Abstract
Apurinic/apyrimidinic endonuclease-1 (APE1) is a multifunctional DNA repair/gene regulatory protein in mammalian cells, and was recently reported to be phosphorylated at Thr233 by CDK5. We here report that ubiquitination of T233E APE1, a mimicry of phospho-T233 APE1, was markedly increased in multiple cell lines. Expression of CDK5 enhanced monoubiquitination of endogenous APE1. Polyubiquitinated APE1 was decreased when K48R ubiquitin was expressed, suggesting that polyubiquitination was mediated mainly through Lys48 of ubiquitin. The ubiquitination activity of MDM2, consistent in its role for APE1 ubiquitination, was increased for T233E APE1 compared to the wild-type APE1. In mouse embryonic fibroblasts lacking the MDM2 gene, ubiquitination of T233E APE1 was still observed probably because of the decreased degradation activity for monoubiquitinated APE1 and because of backup E3 ligases in the cells. Monoubiquitinated APE1 was present in the nucleus, and analyzing global gene expression profiles with or without induction of a ubiquitin-APE1 fusion gene suggested that monoubiquitination enhanced the gene suppression activity of APE1. These data reveal a delicate balance of ubiquitination and phosphorylation activities that alter the gene regulatory function of APE1.
INTRODUCTION
Oxidative stress and alkylating reagents spontaneously generate DNA damage that is mainly repaired by DNA base excision repair (BER) (1). Apurinic/apyrimidinic (AP) endonucleases (APEs) play an essential role in BER. Without APE activity, such damage is sufficient to stall DNA replication and to cause cell death (2–5). In mammals, Apurinic/apyrimidinic endonuclease-1 (APE1) is responsible for the APE activity (6–8). Depletion of APE1 in mouse and cultured cells leads to embryonic lethality and apoptotic cell death, respectively (4,5).
APE1 has at least two types of gene regulatory functions that appear unrelated to its DNA repair functions (9). Known as the redox factor 1 (Ref-1), APE1 activates AP-1, NFkB and other transcription factors important in cancer biology by mediating intracellular redox signaling (10–13). In addition, APE1 acts as a transcriptional co-repressor by binding DNA cis-elements called nCaREs (negative calcium response elements), which are found in upstreams of genes related to calcium dependent-homeostasis (14–17). Histone acetyltransferase (HAT) p300 acetylates APE1 at two N-terminal Lys residues (K6/7) and increases APE1's DNA binding activity (18). Interaction of APE1 with HDAC1 likely enhances the suppression of genes involved in the nCaRE dependent regulation (18). More recently the other Lys residues (K27, 31, 32, 35) in the N-terminal 6-kDa region of which function has not been understood clearly, were found to be acetylated (19).
In addition to acetylation and S-nitrosation (20), two other PTMs on APE1 were recently reported, i.e. ubiquitination (21,22) and phosphorylation (23). APE1 was ubiquitinated, which was enhanced by transient up-regulation of MDM2. Ubiquitin-induced APE1 degradation may sensitize cells to apoptosis. While APE1 gene downregulation is followed by apoptotic cell death (4,5,24,25), a higher expression of APE1 is linked to chemo/radiation-resistant cancer cells (26–29). As researchers attempt to improve the outcomes of non-invasive treatments for tumors (30,31), the mechanism involved in APE1 ubiquitination is an important subject to understand.
APE1 phosphorylation was reported earlier (32–34), but the matter was refreshed by Huang et al. (23) who found that APE1 was phosphorylated in vivo specifically at Thr-233 by CDK5. CDK5 is a paralog of cell cycle-dependent kinases CDK2 and CDK4/6, and its activity is high in post-mitotic neuronal cells (35–37). It has been suggested by numerous studies that CDK5 is critical for cell cycle-associated neuronal cell death (35). Thus, it is intriguing that phosphorylated APE1 and its mimicry T233E APE1 showed decreased APE activity, and that the phosphorylated APE1 was more abundantly found in cells from brains of Parkinson's and Alzheimer's patients (23).
We are interested in the fact that in some conditions APE1 was down-modulated (23). Such immediate downregulation is indicative of protein degradation dependent on ubiquitination. In this report, we show that phosphorylation at T233 of APE1 markedly increased its ubiquitination and that the degradation was regulated by MDM2. A mechanism that links T233 phosphorylation to APE1 ubiquitination is discussed.
MATERIALS AND METHODS
Cell lines
A colon carcinoma cell line HCT116 (38) was grown as described previously (22). A549, a human lung adenocarcinoma cell line was purchased from ATCC. JHU28 and JHU13, human oral squamous carcinoma cell lines, were provided by Dr R. Walvekar (LSUHSC) and Dr R. Ferris (University of Pittsburgh). 2DN cells (p53−/− MDM2−/−) mouse embryonic fibroblasts (MEFs) were originally developed in Dr Lozano's lab and kindly provided by Dr Iwakuma (39). HCT116 and A549 carry the wild-type (wt) p53 gene, and we also confirmed that JHU28 carry the wt p53 gene (data not shown). Stable transfectants of HCT116 that harbor either control vector (pSIREN-RetroQ) or shAPE1 vector (generous gift from Dr Crowe) were established as previously described (40). The 293 T-Rex Flp cell line (Invitrogen) was cultured in DMEM/F12 medium with 10% FBS, 100 µg/ml zeocin and 15 µg/ml blasticidin. Stable 293 transfectants for pOG44 (Flp recombinase) and the tet-on expression vectors (derivatives of pcDNA5/FRT/TO, Invitrogen and Supplementary Figure S2) were selected with 100 µg/ml hygromycin B after transfection. All cells were cultured in the same condition as for HCT116. DNA Transfection was carried out using lipofectamine2000 (Invitrogen) following the vendor's instruction.
DNA and chemicals
The wtAPE1 cDNA was cloned by RT–PCR using total RNA from JHU13. The clone carries identical coding sequence to that in NCBI (GeneID 328) and is regarded as wt in this study. The human CDK5 and p35 genes were obtained from Addgene. Other genes were cloned by PCR using human quick-clone cDNA (Clontech), and confirmed to carry the wt sequences (ACGT, Chicago). Chemicals were purchased from Sigma unless otherwise noted.
Immunoblot assay (western blot)
A standard protocol was used for all immunoblot assays using PVDF membrane (Bio-Rad). Antibodies were purchased from Santa Cruz, including a secondary antibody (sc-2031) and primary antibodies for APE1 (sc-55498), CDK5 (sc-6247), p35 (sc-5614) and β-tubulin (sc-58884).
Ubiquitination assay, co-immunoprecipitation (Co-IP) and fractionation of nuclei and cytosol
The ubiquitination assay was carried out as described previously (22). Co-IP was also carried out as the previous study using Fk-2 (Enzo) and M2-FLAG antibody (Sigma). For separating nuclear and cytosolic fractions, cells expressing T233E APE1 was washed with PBS twice, then extracted with nuclear/cytosol protein fractionation kit (BioVision) with the vendor's protocol. HSP90 and Lap2 antibodies are from SCBT.
Immunocytochemistry of T233E APE1
2DN cells transiently expressing T233E APE1 from IRES-hrGFP1a (Stratagene) were fixed in 3.7% formaldehyde, permeabilized with 0.2% TritonX100, and washed with PBS. After blocking in PBS containing 1% bovine serum albumin (PBS/BSA), the fixed cells were incubated with an APE1 antibody (rabbit polyclonal, raised with full-length hAPE1) for 2 h, and with an anti-rabbit IgG conjugated to rhodamine (Chemicon) for 1 h. Cells were stained with DAPI and mounted on glass plates, and analyzed with Nikon TE2000 epifluorescence microscope.
Total RNA extraction and gene expression array profiling
Three HEK293 derivatives expressing either none (vector), wtAPE1, or Ub-APE1 fusion linked at APE1's 24th Lys, were incubated with/without doxycycline (dox) 2 µg/ml for 16 h. Each −/+ dox cultures were triplicated. Total RNA from were extracted (Qiagen), and then analyzed by LSU's bioinformatics core. The quality and quantity of total RNA were assessed using ND-1000 Spectrophometer (NanoDrop, Wilmington, DE), NanoChip assay and Bioanalyzer 2100 (Agilent). All samples were of high integrity with RIN # >7seven and A260/280 >1.8. Procedures for cDNA synthesis, sense target labeling and hybridization were carried out as described at http://media.affymetrix.com/support/technical/appnotes/wt_appnote.pdf (Affymetrix, Redwood City, CA, USA). All experiments were performed using GeneChip Exon 1.0 ST Arrays and Whole Transcript Sense Target labeling Assay (version 4, Affymetrix). Overnight hybridization of fragmented single-stranded DNA was carried out in an Affymetrix GeneChip Hybridization Oven 640, then washed and stained with streptavidin-phycoerythrin using GeneChip 450 Microfluidics Station (Affymetrix). Chips were scanned with an Affymetrix High Resolution 3000 (G7) scanner. Signal and background intensities were quantitated by pixel intensity using Affymetrix GeneChip Operating Software (GCOS 1.4). Array quality control assessment was carried out using Robust Multi-chip Analysis (RMA) workflow for Core probesets in Expression Console (Affymetrix). Gene-level analysis for differential gene expression was analyzed in GeneSpring 7.3× (Agilent Technologies) using the sample CHP files. The RMA method was used for the background correction, normalization and average expression measures for the probesets. An expression filter (20 percentile cutoff) was applied to remove genes with low signal intensity values. For differential gene expression analysis, the Welch ANOVA test with asymptotic P-value computation was applied and transcripts with P-value (<0.05) and fold change (2.0-fold) were selected.
RESULTS
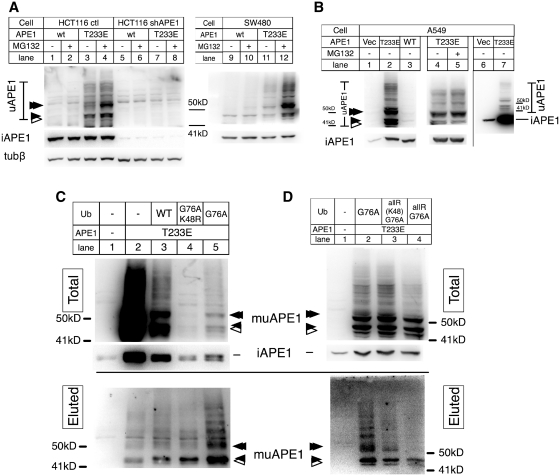
T233E APE1, in which Glu was substituted for Thr at 233th amino acid residue, was recently used to mimic phosphorylated APE1 by CDK5 (23). To test whether phosphorylation of T233 affects APE1 ubiquitination, a plasmid vector encoding human T233E APE1 was transiently expressed in HCT116 sh-ctl, a human colon carcinoma cell lines stably carrying am empty sh-vector (lanes 1–4, Figure 1A). To observe the high molecular weight-bands (HWBs) that were specifically detected with anti-APE1 antibodies, signals of HWBs were separately detected from that of the intact APE1 protein. The amount of HWBs was increased in the cells wit T233E (Figure 1A, lane 3) compared to the wt-APE1 (lane 1). The position of one of HWBs (filled arrow) migrated slightly below β-tubulin (49.7 kDa), above β-actin (41 kDa), approximately matching to the calculated molecular weight (44 kDa) of monoubiquitinated APE1 (muAPE1), although the exact migrating position may be different due to the branched structure of ubiquitin conjugation. Moreover, the band position matched to that of recombinant muAPE1 prepared from Escherichia coli (Supplementary Figure S1A). A band (Figure 1A, open arrow) smaller than the recombinant muAPE1 was likely due to a subsequent truncation of ubiquitinated APE1 (uAPE1). Although it is not clear whether ubiquitination induced the cleavage reaction, it has been known that N-terminal sites are susceptible to spontaneous hydrolytic cleavage. In any case, the band corresponding to monoubiquitinated APE1 was the major product among HWBs. Treatment of the transfected cells with MG132, an inhibitor of the 26S proteasome, increased HWB formation to a much greater extent (Figure 1A, compare lanes 3 and 4), along with the di-ubiquitinated APE1 (double filled arrow, Figure 1A). Although the monoclonal antibody used in this study was highly specific to APE1, to further confirm that the HWB formation was specific to APE1 and not due to some cross-reactivity of the antibody, we transiently expressed the wt and T233E APE1 in HCT116 derivative that expresses APE1 shRNA (HCT116 shAPE1, lanes 5–8, Figure 1A). This cell line shows an extremely low expression of the intact APE1 protein (~3% of the parental cell line). The cells also effectively suppressed the transiently expressed APE1 with the very low amount of unmodified intact APE1 (lanes 5–8 versus 1–4). The HWBs diminished in this cell line, which confirmed that the HWBs were specific to APE1. These observations suggested that the T233E alteration increased muAPE1 and puAPE1 (polyubiquitinated APE1), and that puAPE1 is susceptible to the 26S degradation pathway.
Figure 1.
Enhanced ubiquitination of T233E APE1. (A) HCT116 ctl (1–4), HCT116 expressing shAPE1 (5–8) and SW480 (9–12) transfected with the wtAPE1 (1, 2, 5, 6, 9, 10), T233E APE1 (3, 4, 7, 8, 11, 12). After 24 h, cells of even lanes were treated with MG132 for 3 h, and cell extracts were analyzed with immunoblotting (IB) with a monoclonal APE1 antibody (αAPE1). Intact APE1 (iAPE1) was blotted separately from uAPE1. Reblotted β-tubulin (50 kD) and β-actin (41 kD) were used as size markers. (B) A549 transfected with vector ctl (1, 6), wtAPE1 (3), or T233E (2, 4, 5, 7) with 3 h MG132 treatment (5). Lanes 6 and 7, unlike 1–5, intact and uAPE1 were blotted together to determine the relative amount of uAPE1 compared with the intact APE1. (C and D) His-Ub assay. A549 transfected with the T233E APE1 and indicated ubiquitin (Ub) genes. Total and enriched ubAPE1 by nickel affinity beads (eluted) were detected with αAPE1. Top, uAPE1 and intact APE1 in total fractions; bottom, uAPE1 in eluted fraction. Arrows indicate positions of muAPE1 (closed), puAPE1 (double closed), and truncated muAPE1 (open).
Essentially identical results were obtained with SW480, another colon carcinoma cell line (Figure 1A), and JHU28, an oral squamous carcinoma cell line (data not shown) (41). A549, a human lung carcinoma cell line, showed a substantial difference in its accumulation of HWB (Figure 1B). HWB in A549 was readily visible without MG132 which only had a minimum effect (lanes 4 and 5), suggesting that A549 was particularly tolerant of degradation of puAPE1 and increased the stable existence of muAPE1. In A549, the relative amount of uAPE1 (lane 7) over the total amount of APE1 (intact APE1 and uAPE1 combined) was calculated to be 20.2 ± 12.1% based on three independent experiments where immunoblot signals were not saturated. These results with multiple cell lines suggest that T233E enhancement for uAPE1 formation is a general phenomenon and that the consequence of the uAPE1 protein may differ from one cell type to another. It should be noted that cell lines tested above carry the wild-type p53 gene except for SW480 (42), suggesting that the variability of ubiquitination activity on APE1 was not due to the genetic status of the p53 gene.
To confirm that HWB formation was due to APE1 ubiquitination, a ubiquitination assay was carried out using the ubiquitin gene tagged with a histidine hexamer (His-Ub) (22). In Figure 1C, T233E APE1 was co-expressed in A549 with three kinds of His-Ub genes, namely, the wild-type Ub, G76A Ub, and K48R G76A Ub. The C-terminal Ala substitution (G76A) in ubiquitin increases resistance to intracellular deubiquitination activities (22), whereas the K48R Ub lacks the critical Lys residue necessary for polyubiquitination (43,44). When T233E APE1 was co-expressed with ubiquitin, the amount of intact APE1 and uAPE1 in the total fractions were noticeably decreased (lane 3 compared to 2), suggesting that over-expression of ubiquitin facilitated degradation of uAPE1 (see below). In the eluted fractions, HWB specific to APE1 appeared depending on the His-Ub expression (lanes 3–5). The amount of HWBs (uAPE1) was much higher with the G76A ubiquitin (lane 5) than with the K48R G76A ubiquitin (lane 4), suggesting that puAPE1 was mainly formed via K48. In addition, the amount of uAPE1 by G76A was larger than that of the wt-ubiquitin, suggesting that the presence of an intracellular deubiquitinase activity of which reaction was blocked by the G76A missense.
To examine the nature of the uAPE1 in detail, two additional His-Ub genes were constructed in the background of G76A. The ‘UballR’ has Arg substitutions for all of its seven Lys, and ‘UballRbutK48’ has Arg substitutions in all but 48th Lys residue. We compared ubiquitination of T233E APE1 with the three ubiquitin subforms (Figure 1D). Ub and UballRbutK48 were both enhanced APE1 ubiquitination (Figure 1D, lanes 2 and 3). However, a larger amount of HWBs (puAPE1) was purified with the G76A Ub than with UballRbutK48, possibly implying that K48 was not the sole ubiquitin-acceptor for puAPE1 formation (Figure 1D, lane 3). Amount of puAPE1 with UballR was substantially decreased in the total fraction (lane 4, top), and was negligible in the eluted fraction, compared to the prominent existence of muAPE1 (lane 4, bottom). These results supported our conclusion that the HWB was formed by ubiquitination. Our attempt to detect APE1 conjugation to small ubiquitin like-modifiers, i.e. SUMO, Nedd8 and ISG15, was previously unsuccessful (22). We examined the possibility that the T233 modification might enhance such modifier conjugations as well. Unlike ubiquitin, however, none of the above small molecule modifiers revealed HWBs on T233E APE1 (data not shown). Taken together, we concluded that ubiquitin was the unique small molecule modifier even for T233E APE1.
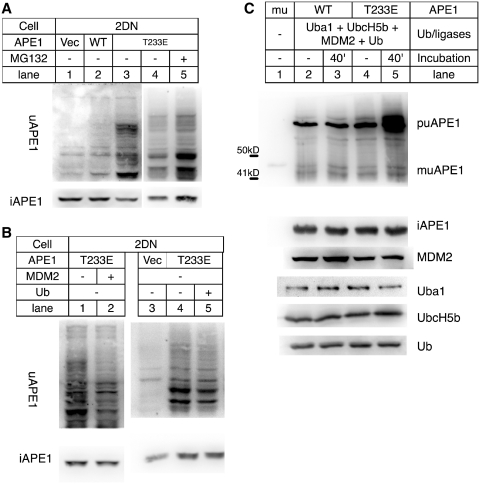
We previously reported the role of MDM2 as an E3 ligase for APE1 ubiquitination (22). Here, T233E APE1 was expressed in 2DN cells, mouse embryonic fibroblasts (MEFs) generated from p53−/− MDM2−/− mice (39). The 2DN cells expressing T233E APE1 showed increased uAPE1 (Figure 2A, lanes 3 versus 2), indicating that there were other E3 ligase(s) that could complement MDM2 for ubiquitination of APE1. We tested the effect of over-expression of MDM2 and ubiquitin genes in the MDM2−/− cells on the amount of uAPE1 generation. The MDM2 gene was co-expressed in the 2DN cells along with T233E APE1 (Figure 2B, lane 2). In this condition, the expression of MDM2 caused a decrease of uAPE1. We also co-expressed the ubiquitin and T233E APE1 in the cells, which also resulted in a reduction of uAPE1 (Figure 2B, lane 5 and also Figure 1C). The amount of uAPE1 in these conditions increased after treating the cells with MG132 (Supplementary Figure S5A), suggesting that expressing either MDM2 or ubiquitin could enhance the degradation process. To ascertain the role of MDM2 in the ubiquitination reaction, an APE1 ubiquitination environment was generated in E. coli (Figure 2C). Five human genes necessary for APE1 ubiquitination were simultaneously expressed in E. coli, i.e. ubiquitin ligases E1 (uba1), E2 (UbcH5b), E3 (MDM2), His-tagged ubiquitin, and APE1 (22,45). While the wtAPE1 led to puAPE1 accumulation at 37°C, the T233E substitution enhanced puAPE1 formation (lane 5 versus 3). The result indicated that MDM2 was capable of ubiquitinating APE1, a reaction that was enhanced with the T233E substitution in APE1. Together with the in vitro reactions previously reported (22) and the increasing interaction between T233E APE1 and MDM2 (Supplementary Figure S1B), we concluded that MDM2 was a functional E3 ligase for APE1, but there were backup E3 ligases in the cells.
Figure 2.
Dependency of HWB appearance on ubiquitin and MDM2. (A) 2DN (p53−/− MDM2−/−) transfected with control vector (1), the WT APE1 (2), or the T233E APE1 cDNA (3), and probed with αAPE1 for uAPE1 (top) and intact APE1 (iAPE1, bot). (B) 2DN expressing T233E alone (1) or T233E + MDM2 (2), ctl vector (3), T233E (4) and T233E + ubiquitin (5). Top: uAPE1; bottom: intact APE1. (C) APE1 ubiquitination assay in E. coli BLR(DE3) with Uba1, UbcH5b, MDM2, ubiquitin (2–5) plus wt APE1 (2 and 3) or T233E APE1 (4 and 5) extracted (2 and 4) and incubated at 37°C for 40 min (3 and 5), and analyzed with αAPE1. (1), purified recombinant muAPE1. Top: uAPE1; rest: intact APE1 and ubiquitination factors in lanes 2–5 detected in separate immunoblot membranes.
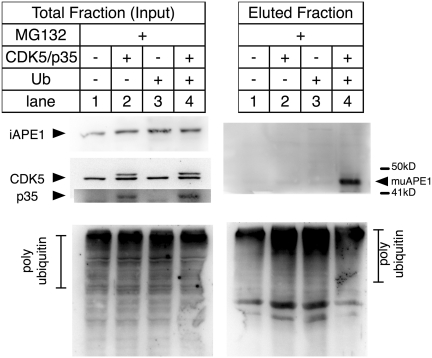
To obtain evidence of enhancement of ubiquitination for endogenous APE1, the CDK5 and its activator p35 genes were co-expressed in the 2DN cells, and ubiquitin-conjugated proteins were immunoprecipitated by a monoclonal antibody specific to Ub-conjugates (46). The immunoprecipitated fractions were then analyzed for uAPE1 (Figure 3). The muAPE1 specific bands were detected only when the CDK5/p35 and ub were expressed in the cells (lane 4). In addition, ubiquitinated APE1 catalyzed by MDM2 was inhibited by treating the cells with roscovitine, a potent inhibitor of the phosphorylation reaction of CDK5 (Supplementary Figure S5B). With these results, we concluded that ubiquitination of endogenous APE1 was triggered with T233 phosphorylation by CDK5/p35. It should be noted that puAPE1 was not detected in the IP fractions, presumably because puAPE1 was sensitive to degradation and was less stable than muAPE1. This inference is consistent with our recombinant muAPE1 purification, where puAPE1 could not be purified (Supplementary Figure S1A), even though puAPE1 was accumulated (Figure 2C).
Figure 3.
Ubiquitination of endogenous APE1 enhanced by CDK5/p35. 2DN cells transfected with vector ctl (1), HA-tagged CDK5 and p35 (2), G76A ub (3), and HA-CDK5/p35 + G76A ub (4), and incubated with MG132. Extracts were immunoprecipitated with Fk2 antibody and analyzed with αAPE1 and monitored for expression of HA-CDK5 and p35 using antibody specific to CDK5 and p35.
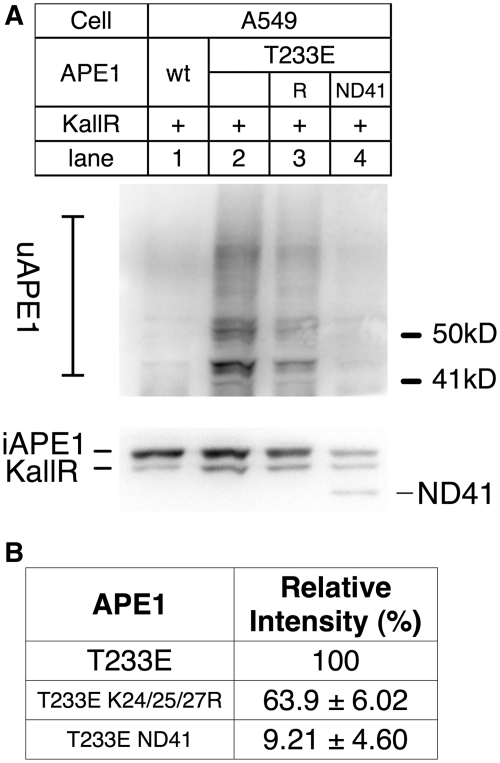
Lys residues 24, 25 and 27 in APE were identified as ubiquitin acceptor sites (22). The K24/25/27R triple Arg substitution mutant was created in the T233E background, and tested if the protein was ubiquitinated (Figure 4A). To calculate the ubiquitination efficiency, we used an internal control to normalize the transfection efficiency: T233E APE1 in which all Lys residues were converted to Arg (KallR, Supplementary Figure S4), as ubiquitination on this Lys-less APE1 should be negligible if any. The amount of uAPE1 was decreased in the K24/25/27R (Figure 4B, ~60% of APE1 with intact Lys). ND41 APE1 (41 amino acid truncation from the N-terminus) in the T233E background resulted in further decrease in ubiquitination (Figure 4B, 9%). We thus concluded that K24, 25, 27 are ubiquitin acceptor sites for T233E APE1, albeit other Lys residues in the N-terminus may be also involved.
Figure 4.
Ubiquitin acceptor Lys in APE1. (A) A549 was transiently transfected with cDNA encoding ‘KallR’ (all Lys residues in APE1 were altered to Arg) as an internal control for transfection (Supplementary Figure S4), along with wtAPE1 (1), T233E (2), T233E K24/25/27R (3) and T233E ND41 APE1 (N-terminal deletion of 41 amino acid residues). (Top) Ubiquitinated APE1; Bot) intact APE1, KallR and ND41 are shown. KallR mutant APE1 migrates slightly faster than the intact APE1 in SDS/PAGE, due to the difference in amino-acid composition from the wtAPE1 (Supplementary Figure S4). (B) Relative intensities of ubiquitinated APE1 were calculated from three independent experiments as in (A), using KallR signals to normalize the transfection efficiency.
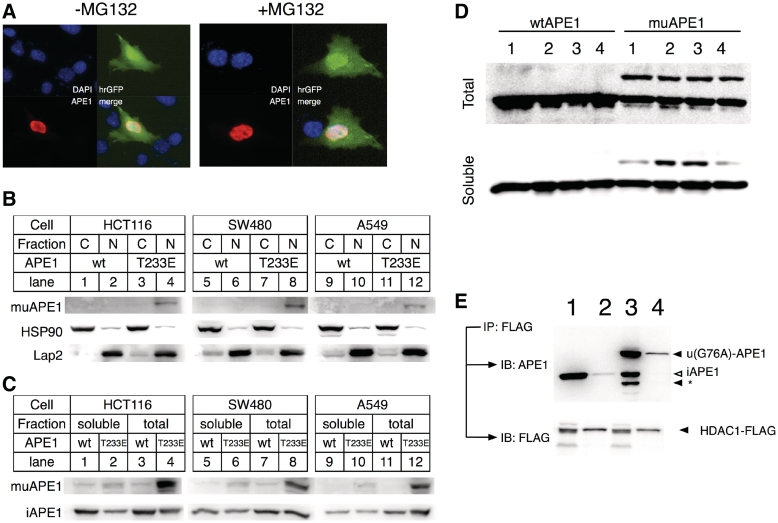
To analyze the subcellular localization of uAPE1, T233E APE1 was expressed in the 2DN cells along with hrGFP using a single IRES containing-vector (Figure 5A). T233E APE1 was found in nuclei exclusively with or without MG132, indicating that ubiquitination of T233E APE1 occurred and remained in the nuclei. To support this observation, HCT116, SW480 and A549 expressing T233E APE1 was extracted after MG132 treatment (Figure 5B). A band of size specific to muAPE1 appeared only with MG132 in the nuclear but not in the cytosol fraction (Figure 5B, lanes 4 versus 2, 8 versus 6, 12 versus 10). These results were in line with the immunocytochemistry above, that uAPE1 was localized in the nuclei. We examined if the muAPE1 in nuclei existed in chromatin assembly by separating the muAPE1 into total and soluble fractions after formaldehyde cross-linking treatment, a procedure commonly carried out during chromatin immunoprecipitation (ChIP) assay (Figure 5C). In this experiment, majority of muAPE1 turned to be insoluble in the nuclear fractions, indicating that the modified APE1 was precipitated along with genomic DNA and chromatin complexes, while the intact APE1 was mostly soluble.
Figure 5.
Presence of uAPE1 in nuclei. (A) Immunocytochemistry of T233E APE1 in 2DN cells. T233E expressed in 2DN cells without (left) or with (right) MG132 treatment, and stained with rhodamine (red). (B) Presence of uAPE1 in nuclear fraction. HCT116 (1–4), SW480 (5–8) and A549 (9–12) were transiently transfected with wt (1, 2, 5, 6, 9, 10) or T233E (3, 4, 7, 8, 11, 12) APE1 and treated with MG132. After 24 h, cell extracts were fractionated to cytosolic (1, 3, 5, 7, 9, 11) and nuclear (2, 4, 6, 8, 10, 12) fractions, and analyzed for muAPE1 along with HSP90 and Lap2 as cytosolic and nuclear marker respectively. (C) Presence of uAPE1 in chromatin assembly. Transiently transfected cells (wtAPE1: 1, 3, 5, 7, 9, 11; T233E: 2, 4, 6, 8, 10, 12) were cross-linked with formaldehyde to trap proteins associated with chromatin. Cells were then sonicated for 5 s to only break cellular membrane. Insoluble proteins, not associated to genomic DNA, were removed from soluble fraction by centrifugation (1, 2, 5, 6, 9, 10). The detailed procedure are described in the Supplementary Data. (D) 293/wtAPE1 (left) or 293/muAPE1 (right), both induced by dox for 16 h (Supplementary Figure S2), were treated with formaldehyde in NaCl at 120 mM (Lanes 1 and 2), 500 mM (3) and 10 mM (4), and sonicated for 10 s once (1, 3, 4) or for 10 times (2). (Top) Total and (bottom) soluble fraction. (E) Enhancement of interaction of APE1 with HDAC1 by monoubiquitination. HCT116 cells co-expressing HDAC1-FLAG with wtAPE1 (1 and 2) or ub-APE1 fusion (3 and 4) were immunoprecipitated with FLAG epitope and analyzed with α-APE1. 1 and 3: total input, 2 and 4: IP fractions. Asterisk (*): truncated APE1.
We developed stable HEK293 T-Rex cell lines that express the intact APE1 or muAPE1 fusion proteins in nuclei (Supplementary Figure S2) with the addition of doxycycline. With the 293 derivatives, solubility of muAPE1 in cells was examined (Figure 5D). After expression of either protein, cells were cross-linked to genomic DNA by formaldehyde (18). After cell lysis by sonication, a large fraction of muAPE1 was found insoluble (Figure 5C, lane 2, top versus bottom). The protein became soluble after multiple cycles of sonication to shear the genomic DNA (Figure 5C lane 2) or by a high salt (0.5 M) concentration (lane 3). In contrast, wtAPE1 was soluble in all tested conditions. It should be noted that recombinant muAPE1 from E. coli was very stable and soluble (Supplementary Figure S3), and thus it is unlikely that the muAPE1 in the cells was intrinsically unstable.
APE1 interacts with histone deacetylase 1 (HDAC1) for its gene suppressor function (18). The interaction of HDAC1 with the intact APE1 and mu-APE1 fusion proteins was examined (Figure 5E). The immunoprecipitated fraction contained more muAPE1 proteins than the intact APE1, suggesting that HDAC1–APE1 interaction was enhanced by ubiquitination of APE1, and possibly contributed to the gene regulatory functions of APE1.
We determined gene expression profiles of the 293 stable transfectants, i.e. 293/ctl, 293/wtAPE1 and 293/ muAPE1, with or without 1 μg/ml dox. Three sets of independent RNA preparations (18) were examined in an Affymetrix GeneChip Exon 1.0 ST Arrays (see Experimental Procedure for details), and pooled about 15 500 genes that showed statistically meaningful results between dox ± conditions (see ‘Materials and Methods’ section).
Investigating an effect of ubiquitination on APE1's gene regulatory function may be complex, because APE1 affects expression of a number of genes in both activation (via its Ref-1 function) and suppression by very different mechanisms (mediated via Ref-1 function or via nCaRE and acetylation, respectively). To answer this question, we examined the gene expression profiles in detail. Histograms revealing the number of down or upregulated genes due to dox addition (Supplementary Figure S6A) indicated that the increased expression of the wtAPE1 gene resulted in more transcriptional downregulation than activation. This tendency was further enhanced when ub-APE1 protein was induced, as the skewness became more negative with a lower kurtosis value (Supplementary Figure S6). In contrast, the control cell line generated a near perfect bell-shape histogram (Supplementary Figure S6) with its skewness near 0 and a much higher kurtosis value than those of the wtAPE1- or ubAPE1-expressing cell lines, indicating that the vector alone did not affect the global gene expression relative to the cases of wtAPE1 and ub-APE1 expression.
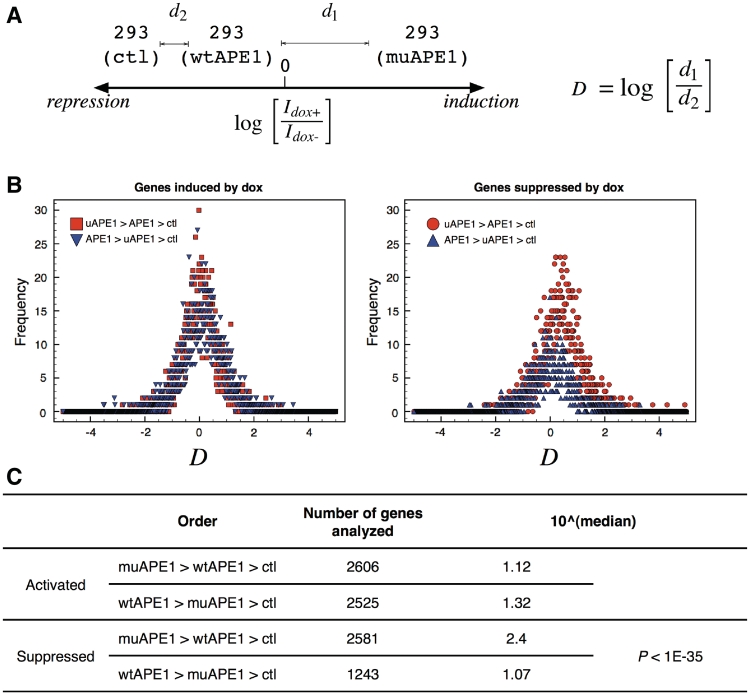
To further elucidate the enhancement of multi-gene suppression by ub-APE1 over intact APE1, we sought for a similar approach to the work by Enard et al. (47). A ‘distance’ value D was defined (Figure 6) to assess whether APE1's activator or repressor function was enhanced by ubiquitination (Figure 6). With the genes that were activated by dox (left, Figure 6B), the distributions of D-values for genes activated by muAPE1 was nearly perfectly bell-shaped with the peak at 0 (where d1 = d2), while a slight right shift was observed with wtAPE1 (median = 1.32, Figure 6C). A right-shift was present with genes suppressed by muAPE1 (circles in right panel, Figure 6B) with median value 2.4 (Figure 6C). Therefore, the results indicated that ubiquitination enhanced the gene suppression activity of APE1 more effectively than the activation.
Figure 6.
Effect of muAPE1 on gene regulation. (A) definition of distance (D) to assess the extent of ‘enhancement’ by muAPE1 (or wtAPE1). Genes were grouped based on their fold increase/decrease by dox for 16 h. In the example, D-value was calculated for a gene that was activated by muAPE1 at highest, then by wtAPE1, and by ctl at least. Idox+ and Idox-: gene expression dosages with or without doxycycline. (B) Histograms of D values. Left: genes that are activated; Right: genes that are suppressed by wtAPE1 and ub-APE1. (C) Median values of the histograms in (B).
DISCUSSION
The T233E amino acid substitution, recently analyzed by Huang et al. (23) was used as a mimicry of T233-phosphorylated APE1 by CDK5. Considering the dramatic change and its effect on APE1 conformation, intracellular levels of APE1 ubiquitination activity might be affected as well. This prediction was correct, as we observed the accumulation of uAPE1 from cells expressing the T233E APE1. Among tested cell lines, A549 and MDM2−/− MEFs were particularly high in generating uAPE1. While understanding the reason behind the variable APE1 ubiquitination in the cells is beyond the goal of this study, these two cells lines helped us elucidate the mechanism of APE1 ubiquitination.
The puAPE1 formation was mediated by ubiquitin conjugation mainly via K48 (Figure 1). Interestingly, purified puAPE1 with UbKallRbutK48 was less than that with the wt-Ub, which may be interpreted that some of polyubiquitin chain on APE1 is mediated via Lys other than K48. More technical consideration is necessary to demonstrate puAPE1 formation via Lys residues other than K48 of Ub. In any case, our conclusion is that HWB formation is thoroughly mediated by ubiquitination, as the UbKallR construct failed to form puAPE1 (Figure 1D). Other small ubiquitin-like modifiers also failed to conjugate T233E APE1.
Eliminating K24/25/27 in APE1 by substituting with Arg decreased the level of both mono- and poly-ubiquitination of T233E APE1. Therefore, while the results indicated the importance of the Lys residues, the data also imply that ubiquitination may take place at Lys other than K24/25/27. This was unexpected, considering that in the previous study ubiquitination on the K24/25/27R mutant was substantially decreased albeit not completely (22). It is possible that modification of T233 alters the local conformation and invokes additional Lys acceptors to be readily accessible by E3 ligases including MDM2. The dependency on K24/25/27 for APE1 ubiquitination may also vary with cell types and growth conditions. Consistent with the previous study (22), deleting the N-terminal 41 amino acid residues decreased the ubiquitination activity further, strongly indicating that APE1 ubiquitination was controlled by the N-terminus. Interestingly, Fantini et al recently reported that the N-terminal Lys residues, including K24, K25, K27, K31 and K32, are critical for its interaction with nucleophosmin (NPM1) (19), which can regulate APE1's endoribonuclease activity (48–50). It is possible that ubiquitination on these Lys residues affects the interaction of APE1 with NPM1, and its function for RNA quality control (48).
The monoubiquitinated form of APE1 was an abundant form of uAPE1, and in the present study we found that the uAPE1 was kept inside the nucleus. The nuclear localization of uAPE1 differs from our earlier observation (22), in which a ubiquitin-APE1 fusion protein was excluded from nuclei. Obviously, the fusion protein encoded in the expression vector in the previous study is translated in cytosol. These studies thus point out that uAPE1 cannot be transported through nuclear membrane. The present data showed that in situ ubiquitination resulted in uAPE1 formation in nuclei. Therefore, ubiquitination may have profound effects on APE1's DNA repair and gene regulatory functions, albeit transiently (51). This concept was supported with the results shown in Figure 6. The HEK293 T-Rex derivatives provides an experimental system ideal for our gene array analysis, as the wt-APE1 and ub-APE1 genes could be integrated in an identical genomic locus and were induced with doxycycline (52). In this tightly controlled system, there was a clear tendency that expression of the mu-APE1 enhance the global gene suppression (Figure 6). Therefore, we propose that once ubiquitinated, APE1 may increase its existence on genomic DNA and its role as a gene repressor. DNA binding assays using fluorescence anisotropy measurement corroborate this scenario, where ub-APE1 fusion protein showed persisting DNA affinity in higher range of salt concentration in the DNA binding assay than the intact APE1 protein (Supplementary Figure S3). Since purified ubiquitin did not bind DNA (Supplementary Figure S3), the increased affinity for DNA must be due to an altered biochemical property introduced to APE1 by the ubiquitin conjugation. Given the fact that interaction of APE1 with HDAC1 was enhanced by ubiquitination (Figure 6), increased affinity for the genomic DNA may enable APE1 to downregulate a number of genes via HDAC1-dependent deacetylation of nearby histones. Such a gene regulatory mechanism likely has a broader influence than that of those involved with nCaRE-dependent genes.
Although the analysis of individual ‘distance’ values provided evidence of the role of ubiquitination in enhancing APE1's gene suppression, we did not categorize genes according to biological functions such as apoptosis. However, we found many genes involved in stress responses were downregulated in the cells expressing ub-APE1 compared to that of wtAPE1, including ferritin and TUBA1 which we confirmed in RT–PCR assay (Supplementary Figure S6B). Interestingly, some genes that were activated with induction of wtAPE1 were among what were previously reported as genes suppressed in APE1-downregulated cells (53) (Supplementary Figure S6C). It should be noted, however, that the experimental strategies, materials, and methods were very different between the previous and our studies, and so comparison between these may be difficult.
Although the present results are more focused on the gene regulatory function of APE1, another interesting question is how ubiquitination alters APE1's repair activity. To know whether mu-APE1 fusion protein can singly repair AP sites, we used an APE-negative E. coli mutant and carried out a complementation assay (survival curve) against methyl methanesulfonate (MMS) that generates AP sites in the cells (Supplementary Figure S3F). There was no noticeable difference in the complementation activity, suggesting that the AP endonuclease activity of ub-APE1 fusion protein is comparable to that of the wt-APE1. It should be noted, however, that we cannot exclude the possibility that ubiquitination may alter the interaction of APE1 with the other repair enzymes in mammalian cells. Dedicated future studies are necessary to answer the question.
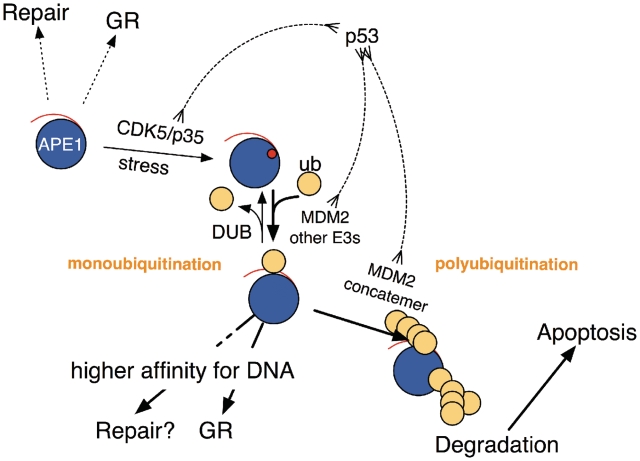
To know the influence of MDM2 on APE1 ubiquitination, MEFs that specifically lack the MDM2 genes was used (Figure 2). Although the present study indicated that other backup E3 enzymes exist in the cells, we have several reasons to believe that MDM2 is a functional E3 ligase and degradation stimulator for APE1. As previously reported, expression of MDM2 enhanced APE1 ubiquitination (22). Also, using the unique ubiquitination assay (Figure 2C), MDM2 was able to ubiquitinate APE1, an activity that was enhanced by the Glu substitution for APE1's T233. Finally, in the MDM2−/− MEF and A549 cells, expression of MDM2 or ubiquitin decreased uAPE1. It is known that forced expression of the wt-ubiquitin can lead to degradation of a target protein in certain cases (54). These observations are in accordance with the positive role of MDM2 for degradation of APE1 via ubiquitination. Based on the present results, we revised the model of the mechanism of APE1 ubiquitination (21) in Figure 7. While MDM2 contributes to APE1 ubiquitination and responds to the phosphorylation event at the T233 position, there must be backup ubiquitin E3 ligase(s) that react on APE1. The availability of other E3 ligases likely depends on tissue types and cell cycle. Once the reaction proceeds beyond the monoubiquitination stage, MDM2 polyubiquitinates APE1 for degradation. It is likely that MDM2 may possess an additional role in facilitating degradation after APE1 polyubiquitination by a similar mechanism proposed for p53 degradation (55–57). The balance between mono- and poly-ubiquitination may be controlled by other factors such as deubiquitinases which need focused investigations. Currently we do not know about the backup E3 ligases for APE1 ubiquitination as there are more than 600 E3 ligases are identified in cells, although CHIP (C-terminus of HSC70-interacting protein) was recently identified as an E3 ligase actor for degradation of DNA polymerase-beta (58). It would be interesting to test the possibility if CHIP also can ubiquitinate APE1, especially after phosphorylation of T233. Regardless, it is possible that T233 phosphorylation need to precede for yet unidentified E3 ligase(s) to act on APE1.
Figure 7.
Model of regulation of APE1 ubiquitination. Under a normal condition, unmodified APE1 functions as a DNA repair protein as well as a general gene regulator. In a stressful condition such as replication stress and DNA damage generation, p53 as well as CDK5/p35 (60,61) are activated and trigger APE1 ubiquitination by phosphorylating T233 in APE1 and by increasing MDM2. Activities of both DNA repair and gene regulation (GR) functions of APE1 may be altered by monoubiquitination which also triggers polyubiquitination and APE1 degradation in the presence of oligomerized MDM2 (62).
Our observation that ubiquitination of endogenous APE1 was enhanced by the expression of CDK5/p35 underscores the physiological and pathophysiological significance of APE1 PTM. While CDK2, CDK4 and CDK6 are essential for normal cell-cycle initiation and checkpoint, specific expression of CDK5 in post-mitotic neuronal cells suggests its unique function in the brain (35,59). Interestingly, any abnormal stimuli to initiate cell growth in neurons invoke activation of CDK5, and result in apoptotic cell death (36). As in studies with other cell types, APE1 has been known to be downregulated before apoptosis, and so it is possible that APE1 phosphorylation occurs in the early time point of apoptosis. Although not systematically analyzed, our preliminary assays indicated that CDK5 was expressed in the entire cancer cell lines analyzed (data not shown). Therefore, it is conceivable that a low level of expression of this protein phosphorylates APE1, which serves as a trigger for ubiquitination. It is important to understand precisely what stimuli activate CDK5 or its cofactors p35/p25 in the cancer cells. Alternatively, CDK4 and CDK6, the closest paralog of CDK5, may also participate in APE1 modification, but in a cell-cycle dependent manner. These questions need to be tested to understand the effect of the APE1 regulation in the cancer biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institute of Health [CA98664 to T.I.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors appreciate Drs Crowe and Chattopadhyay (University of Virginia) for their providing the shAPE1 expression vector. The authors thank Dr T. Iwakuma (LSU, Deptartment of Genetics) for his providing MEF cells and other reagents such as antibodies, and grateful for his and Dr M.W. Lake's discussion of the results and critically reading the manuscript. Dr D. Nguyen at LSU’’s bioinformatics for his expert analysis on gene expression profiling. Ms. Nguyen and Mr. Tran's excellent technical assistance are appreciated.
REFERENCES
- 1.Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saporito SM, Gedenk M, Cunningham RP. Role of exonuclease III and endonuclease IV in repair of pyrimidine dimers initiated by bacteriophage T4 pyrimidine dimer-DNA glycosylase. J. Bacteriol. 1989;171:2542–2546. doi: 10.1128/jb.171.5.2542-2546.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guillet M, Boiteux S. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J. 2002;21:2833–2841. doi: 10.1093/emboj/21.11.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 5.Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl Acad. Sci. USA. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons JL, Dianova II, Dianov GL. APE1-dependent repair of DNA single-strand breaks containing 3′-end 8-oxoguanine. Nucleic Acids Res. 2005;33:2204–2209. doi: 10.1093/nar/gki518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kane CM, Linn S. Purification and characterization of an apurinic/apyrimidinic endonuclease from HeLa cells. J. Biol. Chem. 1981;256:3405–3414. [PubMed] [Google Scholar]
- 8.Izumi T, Hazra TK, Boldogh I, Tomkinson AE, Park MS, Ikeda S, Mitra S. Requirement for human AP endonuclease 1 for repair of 3′-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis. 2000;21:1329–1334. [PubMed] [Google Scholar]
- 9.Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The Many Functions of APE1/Ref-1: Not Only a DNA Repair Enzyme. Antioxid. Redox. Signal. 2008;11:601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao X, Kambe F, Ohmori S, Seo H. Oxidoreductive modification of two cysteine residues in paired domain by Ref-1 regulates DNA-binding activity of Pax-8. Biochem. Biophys. Res. Commun. 2002;297:288–293. doi: 10.1016/s0006-291x(02)02196-4. [DOI] [PubMed] [Google Scholar]
- 11.Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, Kelley MR. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat. Res. 2008;643:54–63. doi: 10.1016/j.mrfmmm.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat. Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 13.Xanthoudakis S, Miao G, Wang F, Pan YC, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs S, Philippe J, Corvol P, Pinet F. Implication of Ref-1 in the repression of renin gene transcription by intracellular calcium. J. Hypertens. 2003;21:327–335. doi: 10.1097/00004872-200302000-00024. [DOI] [PubMed] [Google Scholar]
- 15.McHaffie GS, Ralston SH. Origin of a negative calcium response element in an ALU-repeat: implications for regulation of gene expression by extracellular calcium. Bone. 1995;17:11–14. doi: 10.1016/8756-3282(95)00131-v. [DOI] [PubMed] [Google Scholar]
- 16.Izumi T, Henner WD, Mitra S. Negative regulation of the major human AP-endonuclease, a multifunctional protein. Biochemistry. 1996;35:14679–14683. doi: 10.1021/bi961995u. [DOI] [PubMed] [Google Scholar]
- 17.Okazaki T, Chung U, Nishishita T, Ebisu S, Usuda S, Mishiro S, Xanthoudakis S, Igarashi T, Ogata E. A redox factor protein, ref1, is involved in negative gene regulation by extracellular calcium. J. Biol. Chem. 1994;269:27855–27862. [PubMed] [Google Scholar]
- 18.Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fantini D, Vascotto C, Marasco D, D'Ambrosio C, Romanello M, Vitagliano L, Pedone C, Poletto M, Cesaratto L, Quadrifoglio F, et al. Critical lysine residues within the overlooked N-terminal domain of human APE1 regulate its biological functions. Nucleic Acids Res. 2010;38:8239–8256. doi: 10.1093/nar/gkq691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu J, Liu GH, Huang B, Chen C. Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310. Nucleic Acids Res. 2007;35:2522–2532. doi: 10.1093/nar/gkl1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busso CS, Lake MW, Izumi T. Posttranslational modification of mammalian AP endonuclease (APE1) Cell Mol. Life Sci. 2010;67:3609–3620. doi: 10.1007/s00018-010-0487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busso CS, Iwakuma T, Izumi T. Ubiquitination of mammalian AP endonuclease (APE1) regulated by the p53-MDM2 signaling pathway. Oncogene. 2009;28:1616–1625. doi: 10.1038/onc.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang E, Qu D, Zhang Y, Venderova K, Haque ME, Rousseaux MW, Slack RS, Woulfe JM, Park DS. The role of Cdk5-mediated apurinic/apyrimidinic endonuclease 1 phosphorylation in neuronal death. Nat. Cell Biol. 2010;12:563–571. doi: 10.1038/ncb2058. [DOI] [PubMed] [Google Scholar]
- 24.Fan Z, Beresford PJ, Zhang D, Xu Z, Novina CD, Yoshida A, Pommier Y, Lieberman J. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat. Immunol. 2003;4:145–153. doi: 10.1038/ni885. [DOI] [PubMed] [Google Scholar]
- 25.Robertson KA, Hill DP, Xu Y, Liu L, Van Epps S, Hockenbery DM, Park JR, Wilson TM, Kelley MR. Down-regulation of apurinic/apyrimidinic endonuclease expression is associated with the induction of apoptosis in differentiating myeloid leukemia cells. Cell Growth Differ. 1997;8:443–449. [PubMed] [Google Scholar]
- 26.Bobola MS, Finn LS, Ellenbogen RG, Geyer JR, Berger MS, Braga JM, Meade EH, Gross ME, Silber JR. Apurinic/apyrimidinic endonuclease activity is associated with response to radiation and chemotherapy in medulloblastoma and primitive neuroectodermal tumors. Clin. Cancer Res. 2005;11:7405–7414. doi: 10.1158/1078-0432.CCR-05-1068. [DOI] [PubMed] [Google Scholar]
- 27.Bobola MS, Emond MJ, Blank A, Meade EH, Kolstoe DD, Berger MS, Rostomily RC, Silbergeld DL, Spence AM, Silber JR. Apurinic endonuclease activity in adult gliomas and time to tumor progression after alkylating agent-based chemotherapy and after radiotherapy. Clin. Cancer Res. 2004;10:7875–7883. doi: 10.1158/1078-0432.CCR-04-1161. [DOI] [PubMed] [Google Scholar]
- 28.Bobola MS, Blank A, Berger MS, Stevens BA, Silber JR. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin. Cancer Res. 2001;7:3510–3518. [PubMed] [Google Scholar]
- 29.Koukourakis MI, Giatromanolaki A, Kakolyris S, Sivridis E, Georgoulias V, Funtzilas G, Hickson ID, Gatter KC, Harris AL. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/Ref-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. Int. J. Radiat. Oncol. Biol. Phys. 2001;50:27–36. doi: 10.1016/s0360-3016(00)01561-3. [DOI] [PubMed] [Google Scholar]
- 30.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol. Aspects Med. 2007;28:375–395. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Fantini D, Vascotto C, Deganuto M, Bivi N, Gustincich S, Marcon G, Quadrifoglio F, Damante G, Bhakat KK, Mitra S, et al. APE1/Ref-1 regulates PTEN expression mediated by Egr-1. Free Radic. Res. 2008;42:20–29. doi: 10.1080/10715760701765616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mckenzie JA, Strauss PR. A quantitative method for measuring protein phosphorylation. Anal. Biochem. 2003;313:9–16. doi: 10.1016/s0003-2697(02)00464-5. [DOI] [PubMed] [Google Scholar]
- 33.Fritz G, Kaina B. Phosphorylation of the DNA repair protein APE/REF-1 by CKII affects redox regulation of AP-1. Oncogene. 1999;18:1033–1040. doi: 10.1038/sj.onc.1202394. [DOI] [PubMed] [Google Scholar]
- 34.Yacoub A, Kelley MR, Deutsch WA. The DNA repair activity of human redox/repair protein APE/Ref-1 is inactivated by phosphorylation. Cancer Res. 1997;57:5457–5459. [PubMed] [Google Scholar]
- 35.Zhang J, Herrup K. Cdk5 and the non-catalytic arrest of the neuronal cell cycle. Cell Cycle. 2008;7:3487–3490. doi: 10.4161/cc.7.22.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhariwala FA, Rajadhyaksha MS. An unusual member of the Cdk family: Cdk5. Cell Mol. Neurobiol. 2008;28:351–369. doi: 10.1007/s10571-007-9242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jessberger S, Gage FH, Eisch AJ, Lagace DC. Making a neuron: Cdk5 in embryonic and adult neurogenesis. Trends Neurosci. 2009;32:575–582. doi: 10.1016/j.tins.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 39.Iwakuma T, Parant JM, Fasulo M, Zwart E, Jacks T, de Vries A, Lozano G. Mutation at p53 serine 389 does not rescue the embryonic lethality in mdm2 or mdm4 null mice. Oncogene. 2004;23:7644–7650. doi: 10.1038/sj.onc.1207793. [DOI] [PubMed] [Google Scholar]
- 40.Chattopadhyay R, Bhattacharyya A, Crowe SE. Dual regulation by apurinic/apyrimidinic endonuclease-1 inhibits gastric epithelial cell apoptosis during Helicobacter pylori infection. Cancer Res. 2010;70:2799–2808. doi: 10.1158/0008-5472.CAN-09-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, Koch WM, Ferris RL, Lai SY. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29:163–188. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 42.Rochette PJ, Bastien N, Lavoie J, Guerin SL, Drouin R. SW480, a p53 double-mutant cell line retains proficiency for some p53 functions. J. Mol. Biol. 2005;352:44–57. doi: 10.1016/j.jmb.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 43.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haas AL, Siepmann TJ. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 45.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 46.Fujimuro M, Sawada H, Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- 47.Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 48.Vascotto C, Fantini D, Romanello M, Cesaratto L, Deganuto M, Leonardi A, Radicella JP, Kelley MR, D'Ambrosio C, Scaloni A, et al. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol. Cell. Biol. 2009;29:1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tell G, Wilson DMr, Lee CH. Intrusion of a DNA repair protein in the RNome world: is this the beginning of a new era? Mol. Cell. Biol. 2010;30:366–371. doi: 10.1128/MCB.01174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, Lee CH. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tell G, Fantini D, Quadrifoglio F. Understanding different functions of mammalian AP endonuclease (APE1) as a promising tool for cancer treatment. Cell Mol. Life Sci. 2010;67:3589–3608. doi: 10.1007/s00018-010-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu W, Xiong Y, Gossen M. Stability and homogeneity of transgene expression in isogenic cells. J. Mol. Med. 2006;84:57–64. doi: 10.1007/s00109-005-0711-z. [DOI] [PubMed] [Google Scholar]
- 53.Vascotto C, Cesaratto L, Zeef LA, Deganuto M, D'Ambrosio C, Scaloni A, Romanello M, Damante G, Taglialatela G, Delneri D, et al. Genome-wide analysis and proteomic studies reveal APE1/Ref-1 multifunctional role in mammalian cells. Proteomics. 2009;9:1058–1074. doi: 10.1002/pmic.200800638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadeh R, Breitschopf K, Bercovich B, Zoabi M, Kravtsova-Ivantsiv Y, Kornitzer D, Schwartz A, Ciechanover A. The N-terminal domain of MyoD is necessary and sufficient for its nuclear localization-dependent degradation by the ubiquitin system. Proc. Natl Acad. Sci. USA. 2008;105:15690–15695. doi: 10.1073/pnas.0808373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol. Cell Biol. 2007;27:8284–8295. doi: 10.1128/MCB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Keefe K, Li H, Zhang Y. Nucleocytoplasmic shuttling of p53 is essential for MDM2-mediated cytoplasmic degradation but not ubiquitination. Mol. Cell. Biol. 2003;23:6396–6405. doi: 10.1128/MCB.23.18.6396-6405.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tao W, Levine AJ. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc. Natl Acad. Sci. USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL. CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell. 2008;29:477–487. doi: 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 59.Patzke H, Maddineni U, Ayala R, Morabito M, Volker J, Dikkes P, Ahlijanian MK, Tsai LH. Partial rescue of the p35-/- brain phenotype by low expression of a neuronal-specific enolase p25 transgene. J. Neurosci. 2003;23:2769–2778. doi: 10.1523/JNEUROSCI.23-07-02769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JH, Jeong MW, Kim W, Choi YH, Kim KT. Cooperative roles of c-Abl and Cdk5 in regulation of p53 in response to oxidative stress. J. Biol. Chem. 2008;283:19826–19835. doi: 10.1074/jbc.M706201200. [DOI] [PubMed] [Google Scholar]
- 61.Lee JH, Kim HS, Lee SJ, Kim KT. Stabilization and activation of p53 induced by Cdk5 contributes to neuronal cell death. J. Cell Sci. 2007;120:2259–2271. doi: 10.1242/jcs.03468. [DOI] [PubMed] [Google Scholar]
- 62.Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009;28:3857–3867. doi: 10.1038/emboj.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.