Abstract
A type IIG restriction endonuclease, RM.BpuSI from Bacillus pumilus, has been characterized and its X-ray crystal structure determined at 2.35Å resolution. The enzyme is comprised of an array of 5-folded domains that couple the enzyme's N-terminal endonuclease domain to its C-terminal target recognition and methylation activities. The REase domain contains a PD-x15-ExK motif, is closely superimposable against the FokI endonuclease domain, and coordinates a single metal ion. A helical bundle domain connects the endonuclease and methyltransferase (MTase) domains. The MTase domain is similar to the N6-adenine MTase M.TaqI, while the target recognition domain (TRD or specificity domain) resembles a truncated S subunit of Type I R–M system. A final structural domain, that may form additional DNA contacts, interrupts the TRD. DNA binding and cleavage must involve large movements of the endonuclease and TRD domains, that are probably tightly coordinated and coupled to target site methylation status.
Bacterial restriction of phage infection (1,2) is dependent upon the action of restriction endonucleases (REase) that cleave individual target sites found in invading foreign DNA. Canonical R–M systems consist of an REase that recognizes and cleaves an unmodified nucleotide sequence, and a cognate DNA methyltransferase (MTase) that protects the host DNA from cleavage by methylating an adenine or cytosine nucleotide base within its recognition sequence (3,4). The REase cuts unmethylated DNA targets, but not hemimethylated (the substrate that is transiently generated during DNA replication) or fully methylated DNA. Recently, endonucleases that cleave methylated DNA have been found and characterized (5).
The genes that encode various components of R–M systems are found in virtually all sequenced bacterial and archaeal genomes. Many bacteria contain multiple R–M systems and display the ability to switch between different systems and DNA specificities depending upon conditions (6,7). The total number of R–M systems encoded in various bacterial species varies widely, ranging from single R–M genes in some bacteria to as many as twenty in Helicobacter pylori (8). R–M systems are extremely diverse with respect to their protein subunit composition, the length and symmetry of their DNA recognition sites, the position and pattern of DNA cleavage, requirements for various cofactors [such as divalent metal ions, ATP and S-adenosylmethionine (SAM)] and methylation status (9,10).
The extraordinary diversity in R–M architecture and DNA-recognition patterns has lead to a classification system divided into at least four distinct categories. The most well studied of these are the Type II R–M systems, which usually consist of separate MTase and REase enzymes that independently recognize identical DNA sequences that are 4–8 bp in length; many of their target sites are palindromic. Most REases of this Type IIP subclass combine their DNA recognition and cleavage functions into a single folded protein domain which dimerizes to recognize palindromic sequences.
Variations in the ‘typical’ organization of Type II R–M systems are often observed. For example, Type IIS REase contain separate, independently folded target recognition domains (TRDs) and nuclease domains. This structural architecture allows recognition of asymmetric recognition sequences, with cleavage occurring several base pairs downstream from the recognition site (an activity termed ‘shifted cleavage’) (11). The structures of three Type IIS REases have been determined: [FokI (GGATG N9/N13), BfiI (ACTGGG N5/N4) and BspD6I (GAGTC N4/N6)] (the target site nomenclature denotes the location of cleaved phosphates on each strand, downstream from the 3′-end of the target site sequence) (12–14). A preliminary crystallographic study of Eco57I (CTGAAG N16/N14) has been reported (15), but the actual structure remains to be described.
An evolutionary weakness of R–M systems that comprise separate REase and MTase proteins is that they may not be able to rapidly alter their target site specificity: a change in REase specificity at even a single base pair, without an equivalent change in DNA recognition by the cognate MTase would generally result in host toxicity. One solution to this problem is to have the REase domain relinquish its own specificity determinant and directly couple its DNA-recognition function to that of its corresponding MTase, as is observed for the multi-subunit type I and III R–M systems (16,17). Type I R–M enzymes such as EcoKI, EcoAI and EcoR124I are complex hetero-oligomers of two REase (HsdR) subunits, two MTase (HsdM) subunits and a single DNA sequence-specificity (HsdS) subunit that drives the DNA binding of the entire enzyme assemblage. Depending on the methylation status of the DNA substrate, this complex can function either as an REase or as an MTase. These enzymes recognize an asymmetric, bipartite sequence (usually 12–15 bp) and require ATP hydrolysis to direct cleavage at a distant site.
Type IIG R–M systems also follow this general strategy for target recognition, but instead of encoding separate REase, MTase and specificity subunits, they have evolved to combine them within a single uninterrupted protein chain. A similar architecture is also observed for many type IIB enzymes, which cleave on both sides of their bipartite recognition sequences (although some type IIB enzymes are not organized as R–M fusions). The endonuclease activity for some type IIG enzymes is affected by SAM, a cofactor required for DNA MTase activity (11). The stimulation of cognate endonuclease activity by SAM presents an unexplored regulatory mechanism that may facilitate coordination of the MTase and endonuclease functions. Tight regulation of the MTase and endonuclease activities of some Type IIG enzymes appears to be crucial for their host organisms, particularly in those cases where no obvious additional MTase is found. Examples of such lone R–M fusions include the Tth111II (CAARCA N11/N9) and MmeI (TCCRAC N20/N18) groups of REases (18).
RM.BpuSI (hereafter termed ‘BpuSI’) is a Type IIG REase that displays the DNA-specificity profile and endonuclease cleavage activity that is typical of a Type IIS endonuclease (GGGAC N10/N14, where cleavage is induced downstream at N10 and N14 on the top and bottom strand, respectively) (see ‘Materials and Methods’ and ‘Results’ sections for a description of the identification of BpuSI). Sequence analyses indicated that BpuSI consists of three functional domains: an N-terminal nuclease domain, a γ-type N6mA MTase domain, and a C-terminal specificity domain (or TRD).
Here we report the identification, sequence and characterization of the BpuSI R–M system, the determination of its crystal structure, and verification of its catalytic residues by site-directed mutagenesis. The unbound BpuSI protein contains a nuclease domain that is sequestered in an inactive conformation, an amino-MTase domain, and a shared specificity domain. An α-helical domain (previously postulated based on mutational and structural studies of type I R–M proteins) that connects the endonuclease and MTase domains appears to regulate and physically couple their relative conformations and activities and perhaps establish the cleavage distance from the enzyme's target site.
MATERIALS AND METHODS
Isolation and subcloning of BpuSI M1, M2 and RM fusion genes
BpuSI was initially purified directly from a novel bacterial strain that was isolated from a sacrificed black ant. Sequencing of the 16 rRNA gene from that bacterial strain (Supplementary Figure S1) indicated highest sequence similarity to Bacillus pumilus strains X93 and IMAU80221. This bacterial strain was therefore named B. pumilus S, and the enzyme was named BpuSI (19). Digest experiments conducted on using lambda phage DNA substrate with purified BpuSI indicated that the enzyme appeared to act as an isochizomer of the RM.BsmFI endonuclease (Supplementary Figures S2 and S3).
Genomic DNA was extracted from B. pumilus S strain by phenol–chloroform extractions. The BpuSI genomic DNA was partially digested with ApoI and ligated to EcoRI-digested and CIP-treated pBR322. Similarly, MseI or CviAII partially digested genomic DNA was ligated to NdeI-digested and CIP-treated pBR322 with compatible ends. The ligated DNA was transferred into Escherichia coli strain ER2683 by transformation. A total ~6.4 × 104 transformants were obtained and pooled together for Maxi-plasmid preparation (Qiagen Maxi-prep kit). The plasmid DNA library with genomic DNA inserts was digested with excessive BsmFI, a BpuSI isoschizomer, in order to identify plasmids harboring expressed MTase activities that were specific for the BsmFI and BpuSI target site. Resistant plasmids were recovered by retransformation using the digested plasmid DNA [because the BpuSI MTase-containing plasmids were modified by BpuSI MTase(s) and thus were resistant to BsmFI digestion]. Five BsmFI-resistant plasmids were recovered from the ApoI partial libarary digested by BsmFI and retransformation. From the CviAII and MseI mixed DNA libraries, nine BsmFI-resistant clones were obtained. DNA sequencing of the inserts derived from the 14 resistant clones identified two MTase genes and a partial BpuSI endonuclease gene. Following two rounds of inverse PCR walking, the entire BpuSI R–M system (M1, M2 and an R–M fusion gene) was obtained. The gene organization and coding sequences of the entire BpuSI RM system are shown in Supplementary Figure S4.
Subcloning, expression and purification of recombinant BpuSI
The bpuSIM1M2 genes (flanked by BamHI and SalI sites) were amplified in PCR and cloned into pACYC184 with compatible ends. The pre-modified expression host was ER2683 [pACYC-bpuSIM1M2]. A PCR fragment carrying the bpuSIRM fusion gene was cloned into the EcoRI and SphI sites of pUC19, generating expression strain ER2683 [pACYC-bpuSIM1M2, pUC-bpuSIRM]. To facilitate the purification of BpuSI REase (BpuSI REase = BpuSI RM fusion protein), a 6× His tagged version was also constructed. A PCR fragment with 6× His C-terminal tag was inserted into the XbaI/AgeI sites of pTXB1, generating a T7 expression strain ER2566 [pACYC-bpuSIM1M2, pTXB1-bpuSIRM].
Twelve liters of cells ER2566 [pACYC-bpuSIM1M2, pTXB1-bpuSIRM] were cultured in LB + Cm (30 µg/ml) + Amp (100 µg/ml) to late log phase at 37°C in a shaker and BpuSI expression was induced by addition of 0.3 mM IPTG final concentration. IPTG induction was continued overnight at 37°C. The protein bound poorly on nickel column (data not shown). Therefore, BpuSI REase was purified to over 95% purity by chromatography through heparin Sepharose column and HiTrap Q HP (5 ml; GE Lifesciences) column and used for crystallization trials.
In order to generate BpuSI that was site-specifically substituted with selenomethionine, E. coli host cells (T7 Express Crystal, NEB) were transformed by pACYC-bpuSIM1M2 and pTXB1-bpuSIRM sequentially. The cells were grown at 30°C at 200 r.p.m. overnight in M9 minimal medium (Na2HPO4: 6 g/l; KH2PO4: 3 g/l; NaCl: 0.5 g/l; NH4Cl: 1 g/l; 0.4% glucose, 1 mM MgSO4, 0.3 mM CaCl2, 0.2 ml/l of 1% ferric ammonium citrate) that was supplemented with 50 µg/ml l-methionine, 100 µg/ml ampicillin and 30 µg/ml chloramphenicol. One milliliter of the overnight culture was resuspended in 80 ml of fresh medium and allowed to grow at 30°C, 200 r.p.m. to OD600 = 2.4. Ten milliliters of the culture were inoculated into 1 l of fresh medium. The culture was allowed to grow to OD600 = 1.5 at 30°C, 200 r.p.m. It was then spun down and resuspended in 1 l of fresh medium without methionine and incubated at 30°C, 200 r.p.m. for 3 h. d/l-seleno-methionine (Sigma-Aldrich) was then added to a final concentration of 0.16 mg/l along with IPTG at a final concentration of 0.25 mM. The culture was further incubated at 30°C, 200 r.p.m. for 9 h. It was then harvested and stored at −20°C until lysed for protein purification. Cell pellets from 6 l of culture were used for purification of Se-Met BpuSI.
Purification of the catalytic mutants
BpuSI mutants D60A and E76A were purified from 1 l overnight culture of ER2566 [pACYC-bpuSIM1M2/pUC19-bpuSIRM] using HiTrap Heparin (5 ml; GE Lifesciences) followed by HiTrap Q HP (5 ml; GE Lifesciences) with the same linear gradient of 50–1000 mM NaCl in 20 mM Tris–HCl, pH 8.0, 1 mM EDTA. For direct comparisons, BpuSI WT protein was purified using the same procedures.
Verification of methylation activity of the R–M fusion
Four hundred nanograms of pACYC184, which contains seven BpuSI recognition sites, was treated with 0.8 µg of purified BpuSI E76A (a cleavage deficient mutant) in the presence of 160 µM SAM in NEBuffer 4 at 37°C for 30 min. The enzyme and SAM was then removed from the DNA through purification using Wizard SV gel and PCR clean-up system (Promega). Two hundred nanograms of the treated DNA were subjected to cleavage by 2 U of purified BpuSI in NEBuffer 4 at 37°C for 1 h. The cleavage reactions were then analyzed using agarose electrophoresis.
Comparisons of cleavage kinetics on single-site versus double-site substrates
A double-site BpuSI substrate was generated by amplifying a 274-bp fragment of pACYC184 (from nucleotides 321 to 594) in PCR. The two BpuSI sites are located at Positions 371 and 524 and arranged in a head-to-head orientation. A single-site BpuSI substrate was generated in two-step PCR to mutate the nucleotide 524 site to TTAAA such that a DraI site (TTTAA) is generated. The number of BpuSI sites of the amplified DNA was verified by restriction mapping using BpuSI.
In a 20-µl reaction containing 1× NE Buffer 4, 40 nM of the single-site substrate or 20 nM of the double-site substrate (40 nM BpuSI site) was incubated with 180 nM of BpuSI at 37°C in an Applied Biosystems 2720 thermocycler for the designated period of time. The reactions were stopped by addition of 4 µl of 6× DNA loading buffer containing 1% SDS and 60 mM EDTA. The cleavage reactions were analyzed by electrophoresis through 2.5% agarose gels in 1× TBE followed by staining using SYBR Gold (Invitrogen). The volume of the DNA bands were measured by scanning the gels using the Typhoon 9400 system (GE Lifesciences) at 488 nm for excitation and 520 nm for emission, followed by analysis using ImageQuant (GE Lifesciences). The volume of the substrate band as a fraction of the volume of all DNA bands was plotted against time to obtain an exponential decay curve.
The change of the amount of the substrate over time was used to calculate the first-order rate constant kf where
is given by
| (1) |
[A]t is the amount of substrate at a given time point t, [A]0 is the initial concentration of the substrate, t is time in second and kf is the first-order rate constant for the forward reaction. [A]r is the amount of residual substrate sequestered by cleavage-incompetent form of BpuSI and was not cleaved. Origin 8.5 (OriginLab) was used for curve fitting. The fitted kf values from two independent experiments were averaged to give the final kf values for the single-site and double-site substrates.
Crystallization and structure determination
Initial crystallization trials used a protein concentration of 5 mg/ml. Several conditions produced initial hits, but only one (Hampton Research Index Screen condition A3; 2 M ammonium sulfate in 100 mM Bis–Tris, pH 5.5) yielded crystals reproducibly. The crystals were grown in the presence of synthetic DNA constructs, however while the presence of DNA was strictly required for all subsequent crystal growth, the final structures were found to correspond to unbound protein; therefore the DNA appeared to play a secondary chemical role in crystal formation.
Crystals for X-ray data collection were grown by vapor diffusion using the hanging-drop method with 1.8–2.0 M (NH4)2SO4 as precipitant, in 100 mM Bis–Tris, (pH 5.5) buffer containing 150 mM NaCl, 1 mM sinefungin, 10 mM dithiothreitol and 10 mM MgCl2 or 10 mM MnCl2. All data were collected under cryo-cooled conditions at 100°K. The crystals were initially flash-frozen from mother-liquid containing 20% ethylene glycol. Crystals of both wild-type and SeMet-BpuSI were extremely fragile, diffracted to low resolution (~4 Å), and deteriorated rapidly in the X-ray beam produced at synchrotron beamlines (BL 5.0.1 and 5.0.2; and BL8.2.1 and 8.2.2 at the Advanced Light Source and at Lawrence Berkeley National Laboratory, respectively). It was discovered fortuitously that soaking the crystals with 0.2 M Sodium Iodide drastically improved the stability of the selenomethionyl labeled BpuSI crystals. Eventually, all datasets were collected with NaI soaked selenomethionine-BpuSI crystals after gradual transfers to artificial mother liquors containing 2.0 M (NH4)2SO4 in 100 mM Bis–Tris (pH 5.5), 200 mM NaI and 20% ethylene glycol.
Initial phases (to 3.2Å resolution) were obtained using an in-house dataset (Cu-Kα X-rays generated using a rotating anode generator. Diffraction intensities were measured using an RAXIS-4++ imaging plate detector (Rigaku Inc.) collected from an SeMet-BpuSI crystal soaked with sodium iodide. Phases were calculated using the program SHELXCDE with an HKL2MAP interface. The electron density map calculated with the initial phases were excellent, which allowed manual placement of helices and 8 β-strands in a central β-sheet, totaling 429 residues of a poly-alanine model, with structural elements in three different domains. However, it was difficult to identify a starting point for side-chain registration and unambiguous modeling of the protein sequence.
Two different approaches were used to determine the side-chain registry of the model. First, several models predicted by the automatic fold-recognition server PHYRE each containing a MTase domain were superposed on the starting model, and side chains of the central helix in the MTase core were used as anchoring point for manual side-chain docking. At the same time, the iodide sites from SHELXCDE, together with additional synchrotron-derived datasets from a Ta6Br14-soaked crystal collected at two different wavelengths (1.524 and 0.988Å for the dispersive peak and high-energy remote values of Tantalum, respectively) were used for multiple isomorphous replacement (MIR) by the program suite SOLVE/RESOLVE. This allowed the program to automatically trace 457 residues and to assign 214 unique side chains into the density (Z score = 72 and resolve R-factor 0.23). The main-chain trace of the model superposed well with the manually built model and confirmed most of the manually assigned side chains. To generate a whole molecule, a portion of domain 4 was built into the electron density map of a neighboring asymmetric unit. The partial model and SOLVE/RESOLVE phases were then placed into ARP/WARP for additional autotracing, following by side-chain assignment at 2.6 Å and again at 2.35 Å when higher resolution datasets became available. In the final model, several residues at the N-terminus (1–6) and C-terminus (876–878) and two internal segments, residues (291–292) connecting domains 2 to 3, and residues (549–551) were left out due to high flexibility.
RESULTS
Characterization of BpuSI
The BpuSI enzyme generates pBR322 and λ DNA-digestion patterns identical to that of BsmFI (Supplementary Figure S2). Its recognition sequence and cleavage sites were subsequently determined to be GGGAC N10/N14 by run-off DNA sequencing (Supplementary Figure S3), again corresponding to the cleavage pattern generated by BsmFI. BpuSI is active at 37°C and its cleavage activity can be inactivated by incubation at 65°C for 20 min. BpuSI is active in all four NEB restriction buffers and is most active in New England Biolabs (NEB) buffer 4 (50 mM potassium acetate, 20 mM Tris–acetate, 10 mM magnesium acetate and 1 mM dithiothreitol, pH 7.9). Digestion of λ DNA by BpuSI in buffer 4 using a 20-fold excess of enzyme unit for 1 h did not show ‘star’ activity. Supplementation with SAM in the cleavage reaction does not appear to enhance BpuSI endonuclease activity.
The entire BpuSI R–M system consists of two MTase genes (M1 and M2) and the R–M fusion gene. The organization and coding sequences of the entire BpuSI R–M gene system is shown in Supplementary Figure S4. The MTase domain in the R–M fusion protein displays high sequence similarity to N6A γ-type MTases, and the C-terminus region of the R–M fusion protein was presumed to correspond to the TRD.
Overall structure of BpuSI
The structure of BpuSI was determined to 2.35 Å resolution, with final values for the refinement R-factors (Rwork and Rfree) of 0.21 and 0.26, respectively (Table 1). A total of fourteen residues in the structure are disordered and were not included in the final model: the N-terminus (residues 1–6), the C-terminus (residues 876–878) and two short surface loops (residues 291–292 and residues 549–551). The crystals grew only in the presence of synthetic DNA oligonucleotides. However, no electron density was observed corresponding to the bound DNA; therefore the structure appears to correspond to and is treated as unbound (apo) BpuSI. It is unclear whether the DNA duplex was cleaved and released or if they were not bound under the high-salt conditions used during crystallization. We presume that the high concentration of DNA in the drops affected the pH, free ion concentration or other solution properties in a manner that facilitated crystal growth.
Table 1.
BpuSI Data collection and refinement statistics
| Data set Id Wavelength (Å) | Native-1a 1.5418 | TaBr-1a 1.2542 | TaBr-2a 0.9537 | Native-2a 0.9537 | Native-3a 0.9774 | Se-peak 0.9794 | 11C1 1.5418 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||||
| Space group | C2221 | C2221 | C2221 | C2221 | C2221 | C2221 | C2221 | |||
| a (Å) | 213.1 | 210.5 | 209.9 | 211.84 | 213.47 | 203.1 | 198.81 | |||
| b (Å) | 215.6 | 219.2 | 218.9 | 216.55 | 215.73 | 226.0 | 225.02 | |||
| c (Å) | 73.7 | 74.1 | 73.9 | 73.56 | 73.71 | 75.0 | 76.91 | |||
| Resolution (Å) | 50–3.35 | 50–3.2 | 50–3.2 | 50–2.68 | 50–2.35 | 50–3.5 | 29.9–3.1 | |||
| Unique reflections | 70 485 | |||||||||
| Redundancyb | 6.1 (4.9) | 6.4 (5.6) | 11.5 (10.3) | 3.9 (2.8) | 4.4 (2.0) | 6.2 (5.5) | 3.1 (2.7) | |||
| Completeness (%)b | 99.4 (96.7) | 98.3 (97.9) | 100 (100) | 99.1 (97.8) | 93.2 (63) | 97.4 (93.5) | 99.5 (99.7) | |||
I/ b b
|
16·2 (6.2) | 11·2/(2.1) | 15 (3) | 12·6 (4.5) | 12·2 (1.2) | 17.8 (4.0) | 8·9 (2.0) | |||
| Rmergec (%)b | 9.2 (20.7) | 17.6 (76.2) | 18.2 (69.7) | 8.0 (19.1) | 9.5 (58.1) | 11.3 (39.0) | 11.7 (62.8) | |||
| B(iso) (Å2) | 76.89 | 55.1 | 42.6 | 86.4 | ||||||
| Refinement | ||||||||||
| Protein atomsa | 7110 | |||||||||
| DNA atomsa | none | – | – | – | – | – | ||||
| Heavy atoms | 14I-1 | Ta | Ta | 10 Se | ||||||
| Catalytic Metal ions | Mg+2 | Mg+2 | Mg+2 | Mg+2 | Mn+2 | Mg+2 | Ca+2 | |||
| Cofactor | SAH | SAH | SAH | SAH | ||||||
| Solvent molecules | 316 | |||||||||
| R-factord (%)b | 0.26 | 0.21 (0.32) | ||||||||
| R-freed (%)b | 0.32 | 0.26 (0.36) | ||||||||
| Rmsd | ||||||||||
| Bond length (Å) | 0.009 | |||||||||
| Angles (o) | 1.202 | |||||||||
| Ramachandran (%) | ||||||||||
| Core region | 96.92 | |||||||||
| Allowed region | 2.25 | |||||||||
| Outliers | 0.83 | |||||||||
aCrystals containing SeMet and iodide.
bHighest resolution shell values in parenthesis.
cRmerge = Σ|Ihi − <Ih>|/ΣIh, where Ihi is the ith measurement of reflection h, and <Ih> is the average measured intensity of reflection h.
dR-factor/R-free = Σh|Fh(o) − Fh(c)|/Σh|Fh(o)|. Where R-free was calculated with 5% of the data excluded from refinement.
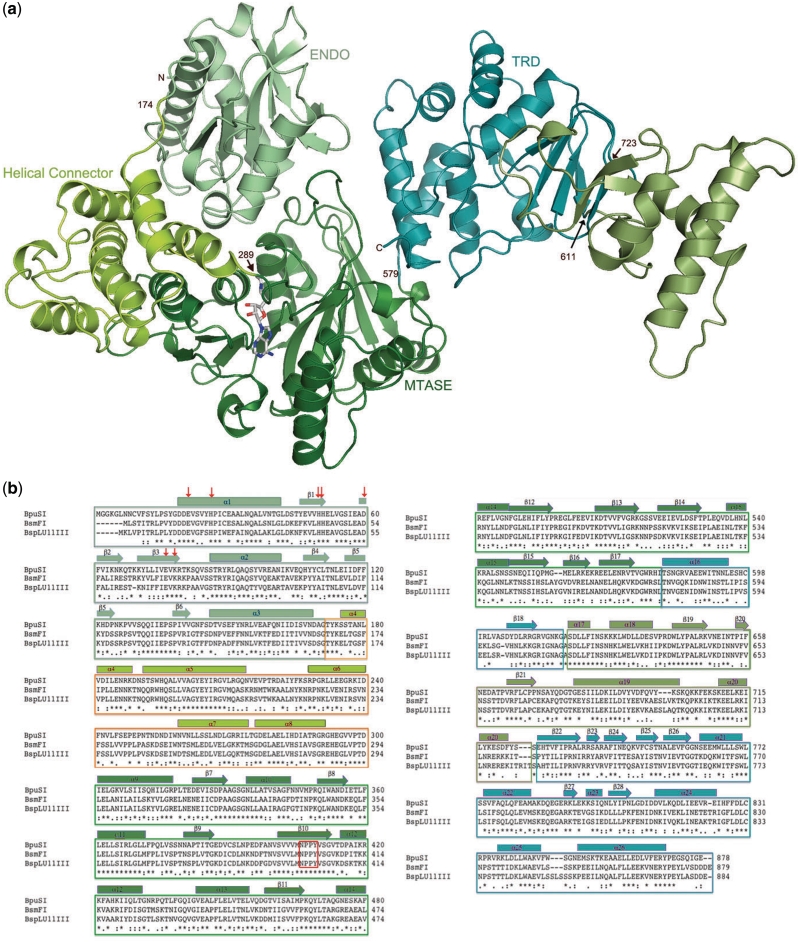
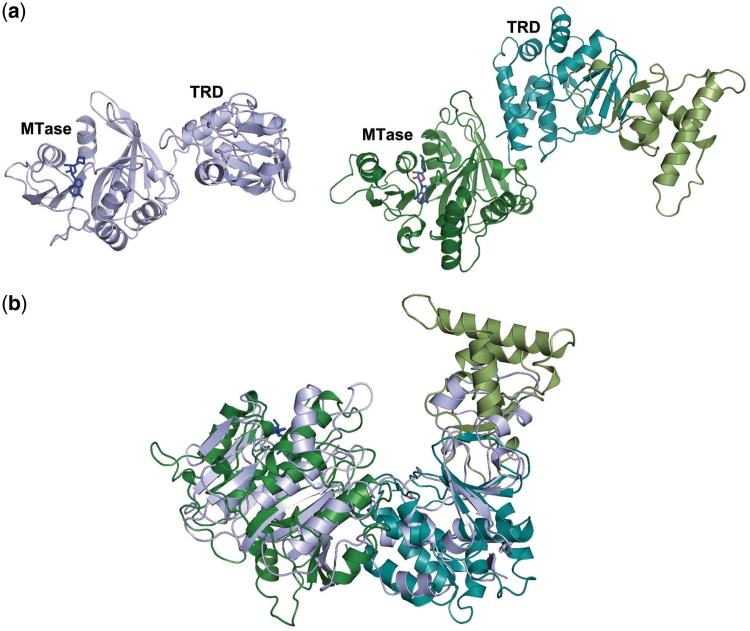
The structure of BpuSI is highly elongated, with overall dimensions of ~115 Å × 75 Å × 45 Å. The protein consists of 878 residues and displays four individually folded domains, each with a putative function (endonuclease, connector, MTase and target recognition), arranged in tandem succession from the N- to C-termini (Figure 1). A fifth-structural domain of unknown function, which has not previously been observed in R–M structures, interrupts the TRD. Based on modeling analyses (described below) we believe that this domain may also contribute to DNA recognition and binding.
Figure 1.
(a) The structure of full-length BpuSI. The domains are labeled and color coded according to individual-folded elements with known functions based on sequence and structural homology. The N-terminal endonuclease domain displays a PD-(D/E)xK nuclease fold and is packed against the MTase domain in a manner that buries its active site and precludes DNA-cleavage activity in the modeled conformation. These two functional domains are connected via a small, helical ‘connector’ domain that displays structural similarity to similar modules found in a wide variety of multi-domain, flexible protein scaffolds. The C-terminal TRD is interrupted by an additional α/β domain that may play a role in establishing additional DNA contacts as discussed in the text. (b) Multi-sequence alignment of BpuSI and two closely related isoschizomers BsmFI and BspLU11III (two frame-shift mutations were introduced into the published sequence in order to correct the reading frame at the N-terminus of BspLU11III). The catalytic residues E21, D60, E76 and K78 and the neighboring histidine triad (H26, 50 and 51) in the REase domain are indicated with red arrows; the NPPY catalytic motif in the MTase domain is indicated with a red box.
The PD-(D/E)xK endonuclease domain spans residues 6–174 in the structure, and coordinates a single manganese ion in the crystal. A small helical connector domain (residues 175–289) follows and is packed directly between the endonuclease domain and the subsequent MTase domain (MTase, residues 293–579). The active site of the MTase domain is occupied by density that appears to be appropriately modeled by a bound S-adenosylhomocysteine molecule (the product formed when a methyl group is transferred from the SAM cofactor). The C-terminal end of the protein is comprised of a TRD (residues 580–876), which is itself interrupted by an intervening folded domain (residues 611–723) that extends from the core TRD fold (Figure 1b). The contact surfaces between the four domains of known or putative functions are relatively small and hydrophilic, and bury ~740, 960, 1050 and 910 Å2 of surface area between domains 1 and 2, 1 and 3, 2 and 3 and 3 and 4, respectively.
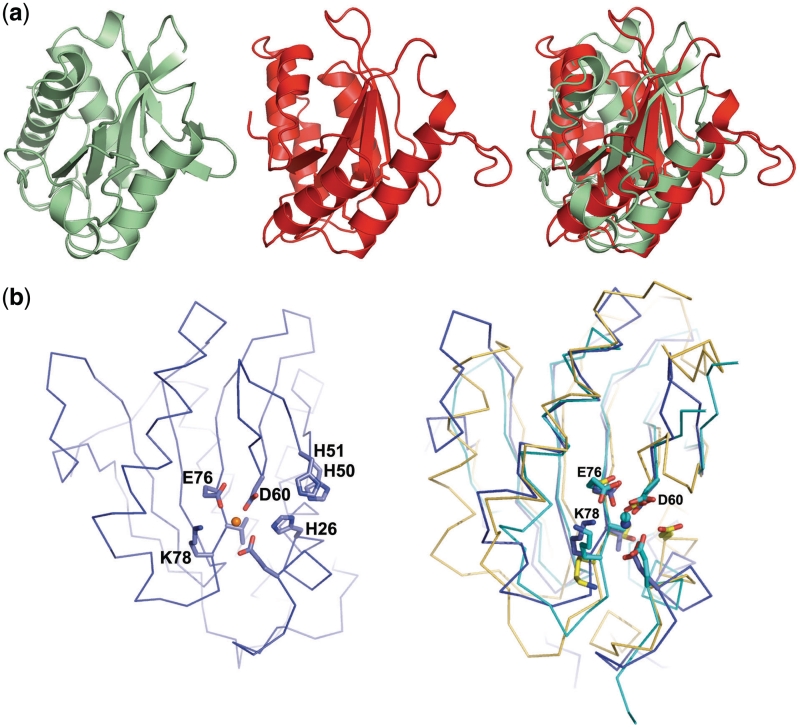
The N-terminal endonuclease domain displays the typical αβα-fold of a type II REase and contains a PD-(D/E)xK nuclease core motif (Figure 2). It contains a single-bound divalent metal ion (modeled as Mn2+ based on the presence of that metal in the crystallization buffer and the presence of anomalous difference density at that position) that is coordinated in an octahedral geometry by the carboxyl group of residues Glu21, Asp60, Glu76, the carbonyl oxygen of Val77, and two structural waters. The structure of this domain is homologous to the nucleolytic domain of the putative HsdR subunit within the Type I R–M system from Vibrio vulnificus [PDB ID 3h1t; (20)] and E. coli pR124 [PDB ID 2w00; (21)] and to the nuclease domain of the type IIS REase FokI [PDB ID 1fok; (14)]. The superposition of the endonuclease domain to the same domain from FokI (Figure 2a) yields an Rmsd value of 3.5Å [calculated for an alignment of 110 Cα carbons, corresponding to a Z-score in the DALI structural similarity server (22) of 6.0]. For complete results of structural similarity searches using online web servers, see Supplementary Table S1.
Figure 2.
(a)Isolated endonuclease domains, shown side-by-side and directly superimposed, from BpuSI (green) and FokI (red). (b) Location of putative BpuSI active site residues (left panel) and superposition of the active-site residues and α-carbon backbones of the catalytic domain of BpuSI (blue), FokI (PDB 1fok; orange) and the HsdR domain of the type I R–M system from Vibrio vulnificus (PDB 3h1t, cyan) (right panel).
The catalytic DDK ‘triad’ found in the PD-(D/E)xK motif of FokI (consisting of residues Asp450, Asp467 and Lys469) is partially conserved in the BpuSI endonuclease active site as Asp60, Glu76 and Lys78. The rmsd for the corresponding three Cα atoms in an alignment of the two endonuclease domains (Figure 2b) is 0.19 Å. In both structures, a single-bound divalent metal ion is coordinated by the two acidic residues. The active site in BpuSI is closely flanked by a group of three histidine residues (H26, 50 and 51) that are conserved with its closest type IIG homologues (isoschizomers BsmFI and BspLU11III).
The active site of the BpuSI endonuclease domain is partially buried in an interface that is formed with the surface of the MTase domain. This interface is relatively small (740 Å2 buried surface area) and is largely comprised of contacts between one negatively charged surface exposed loop within the endonuclease domain that borders its active site (S16-Y17-G18-D19-D20-E21) and a corresponding loop from the MTase (Q534-V535-D536-L537). Combined with a probable ‘hinge point’ located between the endonuclease domain and the intervening domain 2 (located near A170 and G171), it appears likely that the endonuclease domain might undergo significant rotation and movement relative to its position in the crystal structure that would be required in order to cleave its DNA target 10- and 14-bp downstream of its bound DNA-target site.
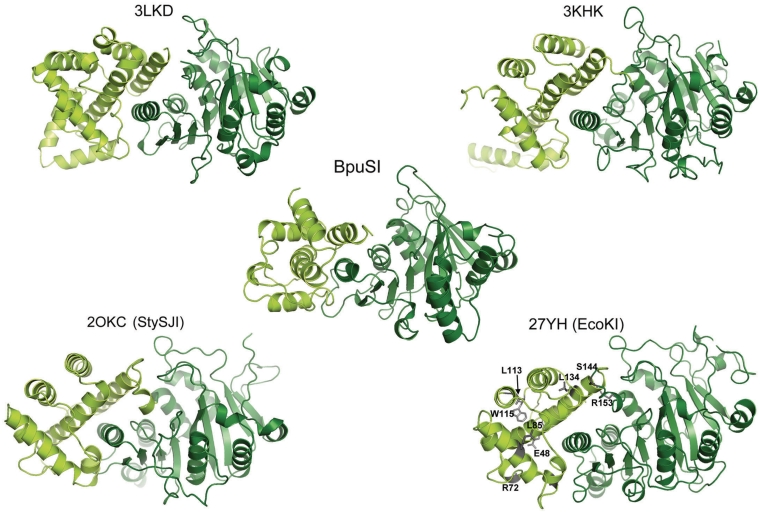
Domain 2 (residues 175–289) is comprised of an all-alpha fold (corresponding to helices α4 to α8) (Figure 3). In this region, α4 is packed against a single-surface loop in the endonuclease domain, while helices α5 and α8 form a four-helix bundle with α11 from the MTase domain (i.e. domain 3). Assuming that the endonuclease domain must exhibit extensive conformational motion as part of the overall function of the R–M enzyme, it is postulated that this domain plays a role in switching between different enzyme conformations and functional states, and that the contacts made by this domain to the endonuclease and MTase regions are transiently formed and broken during catalysis.
Figure 3.
Comparison of structures of domain 2 (the ‘m*’ domain; colored in light green) and MTase domains (dark green) from four type I restriction/modification enzymes and from BpuSI (center panel). The structure of these domains from EcoKI (PDB ID 27YH) is taken from an EM reconstruction of the DNA-bound enzyme, using a high-resolution structure of that subunit bound to the Ocr inhibitor as the initial model.
A role for this domain in switching between functional states had been previously hypothesized as a result of several structural and biological investigations. In particular, a screen for mutations in the EcoKI enzyme that caused a switch in the enzyme's activity from a ‘maintenance’ MTase (corresponding to modification of a hemimethylated target site) to a de novo MTase (corresponding to modification of unmethylated target sites) identified residues that are entirely clustered within domain 2 (23). The positions of those residues (illustrated in Figure 3) led to the designation of this region as the ‘m* domain’ based on their apparent role in establishing the proper balance between restriction and modification activities.
The tertiary structure and relative orientations of domain 2 (the helical connector) and domain 3 (the MTase) in BpuSI closely resembles that of the modification (HsdM) subunit in Type I R–M systems from Methanosarchina mazei, Streptococcus thermophilus and Bacteroides thetaiotaomicron VPI-548 (RSCB PDB entries 3khk, 3lkd and 2okc) as well as structures of the same subunit from the EcoKI endonuclease (RCSB PDB entries 27YH, 27YC) (24) (Figure 3). The peptide loop that connects the two domains (corresponding to residues 289–295) is solvent exposed and also runs directly above the bound cofactor. It is highly flexible, and at least 2 residues (R291 and G292) cannot be traced with confidence. The residues at the same position in at least one of the Type I R–M MTase structures (PDB 3khk) are also disordered.
A search for additional structural homologues to domain 2 using the FATCAT similarity server (25) generated a list of similar helical folded regions that are found primarily within highly flexible multi-domain proteins from a variety of biochemical pathways (Supplementary Figure S5). The rmsd values for these superpositions, calculated across 80–100 aligned α-carbons, range from 1 to 3Å. The top four hits (corresponding to PDB entries 1fpo, 2ido, 2g7o and 1wrd) correspond to structured helical regions found in the bacterial Hsc20 chaperone (26), the RNA pol III epsilon subunit (27) and the F conjugation regulatory protein (28) (all from E. coli) and the human Tom1 ubiquitin transport protein (29). Most of these proteins are notable for their multi-domain structures and the requirement for extensive domain motion and formation of multiple binding interfaces as part of their overall biological function. Therefore, it seems reasonable that the general architecture displayed by this domain is highly adaptable to the requirement for correlated motions between domains of highly modular, multi-domain proteins.
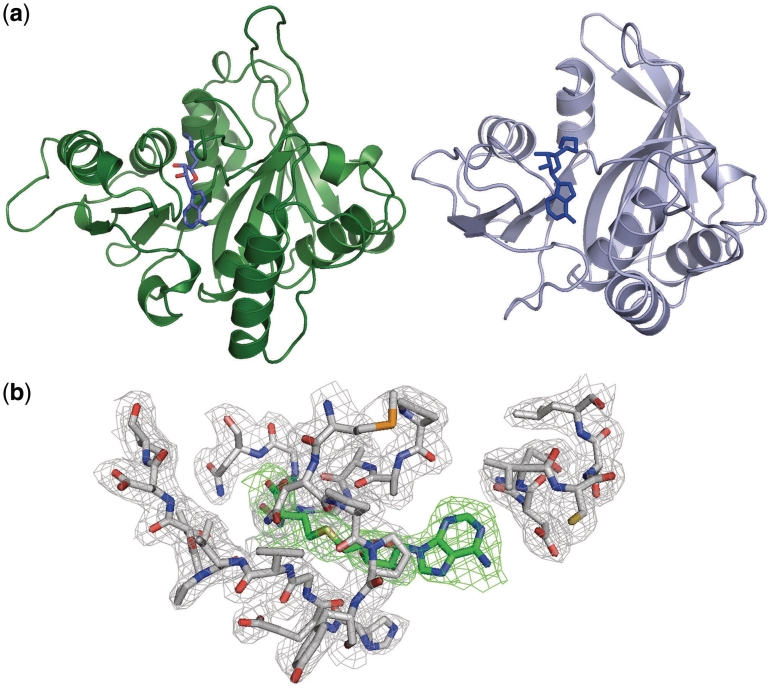
The MTase domain (domain 3) exhibits strong structural similarity to the γ-N6m-Adenine MTase M.TaqI (30) despite a relatively low (~13%) sequence identity (Figure 4A). The arrangement of secondary structural elements within the BpuSI MTase domain is identical to that of M.TaqI with one exception: two short-surface loops in M.TaqI are replaced by two longer α-helix/β-hairpin motifs that are involved in packing interactions with the connector domain and the TRD. Strong features of electron density that can be modeled by a bound S-adenosylhomocysteine molecule (Figure 4b) is observed in contact with the conserved NPPY motif (residues 426–429) in the MTase active site, at a position consistent with previously visualized bound cofactor molecules in structures of related MTase domains.
Figure 4.
MTase domains. (a) MTase domains from BpuSI (left) and M.TaqI (right) with bound cofactor analogues. (b) Unbiased electron density (green mesh) for the S-adenosylhomocysteine cofactor/product (green carbond atoms) bound in the MTase domain of BpuSI. The surrounding grey electrondensity corresponds to a 2Fo − Fc omit map contoured at 2σ around the cofactor binding site.
The C-terminal end of BpuSI contains the TRD (domain 4) and harbors a strong resemblance to the specificity (HsdS) subunits of Type I R–M systems from Mtthanococcus jannaschii (31) and Mycoplasma genitalium (32) (PDB codes 1YF2 and 1YDF, respectively). This TRD domain is interrupted in BpuSI at residue 611 by a fifth and final domain with a mixed α/β structure and unknown function. The interface between this region and domain 4 contains a β-clamshell motif, with two strands within one of the small β-sheets contributed by each domain. Searches for regions of protein structures similar to this final domain are relatively uninformative, with the top hits corresponding to short helical regions from proteins that represent a wide variety of functions and biological contexts.
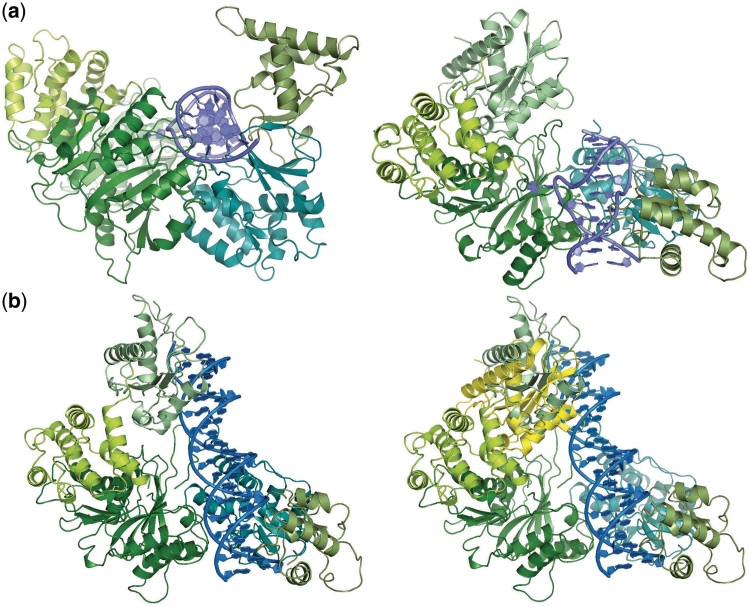
The relative orientation between the MTase domain and the TRD in the structure of BpuSI differs significantly from that observed in M.TaqI (Figure 5). Superposition of the MTase catalytic domains from the two structures places their TRDs in different orientations, with the TRD from M.TaqI corresponding to a ‘closed’ conformation corresponding to its DNA-bound enzyme–substrate complex. The TRD from BpuSI can be closely superimposed on its homologous region from the DNA-bound M.TaqI structure through a straightforward backbone rotation at a hinge point located at or near residue 580. This movement of the TRD in BpuSI corresponds to rotations around the Cartesian xyz axes of ~112°, 22° and 81°, and a corresponding translation along the same axes of ~−14 Å, 66 Å and 20 Å. The structure of unbound BpuSI therefore appears to illustrate an ‘open’ conformation between these two domains that has not been previously observed.
Figure 5.
(a) MTase and TRD domains from DNA-bound M.TaqI (left) and from unbound BpuSI (right). The bound SAM and S-adenosylhomocysteine moieties in each MTase domain are shown as sticks. The MTase domains are shown in the same orientation; relative to this superposition the corresponding TRDs are in dramatically different positions and orientations as described in the text. (b) Rotation of the TRD in BpuSI from its initial location in the unbound crystal structure [as shown in (a), right] into a position corresponding to its DNA-bound position in M.TaqI [(a), left) is easily accomplished by a single-backbone rotation at a discrete hinge point as described in the text, leading to the superposition with M.TaqI as shown.
In a model of BpuSI with the MTase and TRD domains placed into a DNA-bound conformation similar to M.TaqI, and with a bound DNA target located at the same position as was observed in the M.TaqI/DNA complex (Figure 6), two points become clear. First, the intervening fifth domain found on the surface of the TRD may be involved in forming additional contacts with the DNA substrate. Second, the endonuclease domain appears to be positioned in the unbound enzyme structure in a manner that would avoid a significant steric clash with the DNA (therefore allowing binding and methylation by the MTase domain). The endonuclease domain would then need to rotate by ~30° relative to its position in the unbound crystal structure to position its active site appropriately for cleavage 10-bp downstream of the enzyme target site (Figure 6b, right panel).
Figure 6.
Model of DNA-bound BpuSI, generated using the coordinates of DNA-bound M.TaqI as a reference (see Figure 5B). (a) Model of DNA-bound BpuSI with the bound DNA substrate previously visualized for M.TaqI. The left and right panels show two different orientations of this same model. The endonuclease domain (light green, easily seen in right panel) is positioned exactly as observed in the unbound BpuSI structure. (b) The left panel shows a model of DNA-bound BpuSI with a longer, canonical B-form DNA (encompassing the GGGAC target site and the downstream N10/N14 bases, corresponding to the cleavage pattern produced by the enzyme). The endonuclease domain has been rotated as described in the text, to bring its active site into appropriate proximity with the N10 phosphate group. The right panel shows the same model, with the original position of the endonuclease domain (yellow ribbon) superimposed.
Verification of catalytic activity and active-site residues
In the crystal structure, Asp60 and Glu76 coordinate the Mn++ in the catalytic site. We therefore mutated the two residues to Ala separately to verify their role in cleavage activity. Cleavage activity on λ DNA for mutants D60A and E76A showed they did not exhibit observable cleavage activity up to 0.5 or 4.5 µg of protein, respectively (Supplementary Figure S6).
To verify the methylation activity of the MTase domain of BpuSI, the catalytic deficient mutant E76A was incubated with unmodified pACYC184 DNA (which contains 7 BpuSI recognition sites) in the presence of SAM. After removing the BpuSI E76A mutant protein and SAM, the treated DNA was subjected to cleavage by WT BpuSI. Supplementary Figure S6 shows that pACYC184 treated with BpuSI E76A and SAM is partially cleaved by WT BpuSI with bands of complete cleavage as well as partial cleavage (lane 1). Without pre-incubation with BpuSI E76A and SAM, pACYC184 is cleaved to completion (lane 3). Incubation with BpuSI E76A and SAM alone does not result in cleavage of pACYC184 (lanes 2 and 4). This suggests that the γ-N6mA MTase domain of the BpuSI R–M fusion is capable of modifying the recognition site GGGAC albeit partially under the described experimental conditions. It also suggests that the S-adenosylhomocysteine molecule observed in the NPPY motif of the unbound protein structure is a result of the modification activity of the enzyme.
Comparison of cleavage kinetics on single-site versus double-site substrates
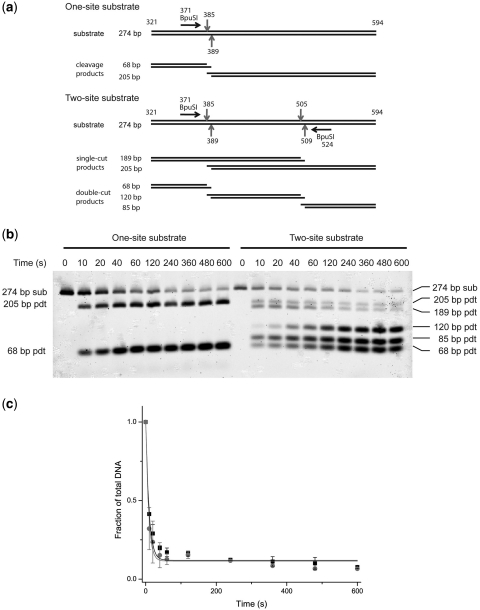
To determine whether BpuSI cleaves DNA more efficiently when presented with multiple target sites, we created two comparable DNA substrates containing either one BpuSI target site or two target sites in a head-to-head arrangement as described in ‘Materials and Methods’ section (Figure 7a). Parallel cleavage reactions were then carried out under single turnover conditions. The rate of the cleavage reaction was monitored by measuring a decrease in substrate over the timecourse of the reaction. The concentration of the double-site substrate is half of that of the single-site substrate to attend an equal molar concentration of BpuSI sites in the reactions. Therefore, the first-order rate constant observed corresponds to the rate of cleavage of the substrate at a single BpuSI site.
Figure 7.
(a) Design of the one-site and two-site substrates and the expected cleavage products. Black horizontal arrows represent the orientation of the BpuSI recognition site (GGGAC) from 5′ to 3′. Red vertical arrows denote the approximate locations of the BpuSI cleavage sites (GGGAC 10/14). The numbers on the DNA are the coordinates on pACYC184 DNA. (b)A cleavage reaction of the single- and double-site substrates as analyzed on a 2.5% agarose gel. The identity of the bands is shown. (c) A plot of the single-site (black squares) and double-site (red circles) substrates as fraction of total DNA versus time. Error bars represent 1 standard deviation.
Figure 7b shows the gel image of a typical cleavage reaction over a period of 10 min. A plot of the relative amount of substrate over time is shown in Figure 7c. By fitting the curves to Equation (1), the kf value of the single-site and double-site substrates were found to be 0.098 ± 0.012/s and 0.129 ± 0.017/s, respectively. The ratio of kf of double-site versus single-site substrate is 1.32, which is significantly lower than over 5-fold of increases from other Type IIS enzymes that are known to cleavage more efficiently with double-site substrates (33,34).
DISCUSSION
The structure of BpuSI appears to roughly correspond to a fusion of half of a heteropentameric complex that is often found in a Type I R–M system. Three characterized Type IIG isoschizomers (BpuSI, BsmFI and BspLU11III) share high-sequence homology (36–49% amino acid sequence identities and 63–84% similarity) among themselves (Figure 1a); additional homologues only display observable homology across the MTase domain (35). In contrast, a BLASTP search using Eco57I, the other known Type IIG REase, as a query produced a list of nine homologs with highly conserved PD-(D/E)xK and NPPY motifs as well as other MTase signatures over the entire length of its sequence. These results suggest that the evolution of Type IIG REases is not unique to the Bacillus strains but rather common among microbial organisms, and that the fusion of these subunits might be a recent event that occurred after the divergence of archaea and bacteria. In fact, more than 1940 type IIG (R–M fusion) enzymes (the majority of which are uncharacterized and are not correlated to known DNA-target site specificities) are found in sequenced microbial genomes [REBASE (19)], which represents a large fraction of R–M systems found in nature.
By definition, type IIG REases are single chain R–M–S fusions, and their endonuclease activities may or may not be stimulated by SAM (11). Many Type IIG enzymes are coupled to one or two companion MTases that are encoded with the R–M–S fusion in a single operon, as part of an overall mechanism for host DNA protection. For example, the BpuSI R–M system contains two additional MTase s (an amino-MTase termed M1, and a C5 MTase M2) within the BpuSI operon. Our biochemical data shows that the MTase activity in the R–M–S fusion protein (the M3 activity) is active but does not completely protect host target sites from BpuSI cleavage activity under standard conditions (Supplementary Figure S7). Without the companion M1 and M2, we were not able to establish a clone of the R–M fusion in E. coli, suggesting that the M3 activity is not sufficient for host DNA protection (data not shown).
In contrast, some Type IIG REases (such as the MmeI-family endonucleases, Tth111II and TaqII) do not seem to have additional MTase enzymes. The endonuclease activity of these lone Type IIG enzymes are strongly dependent on the concentration of free SAM (18), and also appear to require the presence of two target sites arranged in tandem on a DNA substrate in order to generate cleaved products (unpublished results, S.Y.X.). It is possible that the endonuclease activities of Tth111II and TaqII are sufficiently low, or otherwise regulated in a manner that reduces activity towards host genomic targets in the absence of separate MTase enzymes.
Recently, Type I REases with the R, M and S subunits fused into a single polypeptide (termed ‘type ISP’ enzymes) have been found in nature. LlaGI is a single chain restriction-modification enzyme encoded on a naturally occurring plasmid in Lactococcus lactis (36). LlaGI recognizes a asymmetric sequence 5′-CTNGAYG-3′ (where N is A, G, C or T and Y is C or T), but cleaves DNA using a mechanism that requires hydrolysis of ATP or dATP. LlaGI contains an Mrr-like catalytic domain, a superfamily 2 DNA helicase domain, and a γ-family adenine MTase. Many more LlaGI-like fused R–M–S putative endonucleases with helicase domain are found in sequenced microbial genomes and remain to be characterized. The presence of R–M–S fusion proteins across Types I, IIG and IIB suggests that combining all the activity determinants (R, M and S subunits) together in a single polypeptide appears to be a viable evolution strategy for both Types I and II REases.
The most interesting mechanistic question regarding the activity of R–M–S fusion systems such as the Type IIG endonuclease BpuSI appears to be the issue of exactly how the fusion enzyme determines if it is to activate the MTase or the REase activity and how the switch is achieved. In the structure presented here, the endonuclease active site is physically buried, thereby requiring extensive domain movements to position the catalytic nuclease resides near the scissile phosphodiester bond. Several possibilities for the coupling of MTase and REase activities exist; however the simplest model might be that (i) the unbound enzyme sequesters the REase domain in an inactive conformation as illustrated by this structure; (ii) recognition and binding of a hemimethylated target site by the MTase and TRD domains results in the formation of a catalytically active MTase–DNA-target complex, involving a flipped base in the MTase active site and deformation of the DNA backbone that prevents the REase domain from associating with the DNA in a cleavage–competent complex; (iii) a fully methylated target site is either not bound by the enzyme, or is again bound in a conformation that precludes cleavage; and (iv) recognition and binding of a fully unmethylated target produces a different conformation of the bound DNA and protein that results in the formation of a catalytically active complex between the REase domain and the downstream cleavage site. A detailed examination of this model will require crystallographic studies of BpuSI or other Type IIG REases captured at different stages of activity (i.e. before or after DNA cleavage or modification).
As well, the observation that BpuSI cleaves a DNA substrate containing only a single target as efficiently as a substrate with two target sites arranged in a head-to-head orientation (Figure 7) appears to indicate that the kinetic and physical mechanism of double strand break formation by the enzyme differs from at least some type IIS enzymes (such as FokI and BfiI, where transient dimerization of the nuclease domain and cleavage of both DNA strands is clearly facilitated by the presence of two binding sites). Whether BpuSI cleaves the target site as a transient homodimer (with one protein subunit recruited to a single site from solution) or carries out sequential strand cleavage events as a monomer is still to be investigated.
ACCESSION NUMBER
The coordinates and structure factor amplitudes for BpuSI have been deposited at the RCSB PDB database (37) under ID code 3S1S, and designated for immediate release upon publication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (grant R01 GM49857 to B.L.S.); the initial screening work for BpuSI (student internship program at NEB). Funding for open access charge: National Institutes of Health R01 GM49857.
Conflict of interest statement. Several of the authors (D.X., S.H.C., Y.Z., Z.Z. and S.Y.X.) are employees of New England Biolabs; the RM.BpuSI enzyme is a commercial product sold by that company.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Don Comb, Richard Roberts and Jim Ellard for support and encouragement.
REFERENCES
- 1.Bertani G, Seigle JJ. Host controlled variation in bacterial viruses. J. Bacteriol. 1953;65:113–121. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luria SE, Human ML. A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol. 1952;64:557–569. doi: 10.1128/jb.64.4.557-569.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arber W, Dussoix D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J. Mol. Biol. 1962;5:18–36. doi: 10.1016/s0022-2836(62)80058-8. [DOI] [PubMed] [Google Scholar]
- 4.Dussoix D, Arber W. Host specificity of DNA produced by Escherichia coli. II. Control over acceptance of DNA from infecting phage lambda. J. Mol. Biol. 1962;5:37–49. doi: 10.1016/s0022-2836(62)80059-x. [DOI] [PubMed] [Google Scholar]
- 5.Xu SY, Corvaglia AR, Chan SH, Zheng Y, Linder P. A type IV modification-dependent restriction enzyme SauUSI from Staphylococcus aureus subsp. aureus USA300. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr098. 17 March 2011 (doi:10.1093/nar/gkr098; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 7.Dybvig K, Sitaraman R, French CT. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. PNAS USA. 1998;95:13923–13928. doi: 10.1073/pnas.95.23.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin LF, Posfai J, Roberts RJ, Kong H. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl Acad. Sci. USA. 2001;98:2740–2745. doi: 10.1073/pnas.051612298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 10.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grazulis S, Manakova E, Roessle M, Bochtler M, Tamulaitiene G, Huber R, Siksnys V. Structure of the metal-independent restriction enzyme BfiI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease. Proc. Natl Acad. Sci. USA. 2005;102:15797–15802. doi: 10.1073/pnas.0507949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kachalova GS, Rogulin EA, Yunusova AK, Artyukh RI, Perevyazova TA, Matvienko NI, Zheleznaya LA, Bartunik HD. Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit. J. Mol. Biol. 2008;384:489–502. doi: 10.1016/j.jmb.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 14.Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 15.Tamulaitiene G, Grazulis S, Janulaitis A, Janowski R, Bujacz G, Jaskolski M. Crystallization and preliminary crystallographic studies of a bifunctional restriction endonuclease Eco57I. Biochim. Biophys Acta. 2004;1698:251–254. doi: 10.1016/j.bbapap.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Bourniquel AA, Bickle TA. Complex restriction enzymes: NTP-driven molecular motors. Biochimie. 2002;84:1047–1059. doi: 10.1016/s0300-9084(02)00020-2. [DOI] [PubMed] [Google Scholar]
- 17.Szczelkun MD. How to proteins move along DNA? Lessons from type-I and type-III restriction endonucleases. Essays. Biochem. 2000;35:131–143. doi: 10.1042/bse0350131. [DOI] [PubMed] [Google Scholar]
- 18.Morgan RD, Bhatia TK, Lovasco L, Davis TB. MmeI: a minimal Type II restriction-modification system that only modifies one DNA strand for host protection. Nucleic Acids Res. 2008;36:6558–6570. doi: 10.1093/nar/gkn711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uyen NT, Park SY, Choi JW, Lee HJ, Nishi K, Kim JS. The fragment structure of a putative HsdR subunit of a type I restriction enzyme from Vibrio vulnificus YJ016: implications for DNA restriction and translocation activity. Nucleic Acids Res. 2009;37:6960–6969. doi: 10.1093/nar/gkp603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lapkouski M, Panjikar S, Janscak P, Smatanova IK, Carey J, Ettrich R, Csefalvay E. Structure of the motor subunit of type I restriction-modification complex EcoR124I. Nat. Struct. Mol. Biol. 2009;16:94–95. doi: 10.1038/nsmb.1523. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelleher JE, Daniel AS, Murray NE. Mutations that confer de novo activity upon a maintenance methyltransferase. J. Mol. Biol. 1991;221:431–440. doi: 10.1016/0022-2836(91)80064-2. [DOI] [PubMed] [Google Scholar]
- 24.Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DT. The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Res. 2009;37:762–770. doi: 10.1093/nar/gkn988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl 2):ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 26.Cupp-Vickery JR, Vickery LE. Crystal structure of Hsc20, a J-type Co-chaperone from Escherichia coli. J. Mol. Biol. 2000;304:835–845. doi: 10.1006/jmbi.2000.4252. [DOI] [PubMed] [Google Scholar]
- 27.Kirby TW, Harvey S, DeRose EF, Chalov S, Chikova AK, Perrino FW, Schaaper RM, London RE, Pedersen LC. Structure of the Escherichia coli DNA polymerase III epsilon-HOT proofreading complex. J. Biol. Chem. 2006;281:38466–38471. doi: 10.1074/jbc.M606917200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, Edwards RA, Wong JJ, Manchak J, Scott PG, Frost LS, Glover JN. Protonation-mediated structural flexibility in the F conjugation regulatory protein, TraM. EMBO J. 2006;25:2930–2939. doi: 10.1038/sj.emboj.7601151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akutsu M, Kawasaki M, Katoh Y, Shiba T, Yamaguchi Y, Kato R, Kato K, Nakayama K, Wakatsuki S. Structural basis for recognition of ubiquitinated cargo by Tom1-GAT domain. FEBS Lett. 2005;579:5385–5391. doi: 10.1016/j.febslet.2005.08.076. [DOI] [PubMed] [Google Scholar]
- 30.Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 31.Kim JS, DeGiovanni A, Jancarik J, Adams PD, Yokota H, Kim R, Kim SH. Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications. Proc. Natl Acad. Sci. USA. 2005;102:3248–3253. doi: 10.1073/pnas.0409851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calisto BM, Pich OQ, Pinol J, Fita I, Querol E, Carpena X. Crystal structure of a putative type I restriction-modification S subunit from Mycoplasma genitalium. J. Mol. Biol. 2005;351:749–762. doi: 10.1016/j.jmb.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 33.Bath AJ, Milsom SE, Gormley NA, Halford SE. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 2002;277:4024–4033. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 34.Lagunavicius A, Sasnauskas G, Halford SE, Siksnys V. The metal-independent type IIs restriction enzyme BfiI is a dimer that binds two DNA sites but has only one catalytic centre. J. Mol. Biol. 2003;326:1051–1064. doi: 10.1016/s0022-2836(03)00020-2. [DOI] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Smith RM, Diffin FM, Savery NJ, Josephsen J, Szczelkun MD. DNA cleavage and methylation specificity of the single polypeptide restriction-modification enzyme LlaGI. Nucleic Acids Res. 2009;37:7206–7218. doi: 10.1093/nar/gkp790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.