Abstract
Cytosolic 5′-nucleotidase II (NT5C2) is involved in the development of 1-β-d-arabinofuranosylcytosine (ara-C) resistance and has been associated with clinical outcome in patients receiving ara-C-based chemotherapy. NT5C2 inactivates ara-C by dephosphorylating ara-C monophosphate to ara-C. In this study, we sequenced NT5C2 in genomic DNA samples from International HapMap project panels with European [Centre d'Etude du Polymorphisme Humain (CEU); n = 90] or African [Yoruba people in Ibadan, Nigeria (YRI); n = 90] ancestry. We identified 41 genetic variants [one insertion-deletion and 40 single nucleotide polymorphisms (SNPs)], including three nonsynonymous SNPs (Y3A, K47R, and Q136R). Twenty-five SNPs were novel and 16 overlapped with the HapMap data. Subjects with African ancestry had NT5C2 mRNA expression levels that was significantly higher than those with European ancestry (p = 0.005). Furthermore, there was a correlation between NT5C2 mRNA expression and ara-C sensitivity in CEU but not in YRI cell lines. None of the nonsynonymous SNPs demonstrated any effect on NT5C2 activity. The genotypes of several SNPs were significantly associated with NT5C2 mRNA expression and/or ara-C sensitivity in CEU cell lines, but very few were significant in YRI cell lines. Of most interest, SNPs (linkage disequilibrium group CEU.12) in the 5′-untranslated region were associated with NT5C2 expression and ara-C sensitivity in HapMap cell lines and with NT5C2 mRNA expression and ara-C sensitivity in diagnostic leukemic blasts from pediatric patients with acute myeloid leukemia. Functional genomics analysis demonstrated that the promoter SNP rs11191612 was associated with altered luciferase activation in reporter assays and altered DNA-protein binding in gel shift assays. These results suggest that genetic variations in NT5C2 influence its expression and, potentially, cellular responses to nucleoside analogs.
Introduction
Cytarabine (1-β-arabinofuranosylcytosine, ara-C), a deoxycytidine nucleoside analog, is one of the most effective chemotherapeutic agents used in the treatment of acute myeloid leukemia (AML) (Wang et al., 1970). Ara-C is a prodrug that requires activation through intracellular phosphorylation to ara-C triphosphate (ara-CTP). Incorporation of ara-CTP in place of dCTP results in chain termination, thereby blocking DNA and RNA synthesis and causing leukemic cell death, which in turn is associated with the therapeutic response to ara-C (Kufe et al., 1980; Major et al., 1981; Raza et al., 1992; Galmarini et al., 2001b). Thus, the intracellular concentration of ara-CTP is one of the determinants of the clinical efficacy of ara-C chemotherapy. Deoxycytidine kinase is the key enzyme catalyzing the first phosphorylation step, resulting in the formation of ara-CMP. However, cytoplasmic 5′-nucleotidases catalyze dephosphorylation of ara-CMP, thereby reducing the amount of ara-C for conversion to ara-CTP (Amici and Magni, 2002).
Mammalian 5′-nucleotidases are a family of seven known enzymes (five cytosolic, one mitochondrial, and one extracellular) that catalyze the dephosphorylation of ribo- and deoxyribonucleoside monophosphates to their respective nucleoside and inorganic phosphates (Bianchi et al., 1986). Intracellular nucleotidases such as cytosolic 5′-nucleotidase II (NT5C2; also known as cN-II and high Km 5′-nucleotidase) and cytoplasmic 5′-nucleotidase III (NT5C3) are involved in the final step of dephosphorylation before export of nucleosides out of the cell. Although NT5C2 has a preference for 6-hydroxypurine nucleotide monophosphates (such as IMP, dIMP, GMP, dGMP, and XMP) as substrates, NT5C3 only hydrolyzes pyrimidine monophosphates (Pesi et al., 1994; Amici and Magni, 2002).
Multiple reports have suggested involvement of cytosolic nucleotidases in drug resistance to ara-C and/or gemcitabine and their influence on clinical outcome. It has been shown that NT5C2 expression levels correlate with in vitro sensitivity of primary leukemic cells to ara-C (Galmarini et al., 2005). Development of resistance to nucleoside analogs in cancer cell lines has been associated with increased NT5C2 expression (Schirmer et al., 1998; Dumontet et al., 1999). In adult patients with AML, high mRNA levels of NT5C2 as well as a higher ratio of NT5C2/deoxycytidine kinase mRNA expression along with unfavorable karyotype have been associated with shorter overall survival (Galmarini et al., 2001a; Galmarini, 2003). High NT5C2 expression has also been associated with disease-free survival, overall survival, and resistance to ara-C (Galmarini et al., 2005). Suzuki et al. (2007) reported that high NT5C2 mRNA levels in patients with high-risk myelodysplastic syndrome treated with ara-C-containing chemotherapy were associated with shorter median or overall survival.
The interpatient variability in the expression/activity of NT5C2 due to genetic variation could thus contribute to differences in treatment response. We identified genetic variations in NT5C2 by resequencing it in the genomic DNA from HapMap European and African ancestry panels and determined the association of SNPs with NT5C2 mRNA expression levels and ara-C sensitivity in HapMap cell lines. Then, we functionally characterized selected SNPs in vitro and determined the association between potentially significant NT5C2 variants with clinical measures in pediatric patients with AML undergoing treatment including ara-C.
Materials and Methods
Reagents.
The Expand High Fidelity PCR system was obtained from Roche (Indianapolis, IN); the TOPA TA cloning kit, pcDNA3.1 Directional TOPO Expression Kit, and the pET directional cloning kit were obtained from Invitrogen (Carlsbad, CA). Restriction enzymes and JM109 cells were obtained from Promega (Madison, WI). The RNeasy Mini Kit and Plasmid Plus kit were procured from QIAGEN (Valencia, CA). Anti-human antibody was obtained from Abcam Inc. (Cambridge, MA). All other reagents used were of molecular biology grade.
HapMap Cell Lines.
We used Epstein-Barr virus-transformed B-lymphoblastoid HapMap cell lines derived from 30 Centre d'Etude du Polymorphisme Humain (CEU) trios (two parents and a child) (n = 90, European descent) and 30 Yoruba trios (n = 90, African descent, referred to as YRI) to identify genetic variants in NT5C2. The purpose of using the same cell lines that have been used in the International HapMap project was to allow us to use the genotype data generated as part of the HapMap project. In addition, the genome-wide gene expression data using an Affymetrix Exon array was used to extract the expression levels of NT5C2 (GSE7761).
Cells were grown in an RPMI 1640 medium supplemented with 2 mM l-glutamine (Lonza Walkersville, Inc., Walkersville, MD), and 15% heat-inactivated serum at 37°C under 5% CO2. DNA, RNA, and cytoplasmic fractions were extracted from the cell lines using standard protocols. Genomic DNA was used to discover novel genetic variants in the NT5C2 gene. Ara-C sensitivity was determined as described earlier (Hartford et al., 2009). In brief, percentage cell survival values were determined using alamarBlue (BioSource International, Camarillo, CA) after a 72-h exposure to 1, 5, 40, and 80 μM ara-C, and survival curves were generated. The area under the survival curve (AUC) was calculated using the trapezoidal rule, and AUC values were log2-transformed before statistical analysis.
Identification of Sequence Variations in the NT5C2 Gene.
All the 18 coding exons, the flanking intronic sequences, and 1.5 kilobases of the 5′-UTR of the NT5C2 gene were PCR-amplified using primers and conditions listed in Supplemental Table 1. The primers were designed using the PrimerSelect module of Lasergene v6.0 software (DNASTAR, Inc., Madison, WI) and synthesized at University of Minnesota, Biomedical Genomics Center (BMGC). The primer sequences were verified using UCSC BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat) to eradicate the possibility of amplification of any nonspecific DNA sequences. Amplification was performed in a 1× PCR buffer using 10 ng of genomic DNA, 10 pmol each of forward and reverse primers, 0.2 mM dNTPs, and 1.5 units of Taq polymerase (Expand High Fidelity PCR system; Roche). Before sequencing, unincorporated nucleotides and primers were removed by incubation with shrimp alkaline phosphatase and exonuclease I (USB, Cleveland, OH) for 30 min at 37°C, followed by inactivation at 80°C for 15 min. DNA Sequencing was performed with an ABI Prism 3700 automated sequencer (Applied Biosystems, Foster City, CA) at BMGC using the PCR primers or internal primers (sequence available on request). Sequences were assembled using the Phred-Phrap-Consed package (University of Washington, Seattle, WA; http://droog.mbt.washington.edu/PolyPhred.html), which automatically detects the presence of heterozygous single nucleotide substitutions by fluorescence-based sequencing of PCR products (Nickerson et al., 1997) and SeqMan, the Multiple Sequence Alignment module of Lasergene version 6.0 software (DNASTAR, Inc.).
Bioinformatics Analysis.
SNPs identified by resequencing as well as from the HapMap database were analyzed using the following bioinformatic tools:
Splicing, SNPs at exon/intron junctions were screened using splice site prediction programs such as the Berkeley Drosophila Genome Project (http://www.fruitfly.org/seq_tools/splice.html) and the Web-based software Exonic splicing enhancer finder (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process_home), which screens for the potential splice sites and binding affinities for the four main serine/arginine (SR)-rich proteins, namely, SF2/ASF, SC35, SRp40, and SRp55.
Transcription factor-binding sites. Promoter SNPs were evaluated for the loss/gain of known cis-regulatory motif binding sites using TRANSFAC Professional (phttps://portal.biobase-international.com/cgi-bin/portal/login.cgi).
Prediction of consequences on protein structure and/or function. Nonsynonymous single nucleotide polymorphisms were evaluated using prediction programs such as SIFT (Sorting Intolerant From Tolerant, http://blocks.fhcrc.org/sift/SIFT.html) (Ng and Henikoff, 2001, 2003) and PolyPhen (Polymorphism Phenotyping, http://genetics.bwh.harvard.edu/pph/) (Sunyaev et al., 2000, 2001; Ramensky et al., 2002).
Luciferase Reporter Assays.
To study the functional effects of promoter SNPs on NT5C2 transcription, luciferase reporter gene constructs were created for wild-type (WT) and mutant alleles of 2 NT5C2 SNPs (rs11191612 and rs10748839). HapMap genomic DNA samples with a homozygous genotype for these SNPs were amplified using forward and reverse primers designed to capture the SNP loci of interest (listed in Supplemental Table 2). The PCR product was cloned in pCR2.1-TOPO using the TOPO-TA cloning kit (Invitrogen) followed by restriction digestion and cloning into pGL3basic reporter vector (Promega).
Cos7 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Cells were plated at a density of 2 × 105 cells/well in a 24-well plate. Cells were transfected with luciferase reporter constructs (800 ng) and β-galactosidase expression vector (250 ng, pSV-G-Gal) that encoded the β-galactosidase protein using Lipofectamine 2000 (Invitrogen) at 1:2.5 ratio. Forty-eight hours later, the cells were harvested and lysed, followed by luciferase assay (Promega), β-galactosidase enzyme assay system (Promega), and Bradford (Invitrogen) protein estimation, according to the respective manufacturers' protocols. Results are reported as the ratio of firefly luciferase light units to β-galactosidase units or protein units, and values are also expressed as a percentage of the activity of the appropriate WT construct. All assays were performed in triplicate; i.e., three independent transfections were performed, and each experiment was repeated twice for a total of six independent determinations.
Electrophoretic Mobility Shift Assays.
EMSAs were performed for NT5C2 promoter SNPs rs11191612 and rs10748839 to determine their potential effect on transcription regulation. Because the NT5C2 transcript has been reported to be highly expressed in bone marrow and cervix, nuclear extracts from HL60 and HeLa cells were chosen for use in EMSA assays. In brief, oligonucleotides for rs11191612, A allele (′-GCTGTGCGGTaAGAATTCTTA-3′) and G allele (5′-GCTGTGCGGTgAGAATTCTTA-3, and for rs10748839, T allele (5′-CTCCTATACCtGGTGCCCACA-3′) and C allele (5′-CTCCTATACCcGGTGCCCACA-3′), were biotin-labeled using a nonradioactive LightShift Chemiluminescent Assay System (Thermo Fisher Scientific, Waltham, MA) per the manufacturer's instructions; each 20-μl reaction contained 20 fmol of 3′-end-labeled DNA target, 10 μg of nuclear extract, 1 μg of poly(dI-dC), and 1× binding buffer. A 200-fold molar excess of unlabeled specific DNA target was used to perform competition experiments. Reaction mixtures were incubated for 20 min at room temperature followed by electrophoresis on a 6% precast DNA retardation gel (Invitrogen). The binding reactions were then transferred electrophoretically to a Biodyne B modified nylon membrane (Pierce Biotechnology). The transferred DNA was cross-linked at 120 mJ/cm2 for 1 min. Then biotin-labeled DNA-protein interactions were detected using a nonradioactive LightShift Chemiluminescent Assay System and visualized after exposure to X-ray film.
NT5C2 Expression Constructs.
Full-length NT5C2 cDNA was amplified from one of the samples having a WT sequence and cloned into a pET101 expression vector using a champion pET Directional TOPO Expression Kit (Invitrogen), according to the manufacturer's instructions. Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) to create NT5C2 3-mt, 47-mt, and 136-mt expression constructs (numbers refer to amino acid positions). The empty pET101 vector, pET101-NT5C2 WT, pET101-NT5C2 3-mt, pET101-NT5C2 47-mt, and pET101-NT5C2 136-mt expression vectors were then transformed into the BL21 star Escherichia coli provided with the pET101 Directional TOPO Expression Kit, following the manufacturer's instructions. In brief, NT5C2 expression in BL21 cells was induced by 1 mM isopropyl-d-thiogalactoside, and cells were harvested after 5 h. The cell pellet was suspended in 50 mM Tris-HCl buffer, pH 7.4, containing 4 mM dithiothreitol, and the soluble fraction was prepared according to the manufacturer's instructions. The soluble fraction was checked for NT5C2 expression by Western blotting before proceeding to the activity assays.
Western Blotting.
BL21 cell cytosols were subjected to electrophoresis on 10% Tris-HCl acrylamide gel. Proteins were then transferred to nitrocellulose membranes and the membranes were incubated with mouse monoclonal anti-NT5C2 primary antibody (Abcam plc, Cambridge, UK) followed by secondary antibody. Immunoreactive proteins were detected using the ECL Western Blotting System (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) and visualized using a Bio-Rad ChemiDoc XRS+ system.
Diazyme 5′-Nucleotidase Enzymatic Assay.
5′-Nucleotidase activity of the recombinant WT and mutant NT5C2 protein expressed in BL21 was measured using a Diazyme 5′-Nucleotidase Enzymatic Test Kit. The assay is based on the principle that enzymatic hydrolysis of 5′-monophosphate forms inosine, which is converted to hypoxanthine by purine nucleoside phosphorylase, which in turn is converted to uric acid and hydrogen peroxide by xanthine oxidase. This hydrogen peroxide is then quantified by a Trinder reaction, providing average 5′-nucleotidase activity compared against a calibrator of known 5′-nucleotidase activity.
NT5C2 Methylation.
The CpG region sequence of NT5C2 was obtained from the UCSC Golden Path Genome Browser Database. Methyl Primer Express (Applied Biosystems) was used to predict promoter CpG islands and to design primers for methylation-specific PCR (MSP).
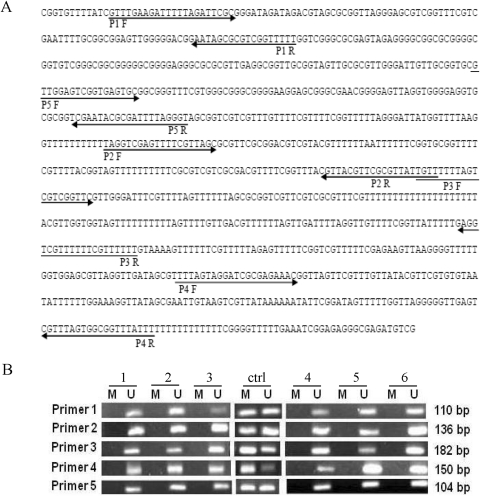
Five different MSP primer sets (P1–P5 in Fig. 6) covering five predicted CpG-rich sites were designed. For every site, a methylated and an unmethylated primer set was synthesized at BMGC. All primers were between 15 and 25 base pairs in length. We used genomic DNA from 10 sensitive and 10 resistant unrelated HapMap cell lines from CEU and YRI panels for MSP. Sodium bisulfite modification of DNA was performed using the EZ DNA Methylation Kit (ZYMO Research, Irvine, CA) according to the manufacturer's protocol with minor modification.
Fig. 6.
A, NT5C2 promoter with CpG sites. Bisulfite-modified genomic DNA sequence of the CpG region of the human NT5C2 gene. The position of the primers that were used for methylation-specific PCR after bisulfite treatment are indicated by forward and reverse arrows for primer 1 to primer 5 covering five different regions of the CpG island. The bisulfite treatment converted the unmethylated cytosine to uracil, which is complementary to adenosine. B, methylation analysis of five CpG sites in the NT5C2 promoter. Representative agarose gels showing methylation patterns in sensitive and resistant HapMap (CEU) cell lines for the NT5C2 primer1 to primer 5. M, band for methylated primers; U, band for unmethylated primers. The positive control (ctrl) methylated and unmethylated bands was used from EpiTect PCR Control DNA Set.
The MSP reaction was performed in a 25-μl reaction mixture containing 2.5 μl of HotStar Taq10× buffer, 0.5 μl of 10 mM dNTP mix, 0.2 μl of HotStar polymerase (QIAGEN), a 0.5 μM concentration of each primer, and 20 to 25 ng of bisulfite-modified genomic DNA. PCR amplification consisted of an initial activation step at 95°C, followed by 34 cycles consisting of denaturation at 94°C for 15 s, annealing at 54 to 57°C (different for different primers) for 30 s, extension at 72°C for 30 s, and final extension at 72°C at 10 min. The MSP products were separated by electrophoresis using 2% agarose gel. The extent of methylation can only be judged qualitatively by this method. Methylation was considered negative (−) when no band was present, and positive (+) when a band was present. We also performed LINE1 methylation analysis on the same samples using pyrosequencing for global methylation.
Samples from Patients with AML.
Primary bone marrow samples were obtained from children with newly diagnosed AML enrolled on St. Jude AML97 (Rubnitz et al., 2009) or St. Jude AML02 (Rubnitz et al., 2010; http://clinicaltrial.gov, NCT00136084) studies, after informed consent was obtained from them or from their parents/guardians, with assent from the patients, as appropriate. Genomic DNA was extracted, and samples were evaluated for NT5C2 SNPs. This study and use of these samples were approved by the institutional review board at St. Jude.
Cohort I: St. Jude AML97.
The eligibility for the enrollment, treatment plan, and clinical outcome of this clinical trial have been published elsewhere (Rubnitz et al., 2009). In brief, patients were randomly assigned to receive either a daily short infusion or a continuous infusion of ara-C. Bone marrow aspirates were obtained at diagnosis in all patients and after day 1 and 2 of ara-C treatment as described previously (Crews et al., 2002). Leukemic cells were separated by Ficoll-Hypaque density-gradient centrifugation and intracellular ara-CTP levels were determined in samples obtained after day 1 and day 2 of ara-C treatment using high-performance liquid chromatography as described earlier (Crews et al., 2002). The patient population included 58% white, 22% black, and 20% with other ethnic backgrounds. Event-free survival (EFS) and overall survival (OS) were estimated as described previously (Rubnitz et al., 2009).
Cohort II: St. Jude AML02.
Details on eligibility of enrollment, study design, and clinical outcome for the AML02 protocol are described elsewhere (Rubnitz et al., 2010). In brief, patients were randomly assigned to receive induction therapy containing either high-dose cytarabine (3 g/m2, given every 12 h on days 1, 3, and 5) or low-dose cytarabine (100 mg/m2) given every 12 h on days 1 to 10 plus daunorubicin and etoposide as described earlier (Rubnitz et al., 2010). The patient population included 69.6% white, 18.7% black, and 11.7% with other ethnic backgrounds. Minimal residual disease (MRD), EFS, and OS were estimated as described earlier (Rubnitz et al., 2010).
In Vitro Sensitivity (MTT) Assay of Primary Leukemic Blast Samples.
In vitro sensitivity of leukemic cells to cytarabine obtained at diagnosis was determined in patients enrolled on the AML02 protocol using the 4-day MTT cytotoxicity assay as described previously (Holleman et al., 2004; Lamba et al., 2011). If necessary, samples were enriched to achieve more than 80% leukemic blasts by the use of magnetic cell sorting (Miltenyi Biotech, Bergisch Gladbach, Germany). The leukemic cells were exposed to six different concentrations of cytarabine (0.002–2.5 ng/μl) or to culture media (untreated) in a 96-well plate format. After a 96-h incubation, MTT was added, and cell viability was measured 6 h later. The LC50 was estimated using nonlinear curve fitting. It was demonstrated previously that 96 h of drug incubation is necessary to achieve a cytotoxic effect in primary leukemic cells.
A cytospin slide for untreated cells was prepared on days 0 and 4. The percentage of leukemic blasts in each sample was determined by Giemsa staining. Samples with 90% or more leukemic cells on day 0, with 70% or more leukemic cells in the untreated wells after 4 days of culture, and with an optical density greater than 0.05 arbitrary unit were considered suitable for evaluation. For samples in which 50% cytotoxicity was not achieved even at the highest tested concentration or ara-C (i.e., complete resistance), the LC50 value was assigned as twice the highest concentration tested (Beesley et al., 2006). For cells with viabilities less than 50% at all tested concentrations (i.e., high sensitivity), the LC50 value was assigned as half of the lowest concentration tested.
NT5C2 mRNA Expression in Clinical AML Blast Samples.
NT5C2 mRNA expression levels in diagnostic leukemic blasts were extracted from Affymetrix U133A array data that were available from 137 patients (AML97, n = 41; AML02, n = 96). NT5C2 SNPs (n = 16) that represented major LD groups and that were potentially significant were genotyped in genomic DNA from AML97 (n = 55) and AML02 (n = 232) patients using the Sequenom platform that uses matrix-assisted laser desorption ionization/time of flight-based chemistry. The SNPs were selected on the basis of analysis of HapMap data and criteria that prioritized TagSNPs, coding, and regulatory SNPs.
Statistical Analysis.
The association of SNP genotype with NT5C2 expression and ara-C sensitivity measured as AUC in HapMap cell lines was explored with an analysis of variance model that uses a Toeplitz correlation matrix (Wolfinger, 1996) to account for correlation of a child's measurements to those of his or her parents. The Kruskal-Wallis test was used to compare expression levels and ara-C LC50 values across groups of patients with AML classified by SNP genotype, clinical risk group, or race. Spearman correlation coefficients were used to characterize the association of the number of minor alleles with MRD, NT5C2 expression, and LC50 values. EFS was defined as the time elapsed from protocol enrollment to induction failure, relapse, secondary malignancy, or failure with patients free of these events censored at the last follow-up. OS was defined as the time elapsed from protocol enrollment to death, with living patients censored at the last follow-up. The statistic of Jung Jung et al. (2005) was used to characterize the association of the number of minor alleles with EFS and OS. Cox regression models were used to explore associations of genotype with EFS and OS while accounting for previously identified prognostically important variables. Pleiotropic effects were identified using projection onto the most interesting statistical evidence (PROMISE; Pounds et al., 2009, 2011). All tests were two-sided, and no multiple-testing adjustments were performed in these exploratory analyses.
Results
Identification of Genetic Variants in NT5C2.
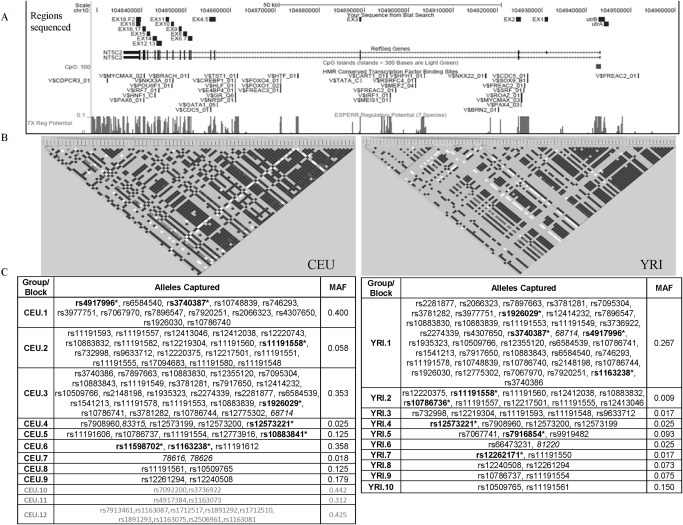
All the coding exons and the proximal promoter were sequenced in CEU (30 trios with European ancestry, n = 90) and YRI (30 trios with African ancestry, n = 90) samples. Figure 1A shows a snapshot from the UCSC genome browser (http://genome.ucsc.edu/index.html?org=Human) of regions resequenced in NT5C2 along with key regions of functional importance (CpG site, 7× regulatory potential, and conserved transcription factor binding sites). A total of 41 genetic variants were identified including 40 SNPs and 1 indel (insertion-deletion) by resequencing. Of these 41 SNPs, 2 were present in the promoter, 5 in exons, 30 in the intron, and 4 in the 3′-UTR (Table 1). Seventeen SNPs were found exclusively in the African ancestry group and 12 exclusively in the European ancestry group; 10 SNPs were common to both groups. Sixteen SNPs overlapped with SNPs present in the HapMap database. Of the exonic SNPs, 3 were nonsynonymous, resulting in Y3A, K47R, and Q136R changes.
Fig. 1.
A, snapshot from the UCSC genome browser for NT5C2 locus. The regions sequenced in HapMap panels are shown by small boxes in the first panel followed by (B) LD plot of NT5C2 in samples with European (CEU) and African (YRI) ancestry. LD plots were generated in Haploview using genotype data from the present study and from HapMap for both CEU and YRI samples. The color scheme is as follows: white when r2 = 0, shades of light gray when 0 < r2 < 1, and black when r2 = 1. C, LD groups within CEU and YRI. SNPs that are linked, r2 > 0.8 (and picked by the tagger program), are categorized in the same groups. SNPs without rs numbers are indicated by number corresponding to Table 1. SNPs genotyped in patients with AML are bold with an asterisk. LD groups CEU 10 to 12 (gray) represent SNPs present upstream of NT5C2 gene. MAF, minimum allele frequency for the group.
TABLE 1.
Genetic variants identified by sequencing NT5C2 in the present study
| Position from Translation Start Site (ATG) as +1 | rs No. | SNP/Indel | Chromosomal Position | Location in the Gene | Amino Acid Change | Bioinformatic Analyses | Allele Frequency |
|
|---|---|---|---|---|---|---|---|---|
| CEU | YRI | |||||||
| −20080 | rs7095304 | G/A | 104944785 | 5′-UTR | Loss of V$E2F_Q4 site | 0.33 | 0.27 | |
| −18832 | rs10748839 | T/C | 104943537 | 5′-UTR | Gain in V$VDR_Q3 site | 0.39 | 0.30 | |
| −237 | rs12261294 | C/T | 104924942 | Intron 1 | 0.00 | 0.07 | ||
| −123 | G/A | 104924828 | Intron 1 | 0.00 | 0.05 | |||
| −69 | C/T | 104924774 | Intron 1 | Loss of BranchSite, loss of SR protein-binding site on the protein surface | 0.00 | 0.01 | ||
| 7 | rs10883841 | T/C | 104924699 | Exon 2 | T3A | 0.12 | 0.00 | |
| 35377 | C/T | 104889329 | Intron 2 | 0.00 | 0.01 | |||
| 35519 | rs72846108 | G/C | 104889187 | Exon 3 | K47N | Possibly damaging (Polyphen), affect protein function (SIFT) | 0.01 | 0.00 |
| 68714 | G/A | 104855992 | Intron 4 | 0.37 | 0.26 | |||
| 73650 | rs12262171 | T/C | 104851056 | Exon 6 | Q136R | Hydrogen bond lost (SNP3D) | 0.00 | 0.01 |
| 73783 | rs41287482 | A/G | 104850923 | Intron 6 | Gain in SR protein-binding site | 0.02 | 0.00 | |
| 74663 | rs2274339 | A/T | 104850043 | Intron 7 | 0.36 | 0.27 | ||
| 75886 | rs73353837 | C/T | 104848820 | Intron 8 | 0.00 | 0.06 | ||
| 77498 | rs10786737 | A/C | 104847208 | Intron 10 | Gain in SR protein-binding site | 0.17 | 0.07 | |
| 78554 | rs12412038 | G/A | 104846152 | Intron 10 | 0.06 | 0.00 | ||
| 78616 | 123193C | T/G | 104846090 | Intron 10 | 0.02 | 0.00 | ||
| 78626 | 123203 | T/G | 104846080 | Intron 10 | 0.01 | 0.00 | ||
| 78639 | T/C | 104846067 | Intron 10 | 0.00 | 0.01 | |||
| 79046 | rs1926029 | C/T | 104845660 | Intron 11 | Loss of SR protein-binding site | 0.34 | 0.27 | |
| 79060 | rs1926030 | A/G | 104845646 | Intron 11 | 0.40 | 0.31 | ||
| 81127 | A/C | 104843579 | Intron 14 | 0.00 | 0.04 | |||
| 81177 | C/T | 104843529 | Intron 13 | 0.00 | 0.01 | |||
| 81213 | rs2274341 | T/A | 104843493 | Intron 13 | 0.00 | 0.08 | ||
| 81220 | A/G | 104843486 | Intron 13 | 0.00 | 0.03 | |||
| 81426 | rs72843997 | T/C | 104843280 | Intron 13 | 0.00 | 0.01 | ||
| 82050 | rs3837340 | 104842656 | Intron 14 | |||||
| 82054 | rs66473231 | G/A | 104842652 | Intron 14 | 0.00 | 0.02 | ||
| 83315 | C/T | 104841391 | Intron 14 | 0.03 | 0.00 | |||
| 83320 | rs11191553 | G/T | 104841386 | Intron 14 | Gain in branch site | 0.32 | 0.27 | |
| 83415 | rs17094683 | G/T | 104841291 | Intron 15 | 0.05 | 0.02 | ||
| 83431 | rs10883830 | C/T | 104841275 | Intron 15 | Loss of SR protein-binding site | 0.35 | 0.26 | |
| 83881 | rs11191551 | T/G | 104840825 | Intron 15 | Loss of branch site, gain in SR protein-binding site | 0.05 | 0.00 | |
| 83893 | T/C | 104840813 | Intron 15 | 0.00 | 0.01 | |||
| 84084 | rs3736922 | C/T | 104840622 | Intron 16 | Gain in 5SS_U2_Human site, change in score | 0.44 | 0.29 | |
| 84096 | rs34758128 | T/G | 104840610 | Intron 16 | Loss of splice site, gain in SR protein-binding site | 0.13 | 0.00 | |
| 84219 | G/T | 104840487 | Exon 17 | E440E | 0.00 | 0.01 | ||
| 85248 | rs3740387 | C/T | 104839458 | Exon 18 | D549D | 0.40 | 0.30 | |
| 85547 | A/G | 104839159 | 3′-UTR | 0.00 | 0.01 | |||
| 85572 | rs12573221 | A/C | 104839134 | 3′-UTR | 0.03 | 0.03 | ||
| 85584 | C/G | 104839122 | 3′-UTR | 0.00 | 0.01 | |||
| 85600 | rs10786736 | G/C | 104839106 | 3′-UTR | 0.09 | 0.00 | ||
Indel, insertion-deletion.
Analysis of linkage disequilibrium on SNPs identified in the current study and from the HapMap database (www.HapMap.org) was performed using Haploview. On the basis of the LD plot and results from tagger (Haploview), we were able to group the strongly linked SNPs (r2 > 0.8) in CEU and YRI into 9 and 10 distinct groups, respectively (Fig. 1, B and C). Eight CEU SNPs and 19 YRI SNPs did not occur in linkage with other SNPs.
Association of NT5C2 SNPs with Its mRNA Expression and Ara-C Sensitivity in HapMap Cell Lines.
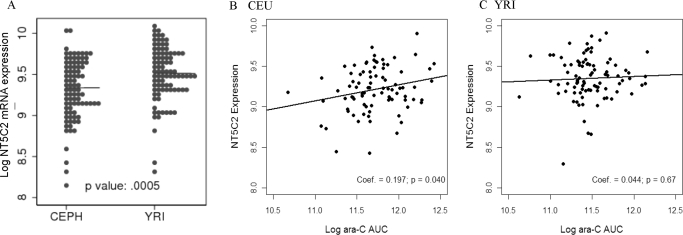
NT5C2 mRNA expression in 87 CEU (representing 29 trios) and 90 YRI (30 trios with African ancestry) samples was obtained from the publicly available database Gene expression omnibus (GSE7761). NT5C2 mRNA expression demonstrated significant interindividual variability (CEU: 8.34–9.90; YRI: 8.29–9.91) and significant ethnic differences with African ancestry having significantly higher NT5C2 mRNA compared with European ancestry (9.35 ± 0.03 versus 9.21 ± 0.031, 0.002) (Fig. 2). NT5C2 expression was positively correlated with ara-C AUC in CEU (coefficient estimate, r = 0.197 p = 0.04) but not in YRI (coefficient estimate, r = 0.044, p = 0.67) panels (Fig. 2, B and C).
Fig. 2.
A, NT5C2 mRNA expression (extracted from GSE7761) in Epstein-Barr virus-transformed lymphoblast cell lines derived from subjects with European (CEU) and African (YRI) ancestry. Median values for log2NT5C2 mRNA levels are indicated by a horizontal line for each ethnic group. B and C, correlation of NT5C2 expression with ara-C cytotoxicity in CEU and YRI cell lines. Ara-C cytotoxicity was determined as described under Materials and Methods. *, p < 0.05; **, p < 0.01.
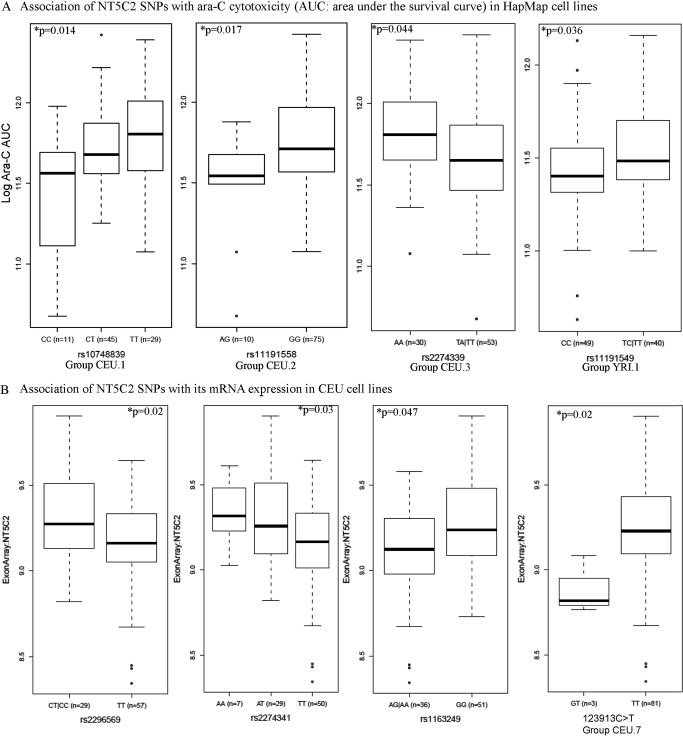
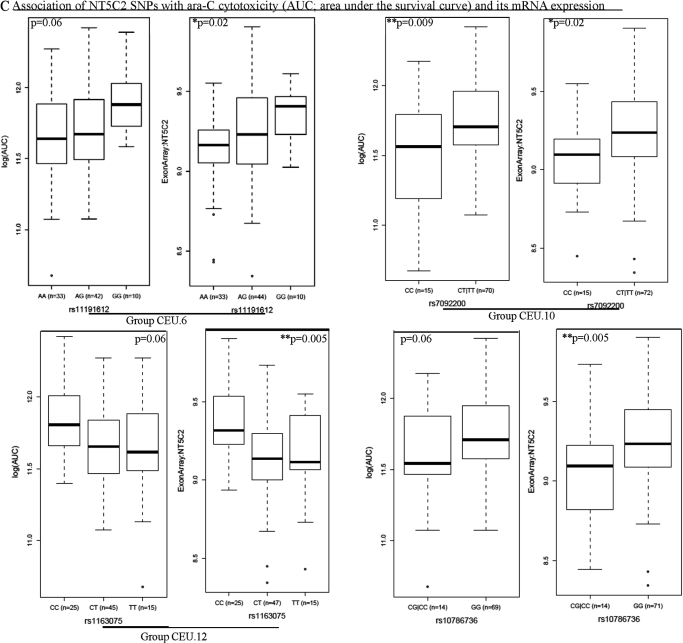
We analyzed the association of NT5C2 SNPs (identified by sequencing as well as from the HapMap database) with its mRNA expression and cellular sensitivity to ara-C in HapMap cell lines (Table 2). Within CEU samples, SNPs represented by groups CEU.1, CEU.2, and CEU.3 (p < 0.05) were significantly associated with ara-C cellular sensitivity measured as AUC (see Materials and Methods) in growth inhibition assays. Table 2 and Fig. 3A demonstrate the association of the representative SNP within each group (rs10748839 for CEU.1, rs11191558 for CEU.2, and rs2274339 for CEU.3). In all these cases, the minor allele was associated with increased sensitivity to ara-C (as depicted by lower AUC). Two SNPs within CEU.1 (rs10748839 and rs4307650) were in the promoter and two (rs943035 and rs943036) were present in the 3′ end of the NT5C2 gene. The rest of the SNPs within CEU.1 were intronic. SNP rs11191548 within group CEU.2 was present in the 3′-UTR of NT5C2.
TABLE 2.
Association of NT5C2 SNPs with its mRNA expression and ara-C cytotoxicity in lymphoblast cell lines from subjects with European (CEU) and African (YRI) ancestry
| Genotype | Phenotype I: Ara-C AUC |
Phenotype II: (Log) NT5C2 mRNA Expression |
|||
|---|---|---|---|---|---|
| Mean ± S.D. | p | Mean± S.D. | P | ||
| European Ancestry: CEU | |||||
| Group CEU0.1 represented by 10748839 C>T | CC (n = 11) | 2895.234 ± 759.255 | CC vs. CT vs. TT, *p = 0.014 | ||
| CT (n = 45) | 3450.095 ± 643.267 | CC vs. CT + TT, *p = 0.005 | |||
| TT (n = 29) | 3659.039 ± 868.523 | ||||
| Group CEU0.2 represented by 11191558 G>A | AG (n = 10) | 2911.575 ± 606.627 | AG vs. GG, p = 0.017 | ||
| GG (n = 75) | 3521.309 ± 763.643 | ||||
| Group CEU0.3 represented by 2274339 A>T | AA (n = 30) | 3704.852 ± 812.523 | AT + TT vs. AA, p = 0.043 | ||
| GT (n = 47) | 3331.494 ± 724.121 | ||||
| TT (n = 6) | 3137.643 ± 788.639 | ||||
| Group CEU0.6 represented by 11598702 C>T | CC (n = 10 | 3980.334 ± 798.856 | CC vs. CT vs. TT, p = 0.05 | 9.371 ± 0.177 | CC vs. CT vs. TT, *p = 0.015 |
| CT (n = 42) | 3458.257 ± 712.271 | CC vs. CT + TT, *p = 0.035 | 9.224 ± 0.299 | CC + CT vs. TT, *p = 0.011 | |
| TT (n = 33) | 3277.691 ± 778.746 | 9.131 ± 0.284 | |||
| Group CEU0.6 represented by rs11191612 A>G | GG (n = 10) | 3980.334 ± 798.856 | AA + AG vs. GG, *p = 0.035 | 9.371 ± 0.177 | AA vs. AG vs. GG, p = 0.02 |
| AG (n = 42) | 3440.149 ± 718.944 | 9.225 ± 0.300 | AA + AG vs. GG, *p = 0.046 | ||
| AA (n = 33) | 3300.738 ± 775.766 | 9.129 ± 0.282 | AA vs. AG + GG, *p = 0.017 | ||
| Group CEU0.7 represented by 78616 T>G | GT (n = 3) | 8.889 ± 0.171 | GT vs. TT, p = 0.026 | ||
| TT (n = 81) | 9.225 ± 0.285 | ||||
| Group CEU0.10 rs7092200 C>T | CC (n = 15) | 3033.064 ± 820.026 | CC vs. CT vs. TT, *p = 0.032 | 9.068 ± 0.286 | CC vs. CT + TT, *p = 0.019 |
| CT (n = 46) | 3492.019 ± 656.968 | CC vs. CT + TT, *p = 0.009 | 9.252 ± 0.292 | ||
| TT (n = 26) | 3628.546 ± 871.160 | 9.203 ± 0.270 | |||
| Group CEU0.11 rs4917384 C>T in LD with rs163703 | CC (n = 37) | 3579.244 ± 783.663 | CC vs. CT vs. TT, *p = 0.041 | 9.310 ± 0.231 | CC vs. CT vs. TT, *p = 0.009 |
| CT (n = 43) | 3306.684 ± 658.351 | 9.127 ± 0.302 | CC vs. CT + TT, *p = 0.002 | ||
| TT (n = 4) | 4027.323 ± 962.587 | 9.195 ± 0.217 | |||
| Group CEU0.12 | |||||
| rs1163087 G>A in LD with 8 other SNPs | AA (n = 15) | 3319.396 ± 890.147 | AA vs. AG vs. GG, p = 0.059 | 9.149 ± 0.315 | AA vs. AG vs. GG, *p = 0.003 |
| AG (n = 46) | 3314.117 ± 679.138 | AA + AG vs. GG, p = 0.017 | 9.135 ± 0.285 | AA vs. AG vs. GG, *p = 0.0006 | |
| GG (n = 25) | 3759.956 ± 799.118 | 9.359 ± 0.224 | |||
| rs2296569 T>C | CC (n = 4) | 9.334 ± 0.241 | CC + CT vs. TT, p = 0.02 | ||
| CT (n = 25) | 9.298 ± 0.269 | ||||
| TT (n = 57) | 9.151 ± 0.291 | ||||
| rs2274341 T>A | AA (n = 7) | 9.339 ± 0.208 | AA vs. AT vs. TT *p = 0.031 | ||
| AT (n = 29) | 9.284 ± 0.276 | AA + AT vs TT, p = 0.012 | |||
| TT (n = 50) | 9.140 ± 0.296 | ||||
| rs1163249 G>A | AA (n = 3) | 9.088 ± 0.032 | AA + AG vs. GG, p = 0.047 | ||
| AG (n = 33) | 9.119 ± 0.315 | ||||
| GG (n = 51) | 9.269 ± 0.265 | ||||
| rs10786736 G>C | CC (n = 1) | 4622.78 | CC vs. CG vs. GG, *p = 0.023 | 9.094 | CC + CG vs. GG, *p = 0.019 |
| CG (n = 13) | 2992.518 ± 664.375 | 9.042 ± 0.332 | CC vs. CG vs. GG, *p = 0.005 | ||
| GG (n = 71) | 3449.576 ± 769.551 | 9.205 ± 0.289 | |||
| African ancestry: YRI | |||||
| Group YRI0.1 represented by rs11191549 | CC (n = 49) | 2787.952 ± 525.570 | CC vs. CT + TT, *p = 0.036 | ||
| CT (n = 36) | 3020.564 ± 642.470 | ||||
| TT (n = 4) | 3026.675 ± 429.907 | ||||
p < 0.05.
Fig. 3.
A, NT5C2 SNPs associated with ara-C cytotoxicity in HapMap cell lines. Box plots for the association of representative NT5C2 SNPs (rs10748839 for CEU.1, rs11191558 for CEU.2, rs2274339 for CEU.3, and rs11191549 for YRI.1) with ara-C AUC in CEU and YRI samples are shown. Plots show medians as a line between boxes, which represent first and the third quartiles; the whiskers represent the range after outliers were excluded. The outliers are defined as data points that fall outside of the first and third quartiles by more than 1.5 times the interquartile range. Circles falling outside the whiskers represent outliers. B, NT5C2 SNPs associated with NT5C2 mRNA expression levels in HapMap cell lines. Box plots for the association of NT5C2 SNPs rs2296569, rs2274341, rs1163249, and 123193 (representing the CEU.7 group) with its mRNA expression in CEU cell lines are shown. C, NT5C2 SNPs associated with its mRNA expression levels and ara-C cytotoxicity in HapMap cell lines. Box plots for the association of NT5C2 SNPs rs11191612 (representing CEU.6), rs7092200 (representing CEU.10), rs1163075 (representing CEU.12), and singleton SNP rs10786736 with its mRNA expression and ara-C AUC in CEU cell lines are shown.
In the 87 CEU samples, NT5C2 mRNA expression was significantly associated with SNPs in group CEU.7 (two new SNPs, 78616 and 78626, identified by resequencing) as well as with SNPs rs2296569, rs2274341, rs1163849, and rs10786736 (p < 0.05) (Table 2; Fig. 3B).
SNPs within groups CEU.6 (includes rs1598702, rs11191612, and rs1163238), CEU.10 (includes rs7092200 and rs3736922), CEU.11 (includes rs4917384 and rs1163073), and CEU.12 (represented by rs1163075 but includes 8 SNPs that occur in LD) were significantly associated with both NT5C2 mRNA expression and sensitivity to ara-C (Table 2; Fig. 3C). Both CEU.11 and CEU.12 groups represent SNPs present in the 5′-UTR of NT5C2. Subjects homozygous for minor allele GG for rs11191612 (a promoter SNP within group CEU.6) had higher NT5C2 expression (p < 0.05) as well as greater resistance to ara-C measured as area under the cell survival curve (p < 0.05) in the cellular growth inhibition assays (Fig. 3C). For rs7092200, a 3′-UTR SNP in group CEU.10 subjects with at least one T allele had higher ara-C AUC (p = 0.009) and greater NT5C2 expression (p = 0.019). Likewise, for both rs1163087 (group CEU.10) and rs10786736 SNPs, the presence of the G allele was also associated with higher NT5C2 expression as well as cellular resistance to ara-C (p < 0.05) (Fig. 3C; Table 2).
Among YRI samples, rs11191549, a 3′-UTR SNP within group YRI.1 was associated with ara-C sensitivity (p = 0.036) (Fig. 3A).
Multiple SNPs demonstrated potential for alternate splicing when analyzed by bioinformatic tools. However, we could not find any evidence of alternate splicing using cDNA samples from selected HapMap cell lines representing different alleles of potential SNPs (data not shown).
In Vitro Luciferase Reporter Assays.
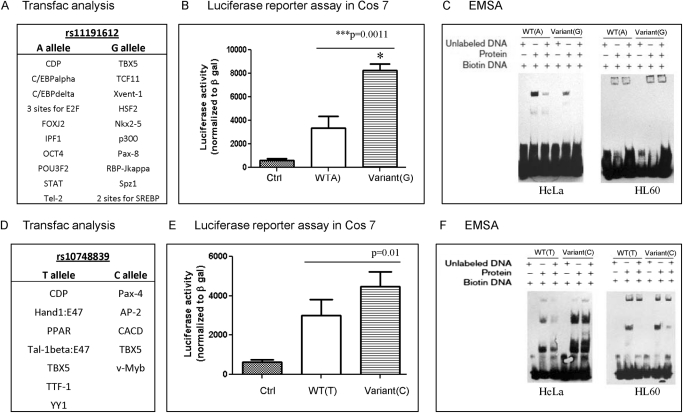
We used TRANSFAC and MatInspector to determine whether the NT5C2 promoter SNPs (rs11191612 and rs10748839) associated with ara-C sensitivity and/or mRNA levels within HapMap cell lines were disrupting or creating any transcription factor binding sites. Results from TRANSFAC (version 10) analysis demonstrating differences in the transcription binding sites for A and G allele of rs11191612 and T and C allele of rs10748839 are depicted in Fig. 4, A and D.
Fig. 4.
Functional characterization of NT5C2 promoter SNPs rs11191612 and rs10748839. A and D, differences in transcription factor binding sites for WT and variant alleles of rs11191612 and rs10748839 identified by Transfac analysis. B and E, luciferase reporter assays comparing transcriptional activation NT5C2-pGL3basic vectors with WT or variant alleles of rs11191612 and rs10748839. Transfection efficiency was normalized to β-galactosidase activity. C and F, representative EMSA gels comparing binding of oligos representing WT and variant alleles of promoter SNPs after incubation with HL60 and HeLa nuclear extracts.
NT5C2-pGL3 reporter plasmids representing rs11191612 and rs10748839 SNPs were transfected into Cos7 cells. The transactivation potentials of rs11191612 A and G alleles and rs10748839 T and C alleles were compared with respect to control pGL3 basic (without any NT5C2 promoter). As is shown in Fig. 4B, the NT5C2 reporter plasmid with variant (rs11191612 G allele) had a significantly increased (4-fold) transactivation potential compared with the WT (rs11191612 A allele) (p = 0.001). This observation complements the earlier observation of higher NT5C2 mRNA expression as well as greater AUC for the ara-C survival curve in subjects homozygous for the rs11191612 GG genotype compared with subjects homozygous for the A allele (Fig. 3C). For rs10748839 polymorphism, the transactivation potential of the variant (C) allele was greater than that for the WT (A) allele (p = 0.01) (Fig. 4E).
EMSAs.
EMSAs were performed to compare the binding efficiencies of the rs11191612 and rs10748839 of the NT5C2 variant allele using nuclear extracts from HL60 and HeLa cell lines as described under Materials and Methods. For the rs11191612 SNP, we observed differential protein binding for the WT and the variant allele. Overall, the variant allele demonstrated weaker binding than the WT allele for the HeLa cell line, but no shift was observed for HL60 (Fig. 4C). For the rs10748839 polymorphism, the variant (C) allele demonstrated stronger binding to factors in the nuclear extracts than the WT (T) allele for both HeLa and HL60 nuclear extracts (Fig. 4F). In addition, EMSA assays were performed for two promoter SNPs in LD block CEU.12 that were also present in a potentially regulatory region as predicted by 7× regulatory potential and DNA hypersensitivity sites (Supplemental Fig. 1A). As shown in Supplemental Fig. 1, B and C, the presence of a variant allele resulted in reduced protein binding upon incubation with HeLa and THP-1 nuclear extracts.
Analysis of NT5C2 Activity.
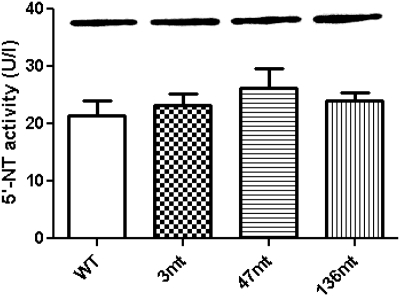
To determine the functional significance of three nonsynonymous coding variants, we expressed NT5C2 WT and variant cDNAs (NT5C2.3-mt, NT5C2.47-mt, and NT5C2.136-mt) in BL21 E. coli using pET101 expression vectors. The recombinant proteins were obtained 5 h after isopropyl β-d-thiogalactoside induction and were analyzed for NT5C2 expression by Western blotting using a NT5C2-C-ter antibody. Recombinant NT5C2 wild-type and mutant proteins were expressed in equivalent amounts, whereas no NT5C2 protein was detected with the empty pET101 vector (inset in Fig. 5). NT5C2 activity was measured using the Diazyme 5′-Nucleotidase Enzymatic Test Kit at different time points. As shown in Fig. 5, the variant NT5C2 isoforms demonstrated no significant difference in NT5C2 activity compared with that of the WT isoform.
Fig. 5.
Activity of recombinant human NT5C2 WT and mutant proteins. NT5C2 WT and amino acid variant (T3A, K47N, and Q136R) isoforms were expressed in BL21 E. coli. The columns represent activity of NT5C2 isoforms that was determined using a Diazyme 5′-Nucleotidase Enzymatic Test Kit. Activities of NT5C2 amino acid variants were compared with that of WT protein, and no difference in activity was observed. Inset shows a representative Western blot when equal amounts of total protein were loaded for WT and mutant NT5C2 proteins; all of the recombinant NT5C2 variants were expressed at equivalent levels.
NT5C2 Methylation.
MSP was performed with all the 5 primer sets (for 5 putative CpG sites) on the 10 most sensitive and 10 most resistant unrelated CEU and YRI cell lines. Commercially available control methylated and unmethylated DNAs were used as positive controls (EpiTect PCR Control DNA Set; QIAGEN) for all the 5 CpG sites. Both positive controls demonstrated amplification with the respective methylated and unmethylated primer sets. None of CEU and YRI cell lines showed any band in the methylated primer set, whereas a band was present in the unmethylated primer set (Fig. 6B), indicating that in HapMap cell lines methylation does not contribute to regulation of NT5C2 gene expression. To assess the global methylation status in these cell lines, we performed LINE1 analysis using Pyrosequencing and found approximately 80% LINE1 methylation in these cell lines.
Association of NT5C2 SNPs with mRNA Expression in Leukemic Cells from Patients with AML.
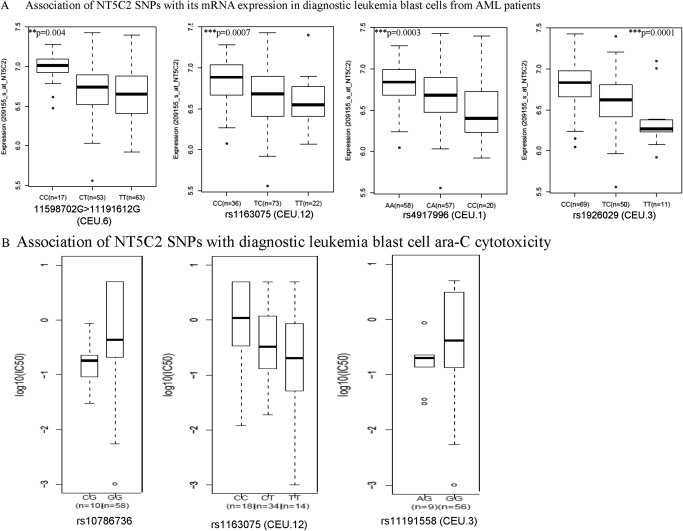
A total of 16 SNPs within NT5C2 were genotyped in DNA samples collected at diagnosis from patients with AML. Four of 16 SNPs were not present in patient samples, and 2 occurred with the minimum allele frequency of <0.005 and were not included in the analysis. Because mRNA expression was determined in patients' diagnostic leukemic blasts as described earlier, the data from two clinical trials were combined. In total, 137 subjects (41 from St. Jude AML97 and 96 from St. Jude AML02) had both NT5C2 genotype and microarray data available. NT5C2 expression levels did not differ significantly according to race (p = 0.59) or risk group (p = 0.26). We observed a significant association of group CEU.6 SNP represented by rs11598702 in Fig. 7A (occurs in LD with promoter SNP rs11191612, described earlier) with expression of NT5C2 (p = 0.0043) (Table 3). In accordance with the results in the HapMap samples, the minor allele (A allele for rs11598702 and G allele for rs11191612) was associated with higher expression of NT5C2 in AML cells. For rs1163075 (SNP within group CEU.12), the C allele demonstrated higher NT5C2 mRNA expression levels in HapMap cell lines as well as in AML patient samples (CC versus CT versus TT, p = 0.0012) (Fig. 7A; Tables 2 and 3). In addition, SNPs rs4917996, (representing CEU.1) and rs1926029 (representing CEU.3) were significantly associated with NT5C2 mRNA expression levels in leukemic cells from patients with AML (CC versus CT versus TT, p < 0.0002) (Table 3).
Fig. 7.
A, association of NT5C2 SNPs with mRNA expression in diagnostic AML leukemic blasts. NT5C2 mRNA expression levels in diagnostic leukemic blasts were extracted from Affymetrix U133A array data from 137 patients (AML97, n = 41; AML02, n = 96) and were analyzed for association with NT5C2 SNPs. Box plots for association of NT5C2 SNPs rs11598702 (representing CEU.6), rs1163075 (representing CEU.12), rs4917996 (representing CEU.1), and rs1926029 (CEU.3) with its mRNA expression in diagnostic leukemic blasts from AML patients are shown. B, association of NT5C2 SNPs with diagnostic blast ara-C cytotoxicity. Ara-C cytotoxicity was determined by treating diagnostic leukemic blasts with varying concentrations of ara-C, and LC50 values were calculated. Box plots represent association of NT5C2 SNPs (rs1078636 and rs1163075 representing CEU.12 and rs11191558 representing CEU.3) in all patients.
TABLE 3.
Association of NT5C2 SNPs with its expression and ara-C cytotoxicity in diagnostic leukemic blasts from patients with AML
| Phenotype I: Ara-C Cytotoxicity of Diagnostic Leukemic Blasts (n = 71, AML02) |
Phenotype II: Log NT5C2 mRNA Expression in Diagnostic Leukemic Blasts (n = 137, AML97; n = 41 and AML02; n = 96) |
|||||
|---|---|---|---|---|---|---|
| Genotype | Mean ± S.D. | p | Genotype | Mean± S.D. | P | |
| CEU0.12 | ||||||
| rs1163075 | CC (n = 18) | 2.109 ± 2.164 | CC vs. CT vs. TT, *p = 0.047 | CC (n = 36) | 6.833 ± 0.287 | CC vs. CT vs. TT, *p = 0.0012 |
| TC (n = 34) | 1.129 ± 1.696 | CC vs. CT + TT, *p = 0.04 | CT (n = 73) | 6.643 ± 0.348 | CC vs. CT + TT, *p = 0.001 | |
| TT (n = 14) | 1.235 ± 2.051 | TT (n = 22) | 6.586 ± 0.297 | |||
| CEU0.1 | ||||||
| rs4917996 | AA (n = 26) | 1.622 ± 1.952 | AA vs. CA vs. CC, p = 0.245 | AA (n = 58) | 6.809 ± 0.262 | AA vs. CA vs. CC, ***p = 0.0001 |
| CA (n = 31) | 1.262 ± 1.903 | CA (n = 57) | 6.669 ± 0.332 | AA + CA vs. CC, **p = 0.0006 | ||
| CC (n = 11) | 1.611 ± 2.212 | CC (n = 20) | 6.458 ± 0.369 | AA vs. CA + CC, **p = 0.0005 | ||
| CEU0.6 | ||||||
| rs11598702 | CC (n = 9) | 1.58 ± 2.018 | CC vs. CT vs. TT, p = 0.3 | CC (n = 17) | 6.974 ± 0.200 | CC vs. CT vs. TT *p = 0.0043 |
| CT (n = 24) | 1.244 ± 1.776 | CT (n = 53) | 6.689 ± 0.323 | CC vs. CT + TT *p = 0.0001 | ||
| TT (n = 30) | 1.414 ± 2.049 | TT (n = 63) | 6.644 ± 0.334 | |||
| CEU0.3 | ||||||
| rs1926029 | CC (n = 28) | 1.167 ± 1.673 | CC vs. TC vs. TT, p = 0.73 | CC (n = 69) | 6.7926 ± 0.279 | CC vs. TC vs. TT, ***p < 0.0001 |
| TC (n = 30) | 1.278 ± 1.933 | TC (n = 50) | 6.6061 ± 0.347 | CC vs. TC + TT, ***p = 0.0001 | ||
| TT (n = 7) | 2.517 ± 2.366 | TT (n = 11) | 6.3808 ± 0.358 | CC + TC vs. TT, **p = 0.0034 | ||
| CEU0.11 | ||||||
| rs4917384 | CC (n = 31) | 2.015 ± 2.153 | CC vs. CT vs. TT, *p = 0.041 | CC (n = 72) | 6.724 ± 0.340 | CC vs. CT vs. TT, p = 0.44 |
| TC (n = 32) | 0.937 ± 1.606 | CT (n = 55) | 6.679 ± 0.327 | |||
| TT (n = 2) | 2.60 ± 3.392 | TT (n = 7) | 6.673 ± 0.308 | |||
| CEU0.2 | ||||||
| rs11191558 | AG (n = 9) | 0.232 ± 0.247 | AG vs. GG, *p = 0.033 | AG (n = 20) | 6.738 ± 0.378 | AG vs. GG, p = 0.68 |
| GG (n = 56) | 1.630 ± 2.019 | GG (n = 111) | 6.689 ± 0.322 | |||
| rs10786736 | CG (n = 10) | 0.218 ± 0.237 | CG vs. GG, **p = 0.0051 | CC (n = 1) | 6.2403 | CC vs. CG vs. GG, p = 0.52 |
| GG (n = 58) | 1.670 ± 2.037 | CG (n = 22) | 6.690 ± 0.388 | |||
| GG (n = 112) | 6.705 ± 0.322 | |||||
p < 0.05.
p < 0.01.
p < 0.001.
Association of NT5C2 SNPs with In Vitro Ara-C Sensitivity of Primary Leukemic Cells from Patients with AML.
Diagnostic blast ara-C cytotoxicity (measured as LC50) was determined in 76 primary leukemic samples and demonstrated significant variation. The median LC50 value was 0.39 ng/μl (range, 0.001–5.0 ng/μl); ara-C LC50 was not associated with race (p = 0.27). Provisional risk group was significantly associated with ara-C sensitivity of leukemic blasts (p = 0.001); hence, we analyzed the association of SNPs in all patients, as well as within the high and standard provisional risk groups (provisional risk group). Of interest, rs1163075 (a group CEU.12 SNP), which was associated with ara-C cytotoxicity and with NT5C2 mRNA expression levels in HapMap cell lines, was a significant predictor of in vitro ara-C sensitivity in AML blasts. The presence of the C allele was associated with higher NT5C2 expression and was also associated with greater ara-C LC50 (p = 0.04) (Fig. 7B). In addition, rs10786736, which was associated with NT5C2 mRNA expression and with ara-C cytotoxicity in HapMap samples (Fig. 3C), was associated with ara-C sensitivity of primary AML leukemic blasts. In consensus with HapMap results, the GG genotype was associated with greater ara-C LC50 versus the CG genotype (p = 0.0051) (Fig. 7B; Table 3). A group CEU.2 SNP rs11191558 that was also associated with ara-C cytotoxicity in HapMap samples (Fig. 3A) was significantly associated with ara-C cytotoxicity in AML leukemic blasts. The median LC50 of leukemic blasts from patients with the GG genotype was higher versus that in patients with the AG genotype (p = 0.03) (Table 3).
Association of NT5C2 SNPs with Clinical Phenotype Measures in Patients with AML.
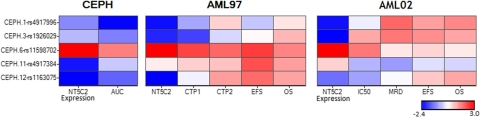
Within the St. Jude AML97 cohort, there was no significant association of NT5C2 SNPs with ara-CTP levels (day 1 and 2) or clinical response. The heat map in Fig. 8 shows the association pattern of the most significant SNPs (from data above) occurring with minimum allele frequency of greater than 0.10 with multiple parameters in patients with AML. Of interest, CEU.12 SNP rs1163075, which was associated with lower NT5C2 expression and ara-C AUC/LC50 in both CEU and primary AML samples, demonstrated a trend toward association with slightly higher ara-CTP levels in the AML97 cohort and with better EFS and OS in both the AML97 and AML02 cohorts. CEU.11 SNP rs4917384 was associated significantly with day 22 MRD levels in the AML02 cohort (p = 0.02). However, SNP rs1163075 did not show a significant association with EFS (p = 0.3) or OS (p = 0.3) in a Cox regression model that included predictors that were identified as clinically or statistically important in an earlier study (Rubnitz et al., 2009).
Fig. 8.
Heat map of association of the five most interesting NT5C2 SNPs with multiple endpoints in HapMap and AML samples. Each row represents a SNP representing different LD groups, and each column represents a phenotype. Blue represents association of a minor allele with reduced levels of phenotype and red represents association of a minor allele with an increased value for the phenotype. The colors are assigned by log10 p value according to the accompanying color scale.
Pleiotropic Effects in HapMap Cell Lines and Patients with AML.
SNPs were screened for pleiotropic effects on NT5C2 expression and ara-C resistance in HapMap cell lines and for pleiotropic effects on NT5C2 expression, clinical response, and event-free survival in patients with AML. In HapMap cell lines, the SNP rs1163075 showed a significant pleiotropic effect (p = 0.0053) characterized by a negative association of the minor allele with both NT5C2 expression and cytarabine resistance. Among patients with AML, this SNP also showed a significant pleiotropic effect (p = 0.0233) characterized by the minor allele showing a negative association with NT5C2 expression and a positive association with both clinical response and event-free survival.
Discussion
NT5C2 is a cytosolic 5′-nucleotidase that has been implicated in the inactivation of ara-C by dephosphorylating ara-CMP to ara-C (Amici et al., 1997; Amici and Magni, 2002). Although reports suggest that expression of NT5C2 is a significant prognostic marker of worse clinical outcome in patients with AML and myelodysplastic syndrome, its role in ara-C response is still unclear (Mazzon et al., 2003; Galmarini, 2007). It has been hypothesized that because NT5C2 is involved in substrate dNTP cycles, survival of highly proliferating NT5C2-expressing cells may favor disease recurrence, thus shortening disease-free and overall survival (Galmarini et al., 2005; Galmarini, 2007).
It is still not clear whether the role of NT5C2 in response is due to its involvement in ara-CMP dephosphorylation or modification of cellular dNTP pools in leukemic cells or perhaps with the leukemic phenotype. Nonetheless, there is evidence in the literature for the association of high NT5C2 expression with worse clinical response as well as with ara-C resistance in multiple cancer cell lines and experimental models. Hence, we sought to determine whether genetic polymorphisms within NT5C2 are associated with its expression and/or activity and, therefore, influence ara-C sensitivity. Because there are no data in the literature on sequencing of NT5C2 for SNP discovery, we sequenced the NT5C2 gene in European (30 CEU trios) and African (30 YRI trios) ancestry HapMap panels to maximize the use of publicly available HapMap genotype data. Forty-one genetic variants including 1 in/del were identified by sequencing the coding and proximal promoter of NT5C2. Five coding SNPs were identified including three nonsynonymous SNPs (Y3A, K47R, and Q136R), which did not have any influence on NT5C2 activity when expressed as recombinant protein.
We observed strong linkage between multiple SNPs at the NT5C2 locus in both CEU and YRI samples (Fig. 1, B and C). We observed that NT5C2 expression was significantly higher in the YRI versus the CEU panel. However, in the CEU cell lines expression of NT5C2 was directly correlated with ara-C cytotoxicity; this relationship was not observed in the YRI cell lines. This might be one of the factors contributing to lack of significant genotype-phenotype association observed in our analysis within the YRI samples (Fig. 2) (Aplenc et al., 2006). Within the CEU panel, NT5C2 SNPs demonstrated significant association with its mRNA expression and in vitro ara-C sensitivity in both CEU HapMap cell lines and diagnostic leukemic blasts from AML patients (Figs. 3 and 7). Table 4 provides a summary of the most interesting genotype-phenotype associations observed in CEU samples and AML patient samples for SNPs occurring with the minimum allele frequency of >0.10. Of interest, the presence of minor alleles for SNPs within groups CEU.1 to 3, 7, and 10 to 12 were associated with lower NT5C2 expression and ara-C sensitivity, whereas CEU.6 group SNPs were associated with increased expression and ara-C resistance. As indicated in Table 4, within each group we have SNPs that are linked and are present in the 5′-UTR or proximal promoter, exon, intron, or 3′-UTR. Therefore, this observed association could be due to any of the SNPs within a group or driven primarily by promoter and 3′-UTR SNPs or due to small effects of multiple SNPs. Functional studies on two 5′-UTR variants and our results indicate that although the group CEU.1 SNP (rs10748839) has no effect on luciferase activation the variant allele (G) for CEU.6 group SNP rs11191612 was associated with increased luciferase activation (Fig. 4). This finding was in agreement with the association of group CEU.6 SNPs with NT5C2 mRNA expression and ara-C cytotoxicity in HapMap cell lines and in AML patient samples (Figs. 3C and 7). Li et al. (2009) had reported in a genome-wide analysis association of CEU.6 group SNP (rs11598702) with ara-C sensitivity.
TABLE 4.
Summary of NT5C2 SNPs showing most significant association in CEU HapMap cell lines and leukemia cells from patients with AML
Downward arrows indicate association of minor allele with reduced expression and in vitro ara-C cytotoxicity. Upward arrows indicate association with increased expression and ara-C cytotoxicity. Double arrows indicate statistically significant association, and single arrows indicate trend not reaching statistical significance.
| LD Group | SNP Location |
HapMap Samples |
AML Patients Diagnostic Blasts |
MAF (HapMap)a | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Promoter/5′UTR | Intronic | Coding SNP | 3′-UTR | mRNA Expression | Ara-C AUC | SNPs Genotyped AML | mRNA Expression | Ara-C IC50 | ||
| CEU0.1 | 4 | 8 | 1 | 0 | ↓ | ⇊ | rs4917996 | ⇊ | ↓ | 0.400 |
| CEU0.3 | 5 | 20 | 0 | 1 | ↓ | ⇊ | rs11191558 | ⇊ | ↓ | 0.353 |
| CEU0.6 | 1 | 2 | 0 | 0 | ⇈ | ⇊ | rs11598702 | ⇊ | 0.358 | |
| CEU0.11 | 2 | 0 | 0 | 0 | ⇊ | ⇊ | rs4917384 | ⇊ | 0.259 | |
| CEU0.12 | 9 | 0 | 0 | 0 | ⇊ | ⇊ | rs1163075 | ⇊ | ⇊ | 0.442 |
MAF, minimum allele frequency.
Distribution of SNPs in gene within each LD group is also included.
The most interesting SNPs in the present study include a group of 5′-UTR SNPs with consistently demonstrated association with mRNA expression and ara-C cytotoxicity in HapMap and AML samples. Three SNPs within this LD group (i.e., rs7913461, rs1891292, and rs1891293) were present in a region of high 7× regulatory potential and ENCODE DNaseI hypersensitivity clusters (UCSC genome browser), thereby indicating potential regulatory significance.
Although our results did not demonstrate strong association of NT5C2 SNPs with clinical phenotype measures in patients with AML, we observed a consistent pattern of association of CEU.12 SNP rs1163075 with clinical outcome in both AML97 and AML02 cohorts. In accordance with these results, CEU.12 SNPs (rs7913461 and rs1891292) also demonstrated reduced binding for the variant oligo in EMSA assays (Supplemental Fig. 1). The nonsignificant association observed with intracellular ara-CTP levels could be due to multiple factors such as small sample size or the contribution of other genes in the ara-C metabolic pathway. An alternative, as suggested in previous studies, is that NT5C2 might not be catalyzing the conversion of ara-CMP to ara-C. Although in the present study we have attempted to explore the impact of NT5C2 genetic variation, the clinical cohorts were not designed to evaluate the pharmacogenetics of ara-C. We also acknowledge the fact that variability in the intracellular levels of ara-CTP is regulated by multiple enzymes in the ara-C activation pathway, and we have previously shown association of deoxycytidine kinase variants (Lamba et al., 2007). To achieve full understanding of the genetic basis for the variability observed in the intracellular concentration of ara-CTP, future studies in our laboratory will be directed toward comprehensive and simultaneous evaluation of other enzymes of relevance in the metabolic pathway of ara-C.
In summary, we have identified novel genetic variants at the NT5C2 locus by sequencing genomic DNA from two ethnic groups. NT5C2 SNPs are predictive of its expression and ara-C cytotoxicity not only in HapMap cell lines but also in patients with AML. We have also performed functional characterization of selected SNPs to further understand the underlying molecular mechanism. In addition, we evaluated the clinical implication of NT5C2 polymorphisms for its association with clinical phenotypes in patients with AML receiving ara-C-based therapy. However, the most significant SNPs identified in this study need to be confirmed in a larger patient population, to confidently arrive at the conclusion that SNPs within NT5C2, at least in part, play a role in predicting drug responsiveness and to guide individualized chemotherapy in patients with AML receiving ara-C or other nucleoside-containing therapies that are activated by the same metabolic pathway as ara-C.
Supplementary Material
Acknowledgments
We acknowledge the support provided by the Biomedical Genomics Center and Minnesota Supercomputing Institute at the University of Minnesota. We also acknowledge help from pediatric oncology education scholars, Brittney Green and Jessica Gresham.
This study was supported by National Institutes of Health National Cancer Institute [Grant R01-CA132946] (to J.K.L.); and the American Lebanese Syrian Associated Charities.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.111.182873.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
- ara-C
- 1-β-d-arabinofuranosyl-cytosine (cytarabine)
- AML
- acute myeloid leukemia
- ara-CTP
- ara-C triphosphate
- ara-CMP
- ara-C monophosphate
- NT5C
- cytoplasmic 5′ nucleotidase
- 5′UTR
- 5′-untranslated region
- SNP
- single nucleotide polymorphism
- PCR
- polymerase chain reaction
- CEU
- Centre d'Etude du Polymorphisme Humain
- YRI
- Yoruba people in Ibadan, Nigeria
- AUC
- area under the survival curve
- UTR
- untranslated region
- BMGC
- Biomedical Genomics Center
- WT
- wild type
- EMSA
- electrophoretic mobility shift assays
- mt
- mutant
- MSP
- methylation-specific polymerase chain reaction
- EFS
- event free survival
- OS
- overall survival
- MRD
- minimal residual disease
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide
- LD
- linkage disequilibrium.
Authorship Contributions
Participated in research design: Mitra and Lamba.
Conducted experiments: Mitra, Crews, Ghodke, Feldberg, and Lamba.
Performed data analysis: Mitra, Pounds, Cao, and Lamba.
Wrote or contributed to the writing of the manuscript: Mitra, Crews, Pounds, Cao, Feldberg, Ghodke, Gandhi, Plunkett, Dolan, Hartford, Raimondi, Campana, Downing, Rubnitz, Ribeiro, and Lamba.
References
- Amici A, Emanuelli M, Magni G, Raffaelli N, Ruggieri S. (1997) Pyrimidine nucleotidases from human erythrocyte possess phosphotransferase activities specific for pyrimidine nucleotides. FEBS Lett 419:263–267 [DOI] [PubMed] [Google Scholar]
- Amici A, Magni G. (2002) Human erythrocyte pyrimidine 5′-nucleotidase, PN-I. Arch Biochem Biophys 397:184–190 [DOI] [PubMed] [Google Scholar]
- Children's Oncology Group, Aplenc R, Alonzo TA, Gerbing RB, Smith FO, Meshinchi S, Ross JA, Perentesis J, Woods WG, Lange BJ, et al. (2006) Ethnicity and survival in childhood acute myeloid leukemia: a report from the Children's Oncology Group. Blood 108:74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beesley AH, Palmer ML, Ford J, Weller RE, Cummings AJ, Freitas JR, Firth MJ, Perera KU, de Klerk NH, Kees UR. (2006) Authenticity and drug resistance in a panel of acute lymphoblastic leukaemia cell lines. Br J Cancer 95:1537–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi V, Pontis E, Reichard P. (1986) Interrelations between substrate cycles and de novo synthesis of pyrimidine deoxyribonucleoside triphosphates in 3T6 cells. Proc Natl Acad Sci USA 83:986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews KR, Gandhi V, Srivastava DK, Razzouk BI, Tong X, Behm FG, Plunkett W, Raimondi SC, Pui CH, Rubnitz JE, et al. (2002) Interim comparison of a continuous infusion versus a short daily infusion of cytarabine given in combination with cladribine for pediatric acute myeloid leukemia. J Clin Oncol 20:4217–4224 [DOI] [PubMed] [Google Scholar]
- Dumontet C, Fabianowska-Majewska K, Mantincic D, Callet Bauchu E, Tigaud I, Gandhi V, Lepoivre M, Peters GJ, Rolland MO, Wyczechowska D, et al. (1999) Common resistance mechanisms to deoxynucleoside analogues in variants of the human erythroleukaemic line K562. Br J Haematol 106:78–85 [DOI] [PubMed] [Google Scholar]
- Galmarini CM. (2007) What does over-expression of cN-II enzyme signify in haematological malignancies? Leuk Res 31:1325–1326 [DOI] [PubMed] [Google Scholar]
- Galmarini CM, Cros E, Thomas X, Jordheim L, Dumontet C. (2005) The prognostic value of cN-II and cN-III enzymes in adult acute myeloid leukemia. Haematologica 90:1699–1701 [PubMed] [Google Scholar]
- Galmarini CM, Graham K, Thomas X, Calvo F, Rousselot P, El Jafaari A, Cros E, Mackey JR, Dumontet C. (2001a) Expression of high Km 5′-nucleotidase in leukemic blasts is an independent prognostic factor in adults with acute myeloid leukemia. Blood 98:1922–1926 [DOI] [PubMed] [Google Scholar]
- Galmarini CM, Mackey JR, Dumontet C. (2001b) Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia 15:875–890 [DOI] [PubMed] [Google Scholar]
- Galmarini CM, Thomas X, Graham K, El Jafaari A, Cros E, Jordheim L, Mackey JR, Dumontet C. (2003) Deoxycytidine kinase and cN-II nucleotidase expression in blast cells predict survival in acute myeloid leukaemia patients treated with cytarabine. Br J Haematol 122:53–60 [DOI] [PubMed] [Google Scholar]
- Hartford CM, Duan S, Delaney SM, Mi S, Kistner EO, Lamba JK, Huang RS, Dolan ME. (2009) Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood 113:2145–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, Kazemier KM, Pei D, Cheng C, Pui CH, Relling MV, et al. (2004) Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med 351:533–542 [DOI] [PubMed] [Google Scholar]
- Jung SH, Owzar K, George SL. (2005) A multiple testing procedure to associate gene expression levels with survival. Stat Med 24:3077–3088 [DOI] [PubMed] [Google Scholar]
- Kufe DW, Major PP, Egan EM, Beardsley GP. (1980) Correlation of cytotoxicity with incorporation of ara-C into DNA. J Biol Chem 255:8997–9000 [PubMed] [Google Scholar]
- Lamba JK, Crews K, Pounds S, Schuetz EG, Gresham J, Gandhi V, Plunkett W, Rubnitz J, Ribeiro R. (2007) Pharmacogenetics of deoxycytidine kinase: identification and characterization of novel genetic variants. J Pharmacol Exp Ther 323:935–945 [DOI] [PubMed] [Google Scholar]
- Lamba JK, Crews KR, Pounds SB, Cao X, Gandhi V, Plunkett W, Razzouk BI, Lamba V, Baker SD, Raimondi SC, et al. (2011) Identification of predictive markers of cytarabine response in AML by integrative analysis of gene-expression profiles with multiple phenotypes. Pharmacogenomics 12:327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Fridley BL, Kalari K, Jenkins G, Batzler A, Weinshilboum RM, Wang L. (2009) Gemcitabine and arabinosylcytosin pharmacogenomics: genome-wide association and drug response biomarkers. PLoS One 4:e7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major PP, Egan EM, Beardsley GP, Minden MD, Kufe DW. (1981) Lethality of human myeloblasts correlates with the incorporation of arabinofuranosylcytosine into DNA. Proc Natl Acad Sci USA 78:3235–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzon C, Rampazzo C, Scaini MC, Gallinaro L, Karlsson A, Meier C, Balzarini J, Reichard P, Bianchi V. (2003) Cytosolic and mitochondrial deoxyribonucleotidases: activity with substrate analogs, inhibitors and implications for therapy. Biochem Pharmacol 66:471–479 [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. (2001) Predicting deleterious amino acid substitutions. Genome Res 11:863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. (2003) SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31:3812–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL. (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesi R, Turriani M, Allegrini S, Scolozzi C, Camici M, Ipata PL, Tozzi MG. (1994) The bifunctional cytosolic 5′-nucleotidase: regulation of the phosphotransferase and nucleotidase activities. Arch Biochem Biophys 312:75–80 [DOI] [PubMed] [Google Scholar]
- Pounds S, Cao X, Cheng C, Yang JJ, Campana D, Pui CH, Evans WE, Relling MV. (2011) Integrated analysis of pharmacologic, clinical and SNP microarray data using Projection Onto the Most Interesting Statistical Evidence with Adaptive Permutation Testing. Int J Data Min Bioinform 5:143–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pounds S, Cheng C, Cao X, Crews KR, Plunkett W, Gandhi V, Rubnitz J, Ribeiro RC, Downing JR, Lamba J. (2009) PROMISE: a tool to identify genomic features with a specific biologically interesting pattern of associations with multiple endpoint variables. Bioinformatics 25:2013–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. (2002) Human non-synonymous SNPs: server and survey. Nucleic Acids Res 30:3894–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza A, Gezer S, Anderson J, Lykins J, Bennett J, Browman G, Goldberg J, Larson R, Vogler R, Preisler HD. (1992) Relationship of [3H]Ara-C incorporation and response to therapy with high-dose Ara-C in AML patients: a Leukemia Intergroup study. Exp Hematol 20:1194–1200 [PubMed] [Google Scholar]
- Rubnitz JE, Crews KR, Pounds S, Yang S, Campana D, Gandhi VV, Raimondi SC, Downing JR, Razzouk BI, Pui CH, et al. (2009) Combination of cladribine and cytarabine is effective for childhood acute myeloid leukemia: results of the St Jude AML97 trial. Leukemia 23:1410–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubnitz JE, Inaba H, Dahl G, Ribeiro RC, Bowman WP, Taub J, Pounds S, Razzouk BI, Lacayo NJ, Cao X, et al. (2010) Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial. Lancet Oncol 11:543–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Stegmann AP, Geisen F, Konwalinka G. (1998) Lack of cross-resistance with gemcitabine and cytarabine in cladribine-resistant HL60 cells with elevated 5′-nucleotidase activity. Exp Hematol 26:1223–1228 [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Bork P. (2000) Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet 16:198–200 [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. (2001) Prediction of deleterious human alleles. Hum Mol Genet 10:591–597 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Sugawara T, Oyake T, Uchiyama T, Aoki Y, Tsukushi Y, Onodera S, Ito S, Murai K, Ishida Y. (2007) Clinical significance of high-Km 5′-nucleotidase (cN-II) mRNA expression in high-risk myelodysplastic syndrome. Leuk Res 31:1343–1349 [DOI] [PubMed] [Google Scholar]
- Wang JJ, Selawry OS, Vietti TJ, Bodey GP., Sr (1970) Prolonged infusion of arabinosyl cytosine in childhood leukemia. Cancer 25:1–6 [DOI] [PubMed] [Google Scholar]
- Wolfinger RD. (1996) Heterogeneous variance-covariance structures for repeated measures. J Agric Biol-Environ Sci 1:205–230 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.