Abstract
Many insects rely on cuticular hydrocarbons (CHCs) as major recognition signals between individuals. Previous research on the genetics of CHCs has focused on Drosophila in which the roles of three desaturases and one elongase were highlighted. Comparable studies in other insect taxa have not been conducted so far. Here, we explore the genetics of CHCs in hybrids of the jewel wasps Nasonia giraulti and Nasonia vitripennis. We analyzed the CHC profiles of pure strain and of F2 hybrid males using gas chromatography coupled with mass spectrometry and distinguished 54 peaks, of which we identified 52 as straight-chain, monounsaturated, or methyl-branched CHCs. The latter compound class proved to be particularly abundant and diverse in Nasonia. Quantitative trait locus (QTL) analysis suggests fixed genetic differences between the two strains in 42 of the 54 studied traits, making Nasonia a promising genetic model for identifying genes involved in CHC biosynthesis. QTL for methyl-branched CHCs partly clustered in genomic regions with high recombination rate: a possible indication for pleiotropic genes that control their biosynthesis, which is largely unexplored so far. Finally, we identified and mapped genes in the Nasonia genome with high similarity to genes that have been implicated in alkene biosynthesis in Drosophila and discuss those that match in their position with predicted QTL for alkenes.
Keywords: Nasonia, cuticular hydrocarbons, quantitative trait loci, candidate genes, desaturases
Introduction
Cuticular hydrocarbons (CHCs) are of fundamental importance for insects, serving both as protection against desiccation and as major recognition signals between individuals (Howard and Blomquist, 2005). Although our knowledge of the biosynthesis of CHCs has moderately increased over the last decades, we still know little about the enzymes that are involved, their genetics and their evolutionary history (Nelson and Blomquist, 1995; Blomquist and Bagneres, 2010). Most of what is known about the genetics of CHCs is based on Drosophila research (Ferveur, 2005). How this data can be applied to other insect taxa, for example, the economically important Hymenoptera, is unclear. Research on the parasitoid wasp genus Nasonia with its four described interfertile species (Darling and Werren, 1990; Raychoudhury et al., 2010), a haplodiploid sex determination and a steadily growing set of genetic tools and genomic resources (Werren et al., 2010) could help to answer this question and improve our comprehension of the evolution and genetics of CHCs in general and in the Hymenoptera in particular.
CHCs are long-chain saturated and unsaturated non-polar waxes that are mainly synthesized by epidermal cells (oenocytes) and secreted via the lymphatic system on the epidermis (Schal et al., 1998). Although assumed to be initially evolved as protection against desiccation (Hadley, 1981), CHCs display a wide variety of other functions: for instance, they are involved in chemical mimicry, are employed as dominance and fertility cues, and serve in eusocial Hymenoptera, like ants, certain bees and wasps, as cues for nestmate and caste recognition (Howard and Blomquist, 2005; Blomquist and Bagneres, 2010). In addition, CHCs have been shown to have a pivotal role in sexual communication as sex attractants and cues for species, gender and individual recognition (Howard and Blomquist, 2005; Blomquist and Bagneres, 2010).
The importance of CHCs for species recognition and sexual communication suggests that CHCs could function as prezygotic hybridization barriers by causing behavioral isolation (Coyne and Orr, 2004). Although difficult to demonstrate, several studies provide evidence for CHCs being involved in prezygotic reproductive isolation (for example, Coyne et al., 1994; Peterson et al., 2007). However, what evolutionary forces cause the divergence of CHCs between populations and how many genes with what functions are involved during this process is still largely unknown. Given the significance of CHCs as cues or signals, answers to the above questions are important in order to understand the rapid diversification of CHCs during speciation and how species-specific CHC profiles originate and are maintained.
Our current knowledge of the genetics of CHC differences stems primarily from research on Drosophila. Species of the Drosophila melanogaster subgroup are coated with a layer of 14–20 different hydrocarbons, with chain-lengths between 23 and 33 carbons, of which alkenes constitute the predominant fraction (Ferveur, 2005). Males and females of Drosophila simulans and Drosophila mauritiana are sexually monomorphic and produce large quantities of the alkene (Z)-7-tricosene (Jallon and David, 1987). In the sexually dimorphic species D. melanogaster and Drosophila sechellia, (Z)-7-tricosene is found predominantly in males while their females typically produce the alkadiene (Z,Z)-7,11-heptacosadiene instead (Jallon and David, 1987). However, females of D. melanogaster occurring in sub-Saharan Africa and the Caribbean synthesize primarily the positional isomer 5,9-heptacosadiene (Coyne et al., 1999). This difference was found to be controlled by a single gene, desat2, which encodes a Δ9-desaturase that acts preferentially on myristate as a substrate, leading to the synthesis of ϖ5-hydrocarbons (Coyne et al., 1999; Dallerac et al., 2000). Although desat2 is present in all D. melanogaster populations, its promoter is missing in those that produce primarily 7,11-heptacosadiene (Takahashi et al., 2001). Intriguingly, another gene, desat1, was found only 3.7 kb downstream of desat2. It also encodes a Δ9-desaturase, but one that preferentially acts on palmitate, thus leading to a ϖ7-fatty acid CHC precursor (Wicker-Thomas et al., 1997; Dallerac et al., 2000). Research showed that desat1, expressed in both males and females of Drosophila, controls the desaturation in 7-alkenes and the introduction of the first double bond in 7,11-alkadienes (Wicker-Thomas et al., 1997; Dallerac et al., 2000). RNA interference-mediated knock-down experiments revealed yet another female-specific desaturase, encoded by the gene desatF, that is responsible for the introduction of the second double bond in the above 5,9- and 7,11-alkadienes (Chertemps et al., 2006; Legendre et al., 2008). Finally, Chertemps et al. (2007) mapped and identified a gene, eloF, which encodes an elongase that controls the elongation of alkadienes and, to a lesser extent, of alkenes in females of D. melanogaster. These results highlight the progress that has been made in understanding the genetics and biosynthesis of CHCs by research on Drosophila. It remains to be investigated, though, whether these insights can be generalized and whether or not genes that are orthologous to those already identified in Drosophila are also involved in the CHC biosynthesis in other insect taxa, for example, the Hymenoptera.
With the recent sequencing of three Nasonia parasitoid wasp genomes (Werren et al., 2010) and the simultaneous inference of a Nasonia high-density linkage map (Niehuis et al., 2010), studying the genetics of CHCs in the large order of Hymenoptera, which include >125 000 species (Grimaldi and Engel, 2005), has come into reach. Nasonia has proven excellent for genetic research because of its haplodiploidy, which allows efficient genotyping without the added complexity of genetic dominance, and the cross-fertility of its four species (Werren et al., 2010). Only a few studies have investigated CHCs in Nasonia so far. Carlson et al. (1999) were the first who showed that the CHC profiles of Nasonia vitripennis males and females differ from each other. Later, Steiner et al. (2006) demonstrated that the CHCs of N. vitripennis females act as short-range sex pheromone on N. vitripennis males. Recently, Raychoudhury et al. (2010) found that Nasonia giraulti differs from its sister species Nasonia oneida in both its male and female CHC profile. This makes Nasonia a promising genetic model to map and characterize genetic factors that are involved in CHC biosynthesis.
Here, we explore the potential of Nasonia as genetic model to study the genetics of CHCs in general and in the Hymenoptera in particular. For this purpose, we analyze and characterize the CHC profiles of pure strain males of N. vitripennis and N. giraulti as well as of N. giraulti × N. vitripennis F2 hybrid males using gas chromatography coupled with mass spectrometry (GC-MS). We collect genotype data for molecular markers distributed throughout the genome of the F2 hybrid males and use it to estimate the number, position, effect and possible interaction of quantitative trait loci (QTL) that could explain the observed variance in the relative abundance of CHCs. Considering the local recombination rate that differs by at least two orders of magnitude in the Nasonia genome (Niehuis et al., 2010), we then search for conspicuous clusters of QTL in regions of high recombination because these could indicate pleiotropy and thus may provide a basis for identifying pathways and regulatory sequences that are involved in CHC biosynthesis. Finally, we screen the genome of Nasonia for predicted genes with high similarity to the four Drosophila genes desat1, desat2, desatF and eloF, which have been implicated in the biosynthesis of alkenes, and we discuss their localization with respect to the position of predicted QTL for Nasonia alkenes.
Materials and methods
Stocks and cross experiment
We studied the Wolbachia-free, highly inbred (that is, homozygous for every marker tested so far) and sequenced strains RV2X(U) (=N. giraulti) and AsymCX (=N. vitripennis) (Werren et al., 2010). Both strains were cultured on pupae of the flesh fly (Sarcophaga bullata) in an incubator at 25 °C under constant light. To obtain haploid F2 hybrid males of N. giraulti and N. vitripennis, we conducted the cross experiment described by Niehuis et al. (2008). Briefly, we mated virgin N. vitripennis females with haploid N. giraulti males and collected their virgin F1 hybrid female offspring. By depriving the virgin F1 hybrid females of males, we forced them to lay unfertilized eggs, which developed into haploid F2 hybrid males. We studied a total of 34 pure strain males (Ng=12; Nv=22) and 120 F2 hybrid males, all collected ∼36 h after their eclosion. Each wasp was transferred to a new glass tube (12 mm × 75 mm; Fischer Scientific, Fair Lawn, NJ, USA) that we sealed with a medium size absorbent cotton ball (Fischer Scientific) and stored at −80 °C.
Gas chromatography–mass spectrometry
The CHCs were extracted from individual pure strain and F2 hybrid males with 10 μl hexane each (⩾99.0% Sigma-Aldrich, St Louis, MO, USA) for 10 min using 1 ml glass vials with a 0.1 ml micro insert (Alltech, Deerfield, IL, USA). Each extract was transferred to a new vial, concentrated to ∼1 μl each by evaporating the hexane under gaseous nitrogen, and the entire residual volume was injected into an Agilent 6890N gas chromatograph coupled to an Agilent 5975 mass selective detector (Agilent, Santa Clara, CA, USA). The GC was operated in splitless injection mode and fitted with a J&W DB-5MS column (30 m–0.25 mm ID; film thickness: 0.25 μm). The oven was programmed from 60 to 200 °C at 40 °C min−1 with an initial 1 min delay and then from 200 to 320 °C at 5 °C min−1. The GC injector temperature was 250 °C, the mass selective (MS) quad temperature 150 °C, the MS source temperature 230 °C, and the mass selective detector (MSD) transfer line temperature 315 °C. Helium with a constant flow of 1 ml min−1 was used as carrier gas. All chromatograms were analyzed with the software Enhanced Chemstation E.01.00.237 (Agilent). Peaks were identified according to their retention indices (Carlson et al., 1998) and their respective mass spectra. To account for differences in the absolute amount of CHC extract between samples, the data were standardized by dividing each peak area through the sum of all integrated peaks.
Molecular procedures
DNA from adult wasps was extracted using the Chelex protocol described by Niehuis et al. (2007). Genotype data were collected for 71 molecular markers (56 length polymorphic; 11 single-nucleotide polymorphic; 4 present–absent) of known position in the nuclear genome of Nasonia (Niehuis et al., 2010) and provided in the online Supplementary information. The haplotype of the mitochondrial genome was verified with the aid of a length polymorphic marker in the LSU (16S) ribosomal RNA gene (online Supplementary information). The genotype of length polymorphic and present–absent markers was inferred by separating their PCR products on a denaturing polyacrylamide gel using the protocols given by Niehuis et al. (2008). Single-nucleotide polymorphic markers were genotyped using the Ecotilling technique (Comai et al., 2004) and following the protocol given by Niehuis et al. (2007). The PCR temperature profiles for each of the 72 studied molecular markers are listed in the online Supplementary Information.
Statistical analyses
All statistical computations were conducted with a 64-bit build of R version 2.10.1 (http://www.R-project.org). Differences in the relative abundance of CHCs between species were assessed with the Wilcoxon rank-sum test with continuity correction and applying a Bonferroni correction for multiple testing. QTL analyses were conducted with the R/qtl package version 1.14–2 (Broman and Sen, 2009). For each trait, we conducted a one-dimensional, one-QTL and a two-dimensional, two-QTL scan, using Haley–Knott regression (Haley and Knott, 1992) with an assumed genotype error probability of 0.001 and a fixed step width of 1 cM. Adjusted genome-wide significance thresholds for the presence of QTL were estimated from 10 000 permutations of the respective phenotype data. For each trait, we additionally conducted a multiple-QTL model search, applying the forward–backward model selection algorithm implemented in R/qtl. The maximum number of QTL for a given trait was set to 10, and the position of the QTL was allowed to be refined after each step of the forward and backward selection. Model selection penalties were calculated on the basis of the permutation data from the two-dimensional, two-QTL genome scan of the same trait and choosing a significance level of 0.01 (Manichaikul et al., 2009). Approximate Bayes credible intervals (Sen and Churchill, 2001) were calculated to specify the uncertainty in the predicted QTL location. Note that these estimates do not account for the uncertainty in the location of other QTL (Broman and Sen, 2009). If the marginal phenotype distribution of a trait was highly skewed, we ran the analyses with the log10- or square root-transformed phenotype data, if this was possible (that is, the phenotype data included no zero values). In two instances, the parametric analyses of traits with highly skewed marginal phenotype distribution that included zero values suggested a single QTL each. In these two cases, we tried to confirm the predicted QTL by re-analyzing the data with a non-parametric interval mapping approach as implemented in R/qtl. As the two QTL were unsupported by the non-parametric approach, we considered them questionable and dropped them from our reported list of QTL.
Search for candidate genes
We retrieved the D. melanogaster protein sequences of desat1, desat2, desatF and eloF from GenBank (accession no. CAB52474, CAB69054, CAO79269 and NP_649956) and used them as query to search the Nasonia official gene set 1.2 (Werren et al., 2010) with the program BLASTp (Altschul et al., 1990). The map position of the corresponding coding loci was inferred by searching the predicted Nasonia protein sequences against the mapped fraction of the Nasonia genome assembly 1.0 (Niehuis et al., 2010; Werren et al., 2010) with the program tBLASTn (Altschul et al., 1990).
Results
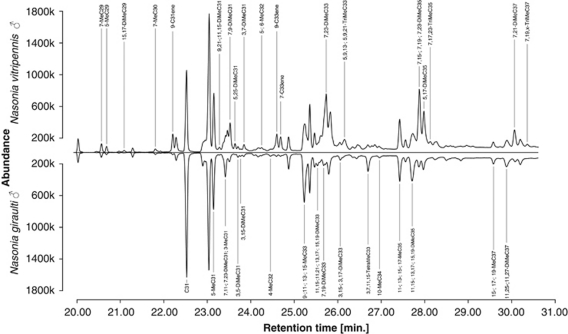
The chromatograms of Nasonia pure strain males allowed us to differentiate 54 CHC peaks with carbon chain lengths ranging from C29 to C37. For 52 of them, we were able to identify the underlying chemical compounds: 35 represented single compounds, 17 likely a mixture of two or more compounds that co-eluted into single peaks (Table 1). Statistical comparison of the 54 CHC peaks between males of N. giraulti and N. vitripennis revealed significant differences in 35 of them when applying a Bonferroni correction for multiple testing and a table-wide significance level of 0.05 (Figure 1; Table 1, retention indices with an asterisk (*) in the first column).
Table 1. CHCs of interspecific Nasonia girauli × Nasonia vitripennis F2 hybrid males and the estimated number, position, interaction and effect of proposed QTL that could explain the observed variance in the relative abundance of the CHCs.
| RIa | CHC(s)b |
CHC abundance (%) |
QTL modelc/LOD score (p)d/explained phenotypic variance | QTL position/LOD score/explained phenotypic variancee | |
|---|---|---|---|---|---|
| N. girauli | N. vitripennis | ||||
| 2900 | C29 | 1.21±0.36 | 1.25±0.34 | log10(y)∼Q1−+Q2−/4.884 (0.009)/20.0% | Q1: c3 (59.7–70–92.1)/3.665/14.6% Q2: c5 (8–23–56)/2.683/10.4% |
| 2934 | 15-/13-/11-/9-MeC29 | 0.01±0.04 | 0.11±0.13 | No QTL found | |
| 2942* | 7-MeC29 | 0.13±0.16 | 0.71±0.3 | log10(y)∼Q+/3.535 (0.004)/14.9% | Q: c4 (28–41–57)/3.535/14.9% |
| 2951* | 5-MeC29 | 0.16±0.13 | 0.42±0.15 | log10(y)∼Q1−+Q2−+Q1Q2−/6.147 (0.008)/24.4% | Q1: c3 (86–92–92.1)/4.820/18.6% Q2: c5 (13–16–23)/4.764/18.3% Q1Q2: 2.678/9.8% |
| 2973 | 3-MeC29 | 0.04±0.07 | 0.13±0.06 | No QTL found | |
| 2982* | 15,17-DiMeC29 | 0.01±0.03 | 0.14±0.08 | y∼Q−/4.180 (0.001)/17.4% | Q: c2 (85–89.8)/4.180/17.4% |
| 3000 | C30 | 0.48±0.1 | 0.37±0.07 | No QTL found | |
| 3040* | 7-MeC30 | 0.14±0.09 | 0.29±0.11 | y∼Q1−+Q2−/5.771 (0.002)/23.1% | Q1: c1 (20–33–45)/2.904/10.9% Q2: c5 (4–11–21)/2.764/10.3% |
| 3049 | 5-MeC30 | 0.01±0.02 | 0.04±0.03 | No QTL found | |
| 3074* | 9-C31ene | 0.08±0.1 | 1.41±0.36 | log10(y)∼Q1++Q2++Q3++Q4+/24.572/67.4% | Q1: c2 (9–23–32)/3.741/5.9% Q2: c3 (0–16)/6.072/10.4% Q3: c3 (73–81–83)/10.220/19.7% Q4: c4 (41–47–49)/9.430/17.9 % |
| 3100* | C31 | 12.84±2.93 | 7.04±1.26 | log10(y)∼Q−/6.833 (<0.001)/26.8% | Q: c3 (63–71–76)/6.833/26.8% |
| 3131 | 15-/13-/11-/9-MeC31 | 1.65±0.71 | 2.32±0.95 | log10(y)∼Q1−+Q2−+Q1Q2+/6.324 (0.004)/25.1% | Q1: c2 (40–44.2–57)/5.368/20.78% Q2: c2 (70–79–84)/5.731/22.38% Q1Q2: 5.058/19.44% |
| 3141 | 7-MeC31 | 10.65±1.77 | 11.46±1.1 | No QTL found | |
| 3149* | 5-MeC31 | 5.61±0.81 | 4.52±0.65 | y∼Q1−+Q2−/8.699 (<0.001)/32.7% | Q1: c2 (5–46–64)/2.682/8.75% Q2: c5 (40–47–56)/7.733/28.44% |
| 3158* | 9,21-/11,15-DiMeC31 | 0.24±0.04 | 0.49±0.15 | log10(y)∼Q1++Q2−/14.499 (<0.001)/48.4% | Q1: c3 (34–44–46)/10.979/33.5% Q2: c5 (37.7–47.6–57)/4.761/12.5% |
| 3171* | 7,11-/7,23-DiMeC31+3-MeC31 | 4.66±0.74 | 3.61±0.37 | log10(y)∼Q1++Q2−+Q3−+Q4++Q5+/22.860/64.7% | Q1: c1 (67–73–88.1)/5.331/9.7% Q2: c2 (86–89.8)/8.397/16.4% Q3: c3 (34–40–55)/6.896/13.0% Q4: c3 (83–88–91.2)/5.713/10.5% Q5: c5 (46–48–56)/7.277/13.9% |
| 3177* | 7,9-DiMeC31 | 1.42±0.16 | 2.23±0.34 | y∼Q1++Q2−+Q3++Q4+/24.313/67.0% | Q1: c1 (74–82–88.1)/7.625/13.7% Q2: c2 (86–89.8/7.327/13.1% Q3: c3 (25–30–34)/7.952/14.4% Q4: c3 (83–88–91.2)/9.217/17.2% |
| 3187* | 5,25-DiMeC31 | 0.19±0.05 | 0.51±0.13 | y∼Q1++Q2++Q3+/20.343/60.4% | Q1: c2 (24–30–41)/4.350/8.7% Q2: c3 (17–27–31)/10.570/24.5% Q3: c3 (81–89–92.1)/6.412/13.4% |
| 3192* | 3,5-DiMeC31 | 0.48±0.08 | 0.38±0.07 | log10(y)∼Q+/4.293 (<0.001)/17.8% | Q: c5 (17–24–35)/4.293/17.8% |
| 3197* | 3,15-DiMeC31 | 0.43±0.12 | 0.21±0.05 | log10(y)∼Q1++Q2−+Q3−/12.615/43.7% | Q1: c1 (27–73–91)/3.121/8.6% Q2: c2 (74–86–89.8)/5.396/15.7% Q3: c5 (27–35–52)/7.544/23.1% |
| 3204* | 3,7-DiMeC31 | 0.41±0.06 | 0.71±0.09 | y∼Q1++Q2++Q3+/16.647/53.2% | Q1: c1 (72–79–90)/5.938/14.6% Q2: c3 (27–33–44)/4.732/11.3% Q3: c3 (87–90–91.2)/8.328/21.6% |
| 3226 | ? | 0.27±0.09 | 0.18±0.08 | y∼Q1−+Q2−/11.345 (<0.001)/40.4% | Q1: c2 (81–85–89)/8.079/26.6% Q2: c5 (23–27–53)/6.601/20.9% |
| 3234 | 7-/8-MeC32 | 0.51±0.12 | 0.4±0.05 | y∼Q1−+Q2−+Q3−+Q4++Q1Q4−/17.270/54.5% | Q1: c1 (56–61–67)/5.699/13.5% Q2: c2 (82–87.0–89.8)/5.642/13.3% Q3: c4 (58–71–81)/4.079/9.3% Q4: c5 (51–58–65)/7.834/19.5% Q1Q4: 2.447/5.4% |
| 3244* | 5-/6-MeC32 | 0.31±0.1 | 0.44±0.07 | y∼Q+/4.102 (0.001)/17.1% | Q: c3 (33–83–90)/4.102/17.1% |
| 3261* | 4-MeC32 | 0.66±0.18 | 0.28±0.08 | No QTL found | |
| 3273* | 9-C33ene | 0.15±0.13 | 1.49±0.27 | log10(y)∼Q1−+Q2++Q3++Q4+/21.655/62.7% | Q1: c1 (77–86–92)/6.615/13.1% Q2: c3 (0–4–17)/7.722/15.7% Q3: c3 (70–75–81)/6.354/12.5% Q4: c4 (46–48–51)/8.099/16.6% |
| 3281* | 7-C33ene | 0.06±0.1 | 0.94±0.2 | log10(y)∼Q1++Q2++Q3++Q4++Q3Q4+/25.550/68.8% | Q1: c3 (2–17–24)/5.862/9.6% Q2: c3 (74–77–82)/10.422/19.0% Q3: c4 (48–49–53)/10.902/20.1% Q4: c4 (84–87.6)/5.752/9.4% Q3Q4: 3.147/4.8% |
| 3288 | ? | 0.17±0.09 | 0.12±0.04 | log10(y)∼Q−/3.220 (<0.003)/13.7% | Q: c2 (77–89.8)/3.220/13.7% |
| 3300 | C33 | 1.37±0.4 | 1.08±0.19 | log10(y)∼Q−/5.788 (<0.001)/23.2% | Q: c3 (74–82–89)/5.788/23.2% |
| 3330* | 9-/11-/13-/15-MeC33 | 7.92±0.9 | 4.58±0.96 | y∼Q1−+Q2−+Q3−/22.943/46.9% | Q1: c1 (51.6–56–59)/7.168/20.5% Q2: c2 (73–79–84)/6.189/17.3% Q3: c4 (60–74.5–82)/3.607/9.5% |
| 3339 | 7-MeC33 | 3.5±0.8 | 3.15±0.65 | log10(y)∼Q+/4.917 (<0.001)/20.1% | Q: c5 (26–47.6–56)/4.917/20.1% |
| 3349 | 5-MeC33 | 1.37±0.4 | 1.4±0.33 | No QTL found | |
| 3355* | 11,15-/11,21-/13,17-/15,19-DiMeC33 | 2.27±0.63 | 0.84±0.38 | log10(y)∼Q1−+Q2−+Q3−+Q4−+Q3Q4−/35.454/80.1% | Q1: c1 (46–48–54)/6.625/7.0% Q2: c2 (47–49–54)/15.327/20.1% Q3: c2 (85–89–89.8)/8.555/9.5% Q4: c5 (47–50) 31.964/65.4% Q3Q4: 3.153/3.0% |
| 3360* | 7,19-DiMeC33 | 1.22±0.21 | 1.91±0.48 | log10(y)∼Q1−+Q2++Q3−+Q4++Q3Q4−/19.627/59.1% | Q1: c1 (48–54–57.1)/6.353/13.7% Q2: c2 (87–89.8)/9.086/21.0% Q3: c3 (0–17–20.4)/5.045/10.6% Q4: c5 (46–48–56)/9.304/21.6% Q3Q4: 3.509/7.1% |
| 3370* | 7,23-DiMeC33 | 3.2±0.38 | 7.18±1.15 | y∼Q1++Q2++Q3++Q1Q2−/26.749/70.5% | Q1: c2 (17–36–42)/5.688/8.7% Q2: c3 (64–69–74)/5.931/9.2% Q3: c5 (50–54–56)/24.382/60.2% Q1Q2/3.975/5.9% |
| 3376 | 5,9-DiMeC33 | 4.9±0.8 | 5.38±0.52 | No QTL found | |
| 3396* | 3,15-/3,17-DiMeC33 | 1.64±0.36 | 0.99±0.2 | No QTL found | |
| 3405* | 5,9,13-/5,9,21-TriMeC33 | 0 | 1.63±0.34 | y∼Q1++Q2++Q3++Q4+/12.954/44.6% | Q1: c1 (0–9–24)/4.066/11.3% Q2: c1 (73–79–89.9)/4.032/11.2% Q3: c2 (49–66–84)/4.531/12.7% Q4: c5 (22–47–59)/3.927/10.9% |
| 3421 | 3,7,11-TriMeC33 | 0.42±0.16 | 0.31±0.06 | No QTL found | |
| 3430 | 8,10-/8,12-/8,14-/8,16-/8,18-DiMeC34 | 0.83±0.21 | 0.76±0.16 | log10(y)∼Q+/2.686 (0.010)/11.5% | Q: c1 (0–10–25)/2.686/11.5% |
| 3453* | 3,7,11,15-TetraMeC33 | 3.73±0.67 | 0.7±0.16 | y∼Q1−+Q2−/8.586 (<0.001)/32.4% | Q1: c1 (48–53.4–58)/3.711/12.5% Q2: c3 (20–28–57)/5.804/20.5% |
| 3477* | 10-MeC34 | 0.51±0.13 | 0.32±0.11 | No QTL found | |
| 3524* | 11-/13-/15-/17-MeC35 | 3.3±0.37 | 2.8±0.24 | log10(y)∼Q1−+Q2++Q3−+Q1Q2+/17.840/55.7% | Q1: c1 (59–60–67)/10.451/27.1% Q2: c2 (87–89.8)/9.916/25.3% Q3: c3 (35–46–56)/5.520/12.7% Q1Q2: 1.369/2.9% |
| 3534 | 7-MeC35 | 0.76±0.12 | 0.8±0.2 | log10(y)∼Q1−+Q2++Q3−+Q4+/17.828/55.6% | Q1: c1 (63–74–86)/4.279/9.6% Q2: c2 (82–86–89.8)/6.571/15.5% Q3: c3 (31–44.3–83)/5.634/13.0% Q4: c5 (26–34–37)/7.288/17.4% |
| 3549* | 11,15-/13,17-/15,19-DiMeC35 | 6.47±0.85 | 3.53±0.66 | log10(y)∼Q1−+Q2−/11.730 (<0.001)/41.4% | Q1: c1 (49–54–80)/4.904/14.7% Q2: c5 (39–51–57)/8.347/27.1% |
| 3563* | 7,15-/7,19-/7,23-DiMeC35 | 2.13±0.28 | 6.21±0.9 | y∼Q1++Q2+/10.044 (<0.001)/36.7 % | Q1: c1 (2–12–23)/3.687/11.6% Q2: c5 (39–46–57)/6.640/22.4% |
| 3572* | 5,17-DiMeC35 | 3.5±0.52 | 4.81±0.5 | log10(y)∼Q1++Q2+/10.138 (<0.001)/37.0% | Q1: c1 (1–12–29)/3.617/11.3% Q2: c2 (83–88–89.8)/6.531/21.9% |
| 3585* | 7,17,23-TriMeC35 | 0.57±0.29 | 0.86±0.14 | No QTL found | |
| 3603 | 5,9,x-TriMeC35 | 1.04±0.33 | 1.41±0.2 | log10(y)∼Q+/4.805 (<0.001)/19.7% | Q: c1 (1–13–23)/4.805/19.7% |
| 3721* | 15-/17-/19-MeC37 | 0.89±0.26 | 0.61±0.15 | y∼Q1−+Q2−/5.504 (0.003)/22.2% | Q1: c1 (56–58–88)/3.342/12.8% Q2: c3 (26–34–91)/3.473/13.3% |
| 3745* | 11,25-/11,27-DiMeC37 | 2.1±0.37 | 1.5±0.44 | y∼Q1−+Q2−+Q3−/14.451/48.3% | Q1: c1 (53.4–58–84)/4.728/12.5% Q2: c3 (23–29–35)/5.778/15.6% Q3: c5 (41–48–57)/8.447/24.3% |
| 3764* | 7,21-DiMeC37 | 0.6±0.16 | 1.97±0.3 | y∼Q1++Q2++Q3+/15.247/50.1% | Q1: c1 (47–48–50)/8.545/23.8% Q2: c2 (63–66–84)/4.381/11.0% Q3: c5 (42–52–59)/7.919/21.7% |
| 3768 | 5,15-/5,17-DiMeC37 | 1.3±0.28 | 1.22±0.18 | y∼Q1−+Q2+/5.062 (0.008)/20.6% | Q1: c3 (18–31–65)/3.251/12.7% Q2: c4 (26–48.2–84)/2.384/9.1% |
| 3779* | 7,19,x-TriMe-C37 | 0.03±0.09 | 0.3±0.08 | y∼Q1++Q2+/8.074 (<0.001)/30.8% | Q1: c1 (0–1–21)/3.197/10.9% Q2: c2 (66–74–82)/5.470/19.6% |
Abbreviations: CHC, cuticular hydrocarbon; LOD, logarithm of the odds; QTL, quantitative trait loci; RI, retention index.
RIs with * indicate a significant difference in the relative abundance of the CHC compound(s) between males of the parental strains, that is, RV2X(U) and AsymCX.
CHC compounds and/or characteristics that have not been reported by Carlson et al. (1999) and Steiner et al. (2006) are printed in bold.
Superscripts indicate the direction of the effect, with minus (−) indicating a decrease of the respective CHC(s). Relative to the total CHC abundance in hybrids with N. vitripennis allele at the QTL.
Currently not reliably calculable for QTL models with more than two QTL.
The QTL position with the highest LOD score is underlined.
Figure 1.
CHC profiles of Nasonia vitripennis (strain AsymCX; above) and Nasonia giraulti (strain RV2X(U); below) males. Labeled are CHC compounds whose relative abundance differs significantly between the two investigated strains (Wilcoxon rank-sum test with continuity correction and Bonferroni correction for multiple testing: Ng=12; Nv=22; α=0.05). CHC labels are shown in the field of the species for which the relative portion of the respective CHC compound was found to be significantly higher.
Of the 120 male F2 hybrids of Nasonia giraulti (♂) and N. vitripennis (♀) that we initially collected, 19 were discarded before the QTL analyses because their GC–MS profiles had an inferior quality and/or >10% of their genotypes were missing. The average percentage of missing genotype information in the remaining 101 F2 hybrid males was 0.9% (min/max: 0.0/8.5%), and none of the 71 nuclear markers had >7.9% missing data. The average recovery rate of the N. vitripennis genotype was 60.5% (min/max: 42.6/71.3%), consistent with the fact that N. giraulti × N. vitripennis F2 hybrid males with N. vitripennis cytoplasm suffer from cytonuclear genic incompatibilities (Breeuwer and Werren, 1995; Niehuis et al., 2008). The genotype of the cytoplasm was confirmed by studying a molecular marker of the mitochondrial genome that differs between the two Nasonia stains.
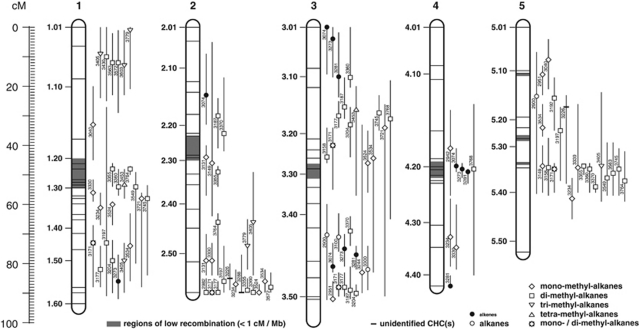
The statistical analyses suggested one or more QTL for 42 of the 54 investigated traits when applying a genome-wide significance level of 0.01 (only approximated for QTL models with more than two QTL; Manichaikul et al., 2009; Table 1). For 31 (74%) of these traits, we had seen significant differences between the parental strains (see above and Table 1). In 8 of the 42 traits with predicted QTL, we also found evidence for QTL interactions (Table 1). The predicted 103 QTL cover all five chromosomes (c1–c5; Figure 2). Although at first glance, the number of QTL per chromosome does not look evenly distributed over the five chromosomes (for example, chromosome 3 has 28 predicted QTL while chromosome 4 has only 8), the observed values are not significantly different from expected values when using the genetic length or the physical length of the chromosomes as basis for estimating the expected values (Fischer's exact test, P>0.09). In some regions of the chromosomes, the QTL for CHCs are clustered. Particularly conspicuous are the clusters on chromosome 1 between 9 and 13 cM (retention index (RI) 3405, 3430, 3563, 3572 and 3603), on chromosome 2 between 85 and 89.8 cM (RI 2982, 3171, 3177, 3197, 3226, 3234, 3288, 3355, 3360, 3524, 3534 and 3572), on chromosome 3 between 27 and 34 cM (RI 3177, 3187, 3204, 3453, 3745, 3721 and 3768) and between 88 and 92 cM (RI 2951, 3171, 3177, 3187 and 3204), and on chromosome 5 between 47 and 52 cM (RI 3149, 3158, 3171, 3339, 3355, 3360, 3405, 3549, 3563, 3745 and 3764) (Figure 2; see also Table 1). Except for the cluster on chromosome 5, the position of the QTL clusters does not coincide with regions of low (<1 cM per Mb) recombination (Figure 2: gray shaded sections).
Figure 2.
Nasonia giraulti (♂) × Nasonia vitripennis (♀) recombination map with estimated positions of predicted QTL for 42 CHCs. For each QTL, the retention index (RI) of the corresponding CHC compound (see Table 1), the position of the maximum LOD score, and the 95% Bayes credible interval are given. The five chromosomes are labeled 1–5. Horizontal lines on the chromosomes indicate the position of the 71 genotyped molecular markers. Landmarks next to the chromosomes (1.1–5.50) correspond to those defined by Niehuis et al. (2010). The same reference was consulted for highlighting regions on the genetic map with low recombination rate (<1 cM per Mb; gray shaded areas).
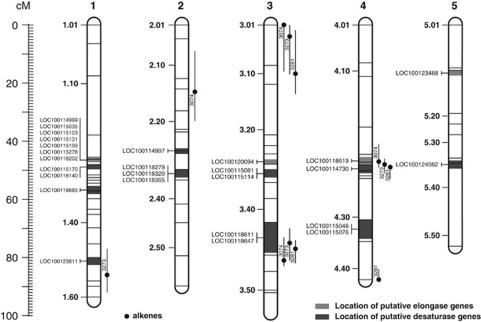
Using D. melanogaster protein sequences of the genes desat1, desat2 and desatF as query for searching the Nasonia official gene set 1.2, we identified 16 predicted Nasonia proteins with high similarity (E values ranging from 10−132 to 4 × 10−47) to the above fruit fly desaturases (Table 2). Four of the corresponding gene loci (that is, LOC100123811, LOC100118647, LOC100118611, and LOC100114730) in Nasonia genome assembly 1.0 map to regions with predicted QTL of alkenes, that is, RI 3074 (9-C31ene), RI 3273 (9-C33ene) and RI 3281 (7-C33ene) (Figure 3). Using the same approach, we identified 11 gene loci in the Nasonia genome with high similarity (E values ranging from 3 × 10−37 to 3 × 10−26) to the elongase-encoding gene eloF of the fruit fly (Table 3). One of them maps to a cluster of QTL for alkenes (Figure 3).
Table 2. Assembly and map position of gene loci in the Nasonia genome with high similarity to the Drosophila genes desat1, desat2 and desatF.
| GenBank accession no. to hit proteins | Associated E values when using protein sequences of desat1 (CAB52474), desat2 (CAB69054) and desatF (CAO79269) as queries | Corresponding coding gene locus in Nasonia genome | Position of coding gene locus in Nasonia genome | Position of coding gene locus on Nasonia linkage map | ||
|---|---|---|---|---|---|---|
| XP_001602592 | 1 × 10−132 | 1 × 10−129 | 1 × 10−111 | LOC100118685 | NW_001815459: 1469902–1479446 | c1: 55.3–58.0 cM |
| XP_001599665 | 1 × 10−120 | 1 × 10−114 | 1 × 10−100 | LOC100115081 | NW_001816015: 38539–41454 | c3: 49.7–52.4 cM |
| XP_001602565 | 1 × 10−119 | 1 × 10−115 | 1 × 10−100 | LOC100118647 | NW_001815459: 1465974–1467302 | c3: 67.9–78.3 cM |
| XP_001600683 | 1 × 10−101 | 3 × 10−95 | 1 × 10−84 | LOC100116140 | NW_001816348: 266783–277924 | c1: 48.0–49.8 cM |
| XP_001602540 | 3 × 10−93 | 3 × 10−85 | 9 × 10−85 | LOC100118611 | NW_001815459: 1460847–1462515 | c3: 67.9–78.3 cM |
| XP_001607533 | 2 × 10−89 | 4 × 10−88 | 7 × 10−84 | LOC100123811 | NW_001815682: 4609881–4618860 | c1: 79.9–82.6 cM |
| XP_001599877 | 3 × 10−87 | 2 × 10−87 | 5 × 10−75 | LOC100115046 | NW_001815459: 537178–548645 | c4: 67.0–73.6 cM |
| XP_001599899 | 2 × 10−84 | 5 × 10−86 | 8 × 10−75 | LOC100115076 | NW_001815459: 557556–559638 | c4: 67.0–73.6 cM |
| XP_001599836 | 2 × 10−84 | 1 × 10−80 | 2 × 10−74 | LOC100114997 | NW_001820538: 49163–51333 | c2: 42.3–44.2 cM |
| XP_001599873 | 2 × 10−84 | 1 × 10−80 | 4 × 10−79 | LOC100115170 | NW_001815070: 1211–4350 | c1: 48.9–49.8 cM |
| XP_001599357 | 4 × 10−75 | 1 × 10−71 | 9 × 10−69 | LOC100114730 | NW_001814915: 81646–83527 | c4: 48.2–51.0 cM |
| XP_001602289 | 4 × 10−75 | 1 × 10−68 | 1 × 10−72 | LOC100118279 | NW_001815681: 339354–341032 | c2: 49.7–52.4 cM |
| XP_001607893 | 4 × 10−74 | 4 × 10−71 | 5 × 10−74 | LOC100124062 | NW_001816793: 6800959–6802527 | c5: 46.7–49.4 cM |
| XP_001602317 | 2 × 10−72 | 8 × 10−75 | 4 × 10−73 | LOC100118320 | NW_001815681: 341425–342772 | c2: 49.7–52.4 cM |
| XP_001602341 | 3 × 10−70 | 3 × 10−71 | 2 × 10−66 | LOC100118355 | NW_001815681: 343163–344570 | c2: 49.7–52.4 cM |
| XP_001599579 | 4 × 10−55 | 1 × 10−48 | 4 × 10−47 | LOC100115114 | NW_001816015: 47776–49768 | c3: 49.7–52.4 cM |
Given are the GenBank accession numbers of protein hits in the Nasonia official gene set 1.2, the associated E values when using the protein sequences of the above genes as query, and the gene locus names with their position in the Nasonia genome assembly 1.0.
Figure 3.
Nasonia giraulti (♂) × Nasonia vitripennis (♀) recombination map with position of genes with high similarity to the Drosophila genes desat1, desat2 desatF and eoF (dark and light grey shaded areas). The map also shows the estimated positions of predicted QTL for three identified alkenes in the Nasonia CHC profile. As in Figure 2, the retention index (RI) of the corresponding CHC compound (see Table 1), the position of the maximum LOD score, and the 95% Bayes credible interval are given. The five chromosomes are labeled 1–5. Horizontal lines on the chromosomes indicate the position of the 71 genotyped molecular markers. Landmarks next to the chromosomes (1.1–5.50) correspond to those defined by Niehuis et al. (2010).
Table 3. Assembly and map position of gene loci in the Nasonia genome with high similarity to the Drosophila gene eloF.
| GenBank accession no. to hit proteins | Associated E value when using protein sequences of eloF (NM_141699) as query | Corresponding coding gene locus in Nasonia genome | Position of coding gene locus in Nasonia genome | Position of coding gene locus on Nasonia linkage map |
|---|---|---|---|---|
| XP_001599867 | 3 × 10−37 | LOC100115035 | NW_001820615: 106812–110218 | c1: 45.3–47.1 cM |
| XP_001599838 | 4 × 10−37 | LOC100114999 | NW_001820615: 98654–99896 | c1: 45.3–47.1 cM |
| XP_001600048 | 1 × 10−35 | LOC100115278 | NW_001820615: 303419–304635 | c1: 45.3–47.1 cM |
| XP_001603768 | 2 × 10−35 | LOC100120094 | NW_001818459: 777457–780553 | c3: 46.1–47.9 cM |
| XP_001599914 | 3 × 10−35 | LOC100115103 | NW_001820615: 124563–126594 | c1: 45.3–47.1 cM |
| XP_001599942 | 5 × 10−35 | LOC100115131 | NW_001820615: 175512–186297 | c1: 45.3–47.1 cM |
| XP_001599996 | 6 × 10−34 | LOC100115199 | NW_001820615: 202550–214182 | c1: 45.3–47.1 cM |
| XP_001600017 | 2 × 10−34 | LOC100115239 | NW_001820615: 253540–269384 | c1: 45.3–47.1 cM |
| XP_001607111 | 5 × 10−31 | LOC100123468 | NW_001820527: 3351012–3357150 | c5: 15.6–17.4 cM |
| XP_001600743 | 6 × 10−29 | LOC100116202 | NW_001816015: 88275–89828 | c1: 45.3–47.1 cM |
| XP_001602545 | 3 × 10−26 | LOC100118619 | NW_001816126: 134952–175847 | c4: 45.4–48.2 cM |
Given are the GenBank accession numbers of protein hits in the Nasonia official gene set 1.2, the associated E values when using the protein sequence of the above gene as query, and the gene locus names with their position in the Nasonia genome assembly 1.0.
Discussion
This study aimed to explore the potential of Nasonia as a genetic model to study the genetics of CHCs in insects in general and in the Hymenoptera in particular. Our analysis of CHC extracts from N. giraulti and N. vitripennis males revealed a plethora (54) of CHC compounds that encompass six different CHC classes (that is, alkanes, mono-, di-, tri- and tetramethylalkanes, as well as alkenes) and CHCs with chain lengths ranging from C29 to C37 (Figure 1). We confirmed the identity of most of the CHC compounds that had previously been reported for N. vitripennis (Carlson et al., 1999; Steiner et al., 2006). In addition, we were able to clarify previously ambiguous characteristics (for example, the exact position of methyl groups) of nine CHCs and identified 11 CHC compounds that had not been previously reported in Nasonia (Table 1, respective changes and new identifications highlighted in second column by bold typeface). Overall, our GC–MS analyses allowed us to precisely identify and classify the majority of CHCs in Nasonia, an important prerequisite for assessing the possible clustering of QTL for structurally similar CHCs. As we found the majority (67%) of CHC compounds to differ in their relative abundance between the two parental species (Figure 1; Table 1), our data provide a promising basis for investigating the genetics of CHCs in Nasonia.
We exploited the particular strengths of the Nasonia genetic model system (that is, availability of highly inbred strains, cross-fertility of Nasonia species, male haploidy) to explore the genetics of CHC in N. giraulti × N. vitripennis F2 hybrid males in more detail. As expected from the large number of CHCs with fixed genetic differences between the two investigated species, statistical analyses suggested one or more QTL for most (78%) of the traits. Only in four traits (4-MeC32; 3,15-/3,17-DiMeC33; 10-MeC34, and 7,17,23-TriMeC35), in which the CHC profile comparison had revealed fixed genetic differences between the parental stains, did the statistical analyses fail to detect QTL, likely because these traits are governed by multiple QTL, each of which with a small effect. Perhaps more interesting, however, is the fact that the statistical analyses suggested QTL for 11 traits (C29; 15-/13-/11-/9-MeC31; 7,8-MeC32; C33; 7-MeC33; 8,10-/8,12-/8,14-/8,16-/8,18-DiMeC34; 7-MeC35; 5,9,x-TriMeC35; 5,15-/5,17-DiMeC37, and two unidentified CHCs) for which we had not found significant differences between the parental strains (Table 1). The two most likely explanations for this outcome are fixed genetic differences between the parental strains that we failed to detect by chance (false negatives) or falsely proposed QTL (false positives). As it is currently not possible to estimate exact significance values for multiple QTL models (that is, models with more than two QTL; Manichaikul et al., 2009), we were unable to apply a sequential Bonferroni correction for multiple testing. We consequently tried to minimize the possible impact of falsely proposed QTL by choosing a conservative genome-wide significance level of 0.01 when calculating model selection penalties and by focusing our discussion on QTL clusters rather than on individual QTL. A completely different possible explanation worth of further investigation is stabilizing selection, but more studies are clearly necessary to corroborate this idea. Overall, the QTL mapping allowed us to provide data necessary for positional cloning and the identification of genes involved in CHC biosynthesis.
Given that >50% of the mapped physical Nasonia genome has an estimated recombination rate <1 cM per Mb but account for <7% of the genetic map (Niehuis et al., 2010), we would have expected many of the predicted QTL to cluster in regions of low recombination, for example, around the centromeres. Contrary to our expectation, however, four of five conspicuous QTL clusters mapped to regions with high recombination rate (>1 cM per Mb; Figure 2). The clustering of QTL in regions with a high recombination rate could be an indication for pleiotropic QTL that account for differences in more than just one trait. A comparison of traits that are affected by a pleiotropic QTL could thus, at least in theory, provide a clue as to what pathway is impacted. The four QTL clusters that map to regions in the genome with high recombination rate indeed partially comprise QTL of CHCs that share particular characteristics, for example, the same position of methyl groups or an equal distance (that is, number of methylene groups) between their methyl groups (Figure 2; Table 1). For example, the QTL cluster on chromosome 1 between 9 and 13 cM contains QTL for dimethylalkanes that have either seven (8,16-DiMeC34 and 7,15-DiMeC35) or 11 (7,19- and 5,17-DiMeC35) methylene groups between their two methyl groups and QTL for trimethylalkanes with three methylene groups between their first two methyl groups (5,9,13-/5,9,21-TriMeC33 and 5,9,x-TriMeC35). The second QTL cluster, on chromosome 2 between 85 and 89.8 cM, also encompasses QTL of structurally related dimethylalkanes: two have one methylene group between their methyl groups (15,17-DiMeC29 and 7,9-DiMeC31), a condition rarely seen in CHCs (Blomquist and Bagneres, 2010), while the others have 3 (11,15-/13,17-/15,19-DiMeC33) or 11 (7,19-DiMeC33 and 5,17-DiMeC35) methylene groups between their methyl groups. The QTL cluster on chromosome 2 further includes QTL of monomethylalkanes whose methyl groups share the same position (7-MeC32 and 7-MeC35). Finally, the first of the two QTL clusters on chromosome 3, between 27 and 34 cM, encompasses QTL for structurally unrelated dimethylalkanes and one mono- and one tetramethylalkane, but the CHCs corresponding to the QTL in the second cluster on chromosome 3, between 88 and 92 cM, are again structurally similar (7,11- and 3,7-DiMeC31; 5-MeC29 and 5-MeC32). Overall, QTL of structurally related and unrelated dimethylalkanes dominate the four conspicuous QTL clusters. Unfortunately, no genes have been identified so far that are involved in the biosynthesis of methyl-branched alkanes (Blomquist and Bagneres, 2010). Our mapping data therefore constitute a promising and valuable basis for unraveling the genetics of methyl-branched alkanes. Ideally, the first targets for future genetic analyses and fine-mapping experiments would be those QTL of structurally related dimethylalkanes that cluster in regions of the genome with high recombination rate.
The only CHC compound class whose biosynthesis has been investigated at the molecular level in greater detail is alkenes. Alkenes and alkadienes constitute the main fraction in the CHC profile of D. melanogaster and most other species of the D. melanogaster subgroup (Jallon and David, 1987; Coyne, 1996). Until now, three desaturases that are involved in the biosynthesis of alkenes have been identified in Drosophila, all of which are species- (Fang et al., 2009), strain- (Dallerac et al., 2000) and/or sex-specifically (Chertemps et al., 2006) expressed. Their enzymatic function has been characterized to regioselectively introduce double bounds in alkene and alkadiene fatty-acid precursors (Dallerac et al., 2000; Chertemps et al., 2006). The CHC profiles of Nasonia also include alkenes, albeit in lesser abundances than in Drosophila (Jallon and David, 1987; Coyne, 1996). The three alkenes identified in Nasonia (9-C31ene, 9-C33ene and 7-C33ene) all differ significantly in their relative abundance between the two investigated strains (Figure 1; Table 1), which could indicate that they function as species-specific cues. Interestingly, several of the QTL of these alkenes cluster in the genome (Figures 2 and 3). Given the significance of desaturases for the biosynthesis of alkenes in Drosophila, we used their sequences to search the Nasonia genome for genes with high sequence similarity. In total, we identified 16 putative desaturase genes that map to nine different regions of the Nasonia genome (Table 2). Intriguingly, three of the predicted desaturase genes map to regions in which the QTL for alkenes are clustered (Figure 3). This coincidence could indicate that one or more of these putative desaturases may have functions similar to that in Drosophila. However, it has to be mentioned that the carbon chain length of alkenes in Drosophila that are affected by the above desaturases (mainly C23, C29) are all shorter than in Nasonia (C31, C33). Desaturases being involved in the biosynthesis of alkenes with chain lengths >C29 have not yet been functionally characterized (Hashimoto et al., 2008), but could occur in insects (Blomquist and Bagneres, 2010). Thus, the data allow prioritizing experiments when considering reverse genetic approaches. For example, it makes sense to start RNA interference-mediated knock-down experiments on those putative desaturase genes (that is, LOC100118611, LOC100118619 and LOC10011847) that map to the same regions as the clustered QTL for alkenes. This would constitute a promising approach to functionally characterize desaturases with different properties than the ones described so far (Blomquist and Bagneres, 2010). However, it has to be stated that a coincidence in the position of the QTL for alkenes and the position of genes that encode putative desaturases does not automatically imply a casual relationship between the latter and the former.
We also searched the Nasonia genome for genes with high similarity to the Drosophila gene eloF. In Drosophila melanogaster, this gene encodes a protein that is responsible for the elongation of alkadienes from 25C to 29C (Chertemps et al., 2007). We identified a total of 11 genes in the Nasonia genome that share a high sequence similarity to eloF (Table 3). However, only one of them coincides in its position with that of QTL for alkenes (that is, LOC100118619; Figure 1). Although at first glance being a reasonable candidate to apply a reverse genetic approach, we have doubts in LOC100118619 being involved in the biosynthesis of alkenes in Nasonia. First, eloF has only a marginal impact on the biosynthesis of alkenes in Drosophila (Chertemps et al., 2007), which could mean that it is specific for alkadienes, a CHC compound class that has not even been found in traces in the CHC profiles of Nasonia males. Second, to the best of our knowledge there is no single report of an elongase that has been shown to elongate hydrocarbons beyond the length of C30 (Hashimoto et al., 2008). For these two reasons, we think that LOC100118619 is not a promising candidate for being functionally involved in the biosynthesis of the alkenes 9-C31ene and 9-C33ene. This example shows how difficult it can be to transfer advances in our knowledge of the biosythesis and genetics of CHCs from Drosophila to other insect taxa.
In conclusion, our study revealed the great advantages of cross-fertile Nasonia strains to rapidly map genomic regions that account for differences in a plethora of CHCs. Especially for investigating the still unknown genetics of methyl-branched alkanes, the main compound class of Nasonia CHC profiles, the mapping data we report here have promising potential. Moreover, using a comparative approach, we were able to identify candidate genes that encode putative desaturases. For some of them, close vicinity to clusters of QTL for alkenes suggest their potential involvement in the biosynthesis of Nasonia alkenes. However, we also exposed how difficult it can be to apply results from studies on Drosophila to Nasonia using the example of eloF. The latter demonstrates the need for more research on the biosynthesis and genetics of CHCs in highly diverged and species rich taxa such as the Hymenoptera. The data from our analyses provide the required basis for these studies.
Acknowledgments
We thank John H Werren for sending us the Nasonia strains and the host puparia for culturing them. Benjamin Allen and Jessica Overson kindly cared for the Nasonia strains in the lab and Bart A Pannebakker and Tosca Koevoets generously provided us with the oligonucleotide primer sequences of two microsatellite markers before their publication. ON acknowledges the Alexander von Humboldt Foundation for a Feodor Lynen Research Fellowship for Postdoctoral Researchers. TS and JB were funded by the Excellence Initiative of the German Research Foundation (GSC-4; Spemann Graduate School).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Blomquist GJ, Bagneres AG. Insect Hydrocarbons: Biology, Biochemistry, and Chemical Ecology. Cambridge University Press: New York, USA; 2010. [Google Scholar]
- Breeuwer JAJ, Werren JH. Hybrid breakdown between two haplodiploid species: the role of nuclear and cytoplasmic genes. Evolution. 1995;49:705–717. doi: 10.1111/j.1558-5646.1995.tb02307.x. [DOI] [PubMed] [Google Scholar]
- Broman KW, Sen S. A Guide to QTL Mapping with R/QTL. Springer: Dordrecht; 2009. [Google Scholar]
- Carlson DA, Bernier UR, Sutton BC. Elution patterns from capillary GC for methyl-branched alkanes. J Chem Ecol. 1998;24:1845–1865. [Google Scholar]
- Carlson DA, Geden CJ, Bernier UR. Identification of pupal exuviae of Nasoniavitripennis and Muscidifurax raptorellus parasitoids using cuticular hydrocarbons. Biol Contr. 1999;15:97–105. [Google Scholar]
- Chertemps T, Duportets L, Labeur C, Ueda R, Takahashi K, Saigo K, et al. A female-biased expressed elongase involved in long-chain hydrocarbon biosynthesis and courtship behavior in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:4273–4278. doi: 10.1073/pnas.0608142104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertemps T, Duportets L, Labeur C, Ueyama M, Wicker-Thomas C. A female-specific desaturase gene responsible for diene hydrocarbon biosynthesis and courtship behaviour in Drosophila melanogaster. Insect Mol Biol. 2006;15:465–473. doi: 10.1111/j.1365-2583.2006.00658.x. [DOI] [PubMed] [Google Scholar]
- Comai L, Young K, Till BJ, Reynolds SH, Greene EA, Codomo CA, et al. Efficient discovery of DNA polymorphisms in natural populations by Ecotilling. Plant J. 2004;37:778–786. doi: 10.1111/j.0960-7412.2003.01999.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA. Genetics of differences in pheromonal hydrocarbons between Drosophila melanogaster and D. simulans. Genetics. 1996;143:353–364. doi: 10.1093/genetics/143.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Crittenden AP, Mah K. Genetics of a pheromonal difference contributing to reproductive isolation in Drosophila. Science. 1994;265:1461–1464. doi: 10.1126/science.8073292. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Associates: Sunderland, MA; 2004. [Google Scholar]
- Coyne JA, Wicker-Thomas C, Jallon JM. A gene responsible for a cuticular hydrocarbon polymorphism in Drosophila melanogaster. Genet Res. 1999;73:189–203. doi: 10.1017/s0016672398003723. [DOI] [PubMed] [Google Scholar]
- Dallerac R, Labeur C, Jallon JM, Knippie DC, Roelofs WL, Wicker-Thomas C. A Δ9 desaturase gene with a different substrate specificity is responsible for the cuticular diene hydrocarbon polymorphism in Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:9449–9454. doi: 10.1073/pnas.150243997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling DC, Werren JH. Biosystematics of Nasonia (Hymenoptera: Pteromalidae): two new species reared from birds' nests in North America. Ann Entomol Soc Am. 1990;83:352–370. [Google Scholar]
- Fang S, Ting CT, Lee CR, Chu KH, Wang CC, Tsaur SC. Molecular evolution and functional diversification of fatty acid desaturases after recurrent gene duplication in Drosophila. Mol Biol Evol. 2009;26:1447–1456. doi: 10.1093/molbev/msp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferveur J-F. Cuticular hydrocarbons: their evolution and roles in Drosophila pheromonal communication. Behav Genet. 2005;35:279–295. doi: 10.1007/s10519-005-3220-5. [DOI] [PubMed] [Google Scholar]
- Grimaldi DA, Engel MS. Evolution of Insects. Cambridge University Press: Cambridge; 2005. [Google Scholar]
- Hadley NF. Cuticular lipids of terrestrial plants and arthropods—a comparison of their structure, composition, and waterproofing function. Biol Rev. 1981;56:23–47. [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Yoshizawa AC, Okuda S, Kuma K, Goto S, Kanehisa M. The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J Lipid Res. 2008;49:183–191. doi: 10.1194/jlr.M700377-JLR200. [DOI] [PubMed] [Google Scholar]
- Howard RW, Blomquist GJ. Ecological, behavioral, and biochemical aspects of insect hydrocarbons. Ann Rev Entomol. 2005;50:371–393. doi: 10.1146/annurev.ento.50.071803.130359. [DOI] [PubMed] [Google Scholar]
- Jallon JM, David JR. Variations in cuticular hydrocarbons among the eight species of the Drosophila melanogaster subgroup. Evolution. 1987;41:294–302. doi: 10.1111/j.1558-5646.1987.tb05798.x. [DOI] [PubMed] [Google Scholar]
- Legendre A, Miao XX, Da Lage JL, Wicker-Thomas C. Evolution of a desaturase involved in female pheromonal cuticular hydrocarbon biosynthesis and courtship behavior in Drosophila. Insect Biochem Mol Biol. 2008;38:244–255. doi: 10.1016/j.ibmb.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Manichaikul A, Moon JY, Sen S, Yandell BS, Broman KW. A model selection approach for the identification of quantitative trait loci in experimental crosses, allowing epistasis. Genetics. 2009;181:1077–1086. doi: 10.1534/genetics.108.094565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Blomquist GJ.1995Waxes: chemistry, molecular biology and functionsIn: Hamilton RJ (ed).Insect Waxes Oily Press: Dundee, Scotland; 1–90. [Google Scholar]
- Niehuis O, Gibson JD, Rosenberg MS, Pannebakker BA, Koevoets T, Judson AK, et al. Recombination and its impact on the genome of the haplodiploid parasitoid wasp Nasonia. PLoS One. 2010;5:e8597. doi: 10.1371/journal.pone.0008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehuis O, Judson AK, Gadau J. Cytonuclear genic incompatibilities cause increased mortality in male F2 hybrids of Nasonia giraulti and Nasonia vitripennis. Genetics. 2008;178:413–426. doi: 10.1534/genetics.107.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehuis O, Judson AK, Werren JH, Hunter WB, Dang PM, Dowd SE, et al. Species-diagnostic single-nucleotide polymorphism and sequence-tagged site markers for the parasitic wasp genus Nasonia (Hymenoptera: Pteromalidae) J Econ Entomol. 2007;100:1033–1036. doi: 10.1603/0022-0493(2007)100[1033:sspass]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Peterson MA, Dobler S, Larson EL, Juarez D, Schlarbaum T, Monsen KJ, et al. Profiles of cuticular hydrocarbons mediate male mate choice and sexual isolation between hybridising Chrysochus (Coleoptera: Chrysomelidae) Chemoecology. 2007;17:87–96. [Google Scholar]
- Raychoudhury R, Desjardins CA, Buellesbach J, Loehlin DW, Grillenberger BK, Beukeboom LW, et al. Behavioral and genetic characteristics of a new species of Nasonia. Heredity. 2010;104:278–288. doi: 10.1038/hdy.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schal C, Sevala VL, Young HP, Bachmann JAS. Sites of synthesis and transport pathways of insect hydrocarbons: cuticle and ovary as target tissues. Am Zool. 1998;38:382–393. [Google Scholar]
- Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S, Hermann N, Ruther J. Characterization of a female-produced courtship pheromone in the parasitoid Nasonia vitripennis. J Chem Ecol. 2006;32:1687–1702. doi: 10.1007/s10886-006-9102-3. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Tsaur SC, Coyne JA, Wu CI. The nucleotide changes governing cuticular hydrocarbon variation and their evolution in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:3920–3925. doi: 10.1073/pnas.061465098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:1577. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker-Thomas C, Henriet C, Dallerac R. Partial characterization of a fatty acid desaturase gene in Drosophila melanogaster. Insect Biochem Mol Biol. 1997;27:963–972. doi: 10.1016/s0965-1748(97)00077-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.