent the visualization of mRNA export defects. Although these data indicate that in the BY-strain Npl3 has no essential function in mRNA export, it is important to note that Npl3 is still part of the exported mRNP. This was shown with a GFP-tagged npl3c-version (GFP–npl3RA8); that is, due to a slowed re-import into the nucleus, cytoplasmic at steady state and can therefore be used to monitor the export requirements of Npl3 (Hacker & Krebber, 2004). In the BY-rat7-1 mutant, GFP–npl3c is, together with the mRNA, trapped in the nucleus at the restrictive temperature (supplementary Fig S1C online). Thus, in the BY background, Npl3 is still involved in the mRNA export from the nucleus to the cytoplasm; however, its deletion does not result in the nuclear accumulation of mRNAs.
Figure 1.
Npl3 mediates the export of the pre-60S ribosomal subunit. (A) Mutations in NPL3 lead to defects in the export of pre-60S ribosomal subunits. Mutants of NPL3 (npl3-1, npl3-3, npl3-17 and npl3-41) in Saccharomyces cerevisiae S288C-background-expressing Rpl25–GFP (green fluorescent protein) were grown to the logarithmic growth phase before they were shifted to 37 °C for 30 min. The localization of Rpl25–GFP and DAPI (4,6-diamidino-2-phenylindole) staining are shown. (B) npl3Δ has no messenger RNA (mRNA) export defects. BY-wild type (WT), BY-npl3Δ, rat7-1 and the BY-npl3Δ rat7-1 double mutant strains were grown at 25 °C to the logarithmic growth phase and then shifted to 37 °C for 30 min. The cellular localization of the poly(A)+RNA using a Cy3-labelled oligo d(T) probe is shown. The DNA was stained with DAPI. (C) Export of the large ribosomal subunit is inhibited in npl3Δ. Localization of the large ribosomal subunit reporter protein Rpl25–GFP is shown in WT, npl3Δ, the ribosomal transport factor mutants nmd3-2, xpo1-1 and mtr2-33, and the mRNA export mutants mtr2-21 and rat7-1 in the BY-strain background at 25 °C and on temperature shift to 37 °C for the indicated times. (D) A new assay tracking single 25S ribosomal RNA (rRNA) molecules shows export defects in npl3Δ. Localization of the DIG-UTP-labelled 25S rRNA is shown in in situ hybridization experiments in WT, npl3Δ, the ribosomal transport factor mutants nmd3-2, xpo1-1 and mtr2-33, and the mRNA export mutant rat7-1 in the BY-strain background at 25 °C and on temperature shift to 37 °C for the indicated times.
Strikingly, we detected significant nuclear mislocalization of the pre-60S reporter protein Rpl25–GFP at 25 °C, which becomes more intense on a temperature shift to 37 °C (Fig 1C). Because Rpl25–GFP is incorporated into ribosomes (supplementary Fig S1D online), it can be used as a reporter for the localization of the large ribosomal subunit. Recently, an impaired pre-60S export was reported for nmd3-2, mtr2-33 and xpo1-1 strains (Bassler et al, 2001; Gadal et al, 2001). These findings were confirmed for the BY background (Fig 1C). It is interesting to note that, although Mtr2 is involved in mRNA export, mtr2-33 has no mRNA export defects, but does have pre-60S export defects, whereas the opposite is true for mtr2-21, indicating different requirements of the protein for both processes (Bassler et al, 2001).
As the mislocalized Rpl25–GFP reporter in npl3Δ is not trapped in the nucleolus (supplementary Fig S2A online), as it would be for pre-60S maturation mutants such as noc1 and rix9 (Gadal et al, 2002; Milkereit et al, 2001), npl3Δ seems unlikely to have significant pre-60S maturation defects that prevent its nucleolar exit. In fact, we detect no obvious rRNA processing defects by using the Bio-Rad-Experion technology (supplementary Fig S2B,C online). This was also concluded by Li et al (2009) who used northern blot analyses to demonstrate few differences in the production of different rRNA species between npl3Δ and the wild type.
In contrast to export defects of the large ribosomal subunit in npl3Δ, localization of the 40S ribosomal subunit reporter Rps2–GFP is unimpaired, indicating that the maturation and transport of the 40S ribosomal subunit is normal (supplementary Fig S3A online).
To provide an independent assay for the localization of the ribosomal subunits, we developed an assay based on the localization of the rRNA to monitor ribosomal subunits. We performed single rRNA localization studies with dioxygenin (DIG)-uracil triphosphate (UTP)-labelled probes against the 25S and the 18S rRNA in wild-type and different mutant strains. Similarly, we found nuclear mislocalization defects of the 25S rRNA in npl3Δ, xpo1-1, nmd3-2 and mtr2-33 cells (Fig 1D), as well as of the 18S rRNA in xpo1-1 (supplementary Fig S3B online). Interestingly, we found no export defects of any ribosomal subunit reporter, not Rpl25–GFP (Fig 1C), 25S or the 18S rRNA mislocalized in the strong mRNA export mutant rat7-1 (Fig 1D; supplementary Fig S3B online), indicating that defects in mRNA export do not lead to ribosomal export defects. Taken together, our results uncovered that Npl3 is required for efficient export of the large ribosomal subunit from the nucleus to the cytoplasm.
NPL3 interacts genetically with pre-60S export factors
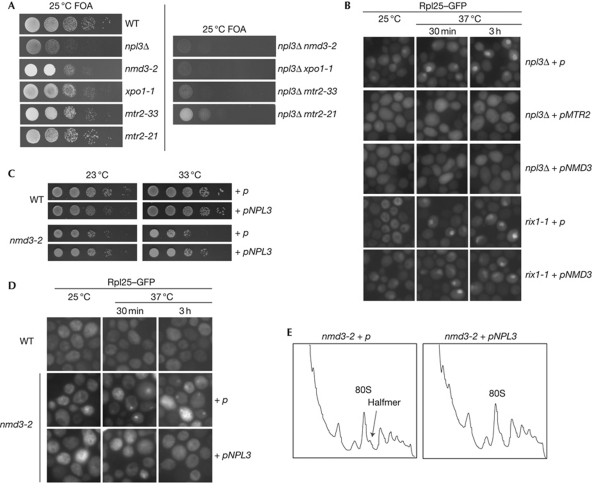
An involvement of Npl3 in the export of the large ribosomal subunit is further supported by several genetic interactions with other pre-60S export receptors (Fig 2A). Growth of the NPL3 deletion strain is significantly impaired in combination with nmd3-2 and mtr2-33 and prevented in combination with xpo1-1. The synthetic lethality of npl3Δ mtr2-33 is especially interesting because mtr2-33 has only pre-60S export defects. By contrast, mtr2-21, defective only in mRNA transport, shows only a slight synthetic growth defect in combination with npl3Δ (Fig 2A). This further argues that Npl3 still participates in the export of mRNA molecules in the BY background, albeit it is less relevant for this process. However, the strong genetic interactions between npl3Δ and pre-60S export mutants further support the fact that Npl3 has an important function in the transport of large ribosomal subunits to the cytoplasm.
Figure 2.
Genetic interactions of NPL3 with pre-60S ribosomal subunit transport factors. (A) Transport factors of the pre-60S ribosomal particle show synthetic growth defects with npl3Δ. Wild type (WT), nmd3-2, xpo1-1, mtr2-33, npl3Δ, the messenger RNA export mutant mtr2-21 and the double npl3Δ mutants carrying 2μ plasmid (p)- encoded NPL3 were spotted in serial dilution onto 5′-fluoroorotic acid (FOA) plates that allow growth only in the absence of the URA3-marked covering NPL3 plasmid. (B) High-copy MTR2 or NMD3 fully restores the pre-60S export defect in npl3Δ, but not in the maturation mutant rix1-1. npl3Δ and rix1-1 cells overexpressing MTR2 or NMD3 from a 2μ vector (p) were cultivated at 25 °C until logarithmic growth, before they were shifted to 37 °C for the indicated times. The cellular localization of Rpl25–GFP (green fluorescent protein) is shown. (C) High-copy NPL3 suppresses the growth defects of nmd3-2. Wild-type or nmd3-2 cells either containing an empty vector (p) or a 2μ plasmid-encoded NPL3 were spotted in serial dilution onto URA plates and incubated for 3 days at the indicated temperatures. (D) High-copy NPL3 restores the mislocalization of the pre-60S ribosomal subunit in nmd3-2 cells. Cells were grown to log phase before they were shifted to 37 °C for the indicated times. The localization of Rpl25–GFP is shown. (E) High-copy NPL3 suppresses the halfmer phenotype in nmd3-2. Cells carrying either an empty vector (p) or 2μ NPL3 were grown to log phase before they were shifted to 37 °C for 1 h. Cell lysates were applied to 6–48% sucrose density gradients, before the polysome profiles were taken by flow-through photometry.
To analyse whether the overexpression of other pre-60S receptors would suppress the export defects in npl3Δ, we introduced 2μ high-copy plasmids encoding either MTR2 or NMD3 into npl3Δ and analysed the localization of Rpl25–GFP at 25 and 37 °C. We found that overexpression of either NMD3 or MTR2 fully restores the pre-60S export defect in npl3Δ (Fig 2B). By contrast, overexpression of NMD3 in the 60S-processing mutant rix1-1 did not restore the cytoplasmic localization of Rpl25–GFP, indicating that the localization defects in npl3Δ are true export defects and not a result of processing defects, and that the full processing is a prerequisite for export. Moreover, npl3Δ and rix1-1 are not synthetically lethal (data not shown) in contrast to other export receptors (Fig 2A). Taken together, these data indicate that Npl3 is part of the Mex67–Mtr2- and Nmd3–Xpo1-associated pre-60S particle and, most importantly, that Npl3 is required for its efficient export. By contrast, single deletions of auxiliary transport proteins such as Arx1 and Ecm1 do not lead to pre-60S export defects, supporting a key role of Npl3 in the export of large ribosomal subunits.
Extensive ribosomal profile analyses have been performed by Li et al (2009), showing defective ribosomal profiles for downregulated MEX67 or MTR2 and npl3Δ. None of the three strains showed the expected formation of halfmers, reflecting subunit joining defects due to 60S export defects. Instead, phenotypes were formed that were described as general defects in translation with high 80S peaks, which might result from additional defects of these proteins in the translation process itself. By contrast, nmd3-2 mutants show the expected phenotype and thus we investigated whether high-copy NPL3 would rescue the nmd3-2 defects. Indeed, overexpression of NPL3 suppressed not only the growth defects and the Rpl25–GFP localization defects but also the halfmer formation (Fig 2C,D,E). This experiment indicates that Npl3 can compensate for the defective export receptor function in nmd3-2.
Npl3 interacts with the export-competent pre-60S subunit
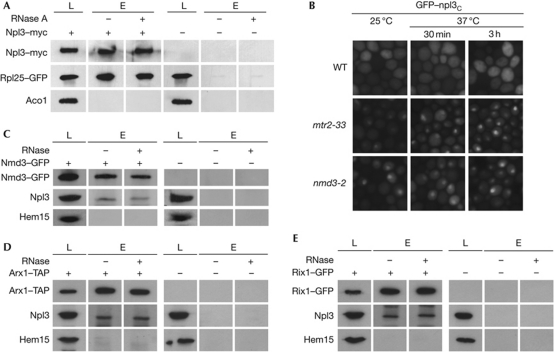
Besides these genetic interactions, Npl3 also interacts physically with the pre-60S ribosomal subunit as shown in co-immunoprecipitation experiments of Npl3 with Rpl25 (Fig 3A). This interaction is RNase-insensitive in contrast to the known RNA-sensitive interaction between Dbp5 and Pab1 (supplementary Fig S4A,B online) and not restricted to the BY-strain background, as we detect the same interaction also in the S288C-strain background (supplementary Fig S4C online).
Figure 3.
Nuclear association of Npl3 with the pre-60S ribosomal subunits, bound to Nmd3, Mtr2 and/or Arx1. (A) Npl3 interacts physically with the large ribosomal subunit. Co-immunoprecipitations of Rpl25–GFP (green fluorescent protein) were performed with Npl3–myc in wild-type or npl3Δ cells. Lysates were split and incubated with or without RNase. The mitochondrial protein Aco1 acts as a negative control. (B) The export of Npl3 is coupled to the export of the Mtr2- and Nmd3-bound pre-60S ribosomal subunit. Nuclear mislocalization of the ‘at-steady-state’ cytoplasmic Npl3 reporter protein GFP–npl3c is shown in pre-60S export mutants. BY-wild type, BY-mtr2-33 and BY-nmd3-2 were grown to log phase at 25 °C before the cells were shifted to 37 °C for 30 min or 3 h. (C,D) Npl3 interacts physically with the export-competent pre-60S ribosomal subunit. Co-immunoprecipitations of Npl3 were performed with GFP-tagged Nmd3 or Arx1–TAP either with or without the addition of RNase. (E) Npl3 interacts physically with the nuclear pre-60S ribosomal subunit. Co-immunoprecipitations of Npl3 were performed with Rix1–GFP. Lysates were split and either incubated with or without the addition of RNase. Western blot analyses with antibodies against Npl3, GFP, TAP and the mitochondrial control protein Hem15 or Aco1 are shown (A–E). L, lysate; E, eluate.
Two obvious questions arise: first, does Npl3 associate with a separate pool of pre-60S particles or does it interact with the Nmd3–Xpo1- and Mex67–Mtr2-associated pool? and second, when does the nucleocytoplasmic shuttling protein Npl3 contact the pre-60S particle? To address these questions, we first performed cellular localization studies of the ‘at-steady-state’ cytoplasmic version of Npl3 (GFP–npl3c) in mtr2-33 and nmd3-2 cells. On the temperature shifts, GFP–npl3c was trapped in the nucleus of both mtr2 and nmd3 mutant cells, showing the intimate coupling of the export of Npl3-bound and Nmd3–Xpo1- or Mex67–Mtr2-bound pre-60S ribosomal subunits (Fig 3B).
Second, we conducted co-immunoprecipitation studies of Npl3 with Nmd3 and Arx1, representing the export-competent pre-60S ribosomal subunit. We found physical interactions of Npl3 with both Nmd3 and Arx1, indicating a stable association of Npl3 with the export-competent pre-60S particle that is not RNase-sensitive (Fig 3C,D). To exclude that Npl3 might only associate with the cytoplasmic pool of the export-receptor-associated pre-60S ribosomal subunit, we investigated the interaction of Npl3 with Rix1, a nuclear 60S-processing protein, which dissociates from the pre-60S particle before export. In co-immunoprecipitation experiments, we detected a physical interaction between both proteins, suggesting a nuclear recruitment of Npl3 to the receptor-bound pre-60S ribosomal subunit (Fig 3E).
Npl3 is an export receptor for the pre-60S subunit
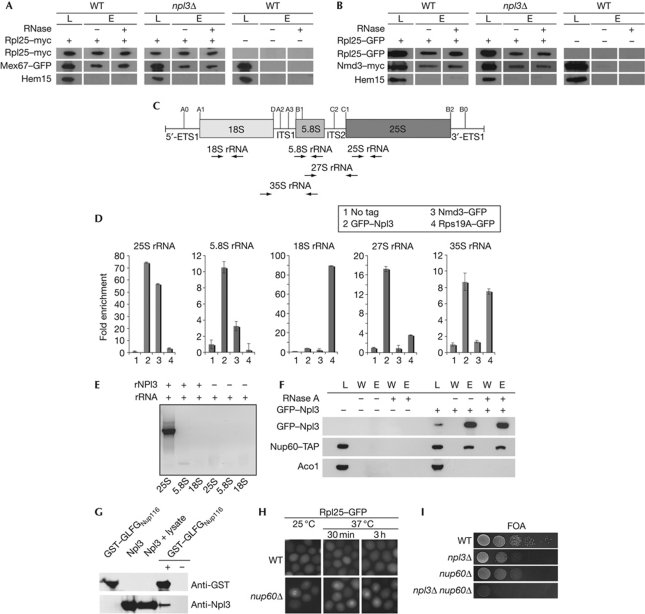
In mRNA export, Npl3 bridges the interaction between mRNA and the export receptor Mex67 (Gilbert & Guthrie, 2004). Therefore, it is conceivable that this might also be the case for the export of the large ribosomal subunit, although it might be less likely because Mex67 has been shown to directly contact the 5S rRNA (Yao et al, 2007). Nevertheless, we addressed this question by co-immunoprecipitation studies of Rpl25 with Mex67 or Nmd3 in wild-type compared with npl3Δ cells. As shown in Fig 4A,B, we found no decreased association of either receptor with the pre-60S ribosomal subunit in npl3Δ, indicating that Npl3 is not required for their recruitment. Instead, it seems likely that Npl3 might contact the pre-60S particle on its own. To test this, we performed in vivo co-immunoprecipitation analyses with GFP–Npl3, Nmd3–GFP and Rps19A–GFP and the 25S, 5.8S, 18S, 27S and 35S rRNA. As shown in Fig 4C,D, we detect strong interactions of Npl3 with the 25S rRNA and lesser but significant interactions with the 5.8S, 27S and 35S rRNA, which might reflect a direct interaction of Npl3 with the 25S rRNA. It further indicates an early recruitment of the SR protein to the pre-90S ribosomal particle, which is supported by our finding that Npl3 interacts with the Rix1-containing pre-60S particle, a protein involved in the pre-60S maturation that dissociates before export (Tschochner & Hurt, 2003).
Figure 4.
Npl3 functions as an independent export adaptor for the pre-60S ribosomal subunit. (A) Deletion of NPL3 does not impair the binding of Mex67 to the large ribosomal subunit. Co-immunoprecipitations of Mex67–GFP (green fluorescent protein) with Rpl25–myc were performed in wild-type (WT) and npl3Δ cells. (B) Deletion of NPL3 does not influence the binding of Nmd3 to the pre-60S subunit as shown in co-immunoprecipitations of Nmd3–myc with Rpl25–GFP. Total lysates shown in A and B were split and incubated with or without RNase before elution. Western blot analyses with antibodies against GFP, myc and the control protein Hem15 are shown. (C) Scheme of the 35S ribosomal RNA (rRNA) precursor and location of the processing sites. The pre-rRNA encodes the 18S, 5.8S and the 25S rRNAs, which are flanked by the 5′ and 3′ external transcribed spacers (5′-ETS and 3′-ETS) and separated by internal transcribed spacers 1 and 2 (ITS1 and ITS2). From the 35S precursor, rRNA cleavage generates the 27S and the 20S pre-rRNA, from which the 5.8S+25S and the 18S rRNAs are generated, respectively. The positions for the primers used in the quantitative reverse transcription PCR (RT–PCR) are shown as arrows. (D) Npl3 binds to the 35S, 27S, 25S and 5.8S rRNA in vivo. Wild-type cells were used as ‘no tag’ control. npl3Δ carrying a CEN GFP–NPL3 plasmid, nmd3-2 carrying a CEN NMD3–GFP plasmid and genomically encoded RPS19A–GFP were grown to log phase before GFP immunoprecipitations were performed. Furthermore, the associated RNA was extracted and used for quantitative RT–PCR experiments. (E) In vitro experiments showing that Npl3 binds directly to the 25S rRNA. Regions of the 25S, 5.8S and the 18S rRNAs were in vitro transcribed and incubated with recombinant His–Npl3 (rNpl3). Co-immunoprecipitations were performed and the purified rRNA was used for RT–PCR with specific primers, shown on an agarose gel. (F) Co-immunoprecipitations of Nup60–TAP with GFP–Npl3 were performed in WT cells. Total lysates were split and incubated with or without RNase before elution. Western blot analyses with antibodies against GFP, TAP and the control protein Aco1 are shown. (G) Npl3 interacts with FG repeats in vitro. Purified recombinant His–Npl3 (rNpl3) and a glutathione S-transferase (GST)-tagged GLFG-repeat-containing region of Nup116 were investigated for an in vitro interaction. Co-purification of rNpl3 was performed with GST pull-down in the presence of whole bacterial lysate. Western blot analyses are shown with antibodies against Npl3 and GST. (H) Deletion of NUP60 leads to export defects of the large ribosomal subunit. Wild-type and nup60Δ cells were grown to log phase and shifted to 37 °C for the indicated times. Localization of Rpl25–GFP is shown. (I) npl3Δ and nup60Δ are synthetically lethal. Wild-type cells or cells deleted for either indicated gene alone or in combination are shown on 5′-fluoroorotic acid (FOA) plates that select for the loss of a covering NPL3 plasmid on growth for 3 days at 25 °C. E, eluate; L, lysate; W, wash.
In contrast to the fact that Nmd3 shows no interaction with the 27S or 35S rRNA, however, significant interaction with the 25S rRNA was also published earlier (Sengupta et al, 2010), indicating a late recruitment of the protein to the pre-60S ribosomal subunit. The 18S and the 35S rRNA were co-immunoprecipitated with Rps19A. The in vivo experiments indicated that Npl3 interacts directly with the 25S rRNA. To verify that, we performed in vitro co-immunoprecipitation analyses with recombinantly expressed His–Npl3 (rNpl3) that was incubated with in vitro transcribed 25S, 5.8S and 18S rRNAs. As shown in Fig 4E, Npl3 specifically binds to the 25S rRNA but not the 18S rRNA. These results indicate that Npl3 is able to contact the pre-60S ribosomal subunit by direct interaction with the 25S rRNA. In fact, all temperature-sensitive mutants of NPL3 that mislocalize the pre-60S ribosomal subunit (Fig 1A) also mislocalize mRNA and contain mutations in the RNA-binding domains of the protein (Lee et al, 1996), indicating that RNA binding is involved in both transport functions of Npl3 and would be consistent with a model in which Npl3 mediates ribosomal export by binding to the 25S rRNA. In addition, Npl3 interacts with the ribosomal protein Rpl25 (Fig 3A), which might stabilize or guide its rRNA association. Recently, the crystal structure of the 60S ribosomal subunit was solved (Ben-Shem et al, 2010) and it showed that Rpl25 binds close to the regions 1,516–1,524 and 1,826–1,843 of the 25S rRNA to the 60S particle, which might also be the contact area for Npl3. However, it is now unclear whether only one molecule of Npl3 binds to the large ribosomal subunit or whether there are several contact points.
In the mRNA export from the nucleus to the cytoplasm, Npl3 has not been suggested to directly contact the NPC, as several interactions of the receptor Mex67 with the NPC have been described (Strawn et al, 2001). However, in the case of pre-60S export, Npl3 does not operate through Mex67–Mtr2 or Nmd3–Xpo1 (Fig 4A,B). Therefore, we investigated whether Npl3 is able to interact with the NPC, and found a physical interaction of this RNA-binding protein with Nup60 (Fig 4F). It is now unclear whether this interaction requires other proteins; however, the interaction is not sensitive to the addition of RNase, emphasizing the fact that Npl3 interacts physically with the NPC. Indeed, in vitro studies reveal a direct interaction of Npl3 with the FG/GLFG repeats of Nup116 (Fig 4G; supplementary Fig S5 online), uncovering the ability of Npl3 to directly contact FG nucleoporins of the NPC. An involvement of Nup60 in the export of the large ribosomal subunit was further verified by the mislocalization of Rpl25–GFP, either at the rim or within the nucleus of nup60Δ (Fig 4H) and the synthetic lethality of npl3Δ nup60Δ (Fig 4I).
Taken together, we propose a model (supplementary Fig S6 online) in which Npl3 not only mediates the export of mRNAs through Mex67, but also that of pre-60S ribosomal subunits, independently of Mex67 or other known transport receptors. These findings uncover the multifunctional nature of the shuttling RNA-binding protein Npl3 and show an elegant example of high efficiency in nature.
Methods
All experiments shown in Figs 1, 2, 3, 4 and supplementary Figs online have been conducted at least three independent times. A detailed methods description can be found in supplementary information online.
Yeast strains, plasmids and oligonucleotides. All yeast strains used in this study are listed in supplementary Table S1 online, plasmids in supplementary Table S2 online and oligonucleotides in supplementary Table S3 online. All experiments, unless indicated differently, were conducted in the BY series of the S. cerevisiae strain background. Details on strain constructions and cloning strategies are listed in supplementary information online.
In situ poly(A)+RNA and rRNA hybridization. Localization of poly(A)+RNA by in situ hybridization was performed as described (Gross et al, 2007). rRNA visualization is described in supplementary information online.
Co-immunoprecipitation experiments. The experimental procedure was essentially conducted as described in Gross et al (2007). Co-immunoprecipitations were performed using Protein-G sepharose beads (Amersham Biosciences) or Protein-G agarose beads (Santa Cruz) conjugated to c-Myc (9E10)- or GFP-specific antibodies (mouse and/or rabbit) or immunoglobulin-G sepharose beads in the presence or absence of 200 μg/ml RNase.
In vivo RNA co-immunoprecipitation experiments. Immunoprecipitation experiments were essentially conducted as described above. The associated RNA was extracted and used in quantitative reverse transcription PCR analyses.
Recombinant protein expression and rRNA co-immunoprecipitation, followed by reverse transcription PCR analyses. His–Npl3 was expressed in Escherichia coli as described in the supplementary information online. Recombinant His–Npl3 was equilibrated with RNA-binding buffer before incubation with fragments of the in vitro transcribed 25S, 18S or 5.8S rRNA.
In vitro protein–protein binding assay. Recombinant proteins were incubated with E. coli lysate and purified by glutathione S-transferase pull-down.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We are grateful to G. Braus, E. Hurt, R. Lill, P.A. Silver and S. Wente for providing strains and/or plasmids and/or antibodies. We thank R. Ficner and S. Khoshnevis for support, and H. Bastians for critically reading the manuscript. This work was funded by grants from the Deutsche Forschungsgemeinschaft and the SFB593 and SFB860 to H.K.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E (2001) Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol cell 8: 517–529 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M (2010) Crystal structure of the eukaryotic ribosome. Science 330: 1203–1209 [DOI] [PubMed] [Google Scholar]
- Bradatsch B et al. (2007) Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol Cell 27: 767–779 [DOI] [PubMed] [Google Scholar]
- Erkmann JA, Kutay U (2004) Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp Cell Res 296: 12–20 [DOI] [PubMed] [Google Scholar]
- Fried H, Kutay U (2003) Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci 60: 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E (2001) Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21: 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Petfalski E, Gleizes PE, Gas N, Tollervey D, Hurt E (2002) Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J Cell Biol 157: 941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Guthrie C (2004) The glc7p nuclear phosphatase promotes mRNA export by facilitating association of mex67p with mRNA. Mol Cell 13: 201–212 [DOI] [PubMed] [Google Scholar]
- Gorsch LC, Dockendorff TC, Cole CN (1995) A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol 129: 939–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H (2007) The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 315: 646–649 [DOI] [PubMed] [Google Scholar]
- Hacker S, Krebber H (2004) Differential export requirements for shuttling serine/arginine-type mRNA-binding proteins. J Biol Chem 279: 5049–5052 [DOI] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW (2000) Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 151: 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Steitz JA (2005) SRprises along a messenger's journey. Mol Cell 17: 613–615 [DOI] [PubMed] [Google Scholar]
- Hung NJ, Lo KY, Patel SS, Helmke K, Johnson AW (2008) Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol Biol Cell 19: 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E (2007) Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA (1996) A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev 10: 1233–1246 [DOI] [PubMed] [Google Scholar]
- Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM (2009) Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol 7: e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H (2001) Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105: 499–509 [DOI] [PubMed] [Google Scholar]
- Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J (2010) Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol 189: 1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage-Zimmermann T, Schmidt U, Silver PA (2000) Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol Biol Cell 11: 3777–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Wente SR (2001) The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J Biol Chem 276: 6445–6452 [DOI] [PubMed] [Google Scholar]
- Thomas F, Kutay U (2003) Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 116: 2409–2419 [DOI] [PubMed] [Google Scholar]
- Tran EJ, Wente SR (2006) Dynamic nuclear pore complexes: life on the edge. Cell 125: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Tschochner H, Hurt E (2003) Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H (2004) Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol 24: 10479–10491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA et al. (1999) Functional characterization of the S cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Yao W, Roser D, Kohler A, Bradatsch B, Bassler J, Hurt E (2007) Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol Cell 26: 51–62 [DOI] [PubMed] [Google Scholar]
- Yao Y, Demoinet E, Saveanu C, Lenormand P, Jacquier A, Fromont-Racine M (2010) Ecm1 is a new pre-ribosomal factor involved in pre-60S particle export. RNA 16: 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I, Kutay U (2007) Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett 581: 2783–2793 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.