Background: Respiratory complex I accepts electrons from NADH.
Results: Mutation of a single amino acid residue leads to a physiological oxidation of NADPH, however, coupled with the production of reactive oxygen species.
Conclusion: The NADH-binding site of complex I evolved to discriminate NADH from NADPH and to reduce the production of reactive oxygen species.
Significance: The mode of nucleotide binding determines the production of reactive oxygen species in complex I.
Keywords: Bioenergetics-Electron Transfer Complex, Dehydrogenase, Electron Transport, Enzyme Mechanisms, NADH, Complex I, Escherichia coli, NAD(P)H Binding, NADH Dehydrogenase, Reactive Oxygen Species
Abstract
The respiratory complex I couples the electron transfer from NADH to ubiquinone with a translocation of protons across the membrane. Its nucleotide-binding site is made up of a unique Rossmann fold to accommodate the binding of the substrate NADH and of the primary electron acceptor flavin mononucleotide. Binding of NADH includes interactions of the hydroxyl groups of the adenosine ribose with a conserved glutamic acid residue. Structural analysis revealed that due to steric hindrance and electrostatic repulsion, this residue most likely prevents the binding of NADPH, which is a poor substrate of the complex. We produced several variants with mutations at this position exhibiting up to 200-fold enhanced catalytic efficiency with NADPH. The reaction of the variants with NAD(P)H is coupled with proton translocation in an inhibitor-sensitive manner. Thus, we have created an energy-converting NADPH:ubiquinone oxidoreductase, an activity so far not found in nature. Remarkably, the oxidation of NAD(P)H by the variants leads to an enhanced production of reactive oxygen species.
Introduction
The NADH:ubiquinone oxidoreductase, also known as respiratory complex I,2 couples the transfer of electrons from NADH to ubiquinone with the translocation of protons across the membrane (1–6). In doing so, it contributes to the generation of the protonmotive force required for energy-consuming processes. In eukaryotes, complex I consists of 45 different subunits resulting in a molecular mass of about 1 MDa (7). The bacterial homologue generally consists of 14 subunits named NuoA to NuoN (or Nqo1–14). Both the mitochondrial and the bacterial complex contain the same cofactors for the electron transfer reaction, namely one flavin mononucleotide (FMN) and up to 10 iron-sulfur (Fe/S) clusters (8). Because of this, the bacterial complex I is regarded as a simple structural and functional model for the eukaryotic enzyme (9). Electron microscopy revealed the two-part structure of the complex consisting of a peripheral and a membrane arm (10, 11). In bacteria, the peripheral arm is made up of seven globular subunits, and the membrane arm consists of seven polytopic subunits comprising 63 transmembrane helices. All known cofactors and the NADH-binding site are located within the peripheral arm, the structure of which was resolved at 3.3 Å resolution (12, 13). The quinone reduction site is proposed to be located at the interface between the peripheral and the membrane arm (14, 15). The membrane arm lacks any known cofactor but has to be involved in proton translocation. This is supported by the homology of the three major subunits of the arm to subunits of cation/proton antiporters (16, 17). Mechanisms based on a conformational link between the redox reaction and the proton translocation have been discussed (4, 5, 18, 19). The recently published crystal structures of complex I from Thermus thermophilus (20) and Yarrowia lipolytica (21) imply that the complex contains a coupling site directly linked to electron transfer as well as another site indirectly coupled with electron transfer.
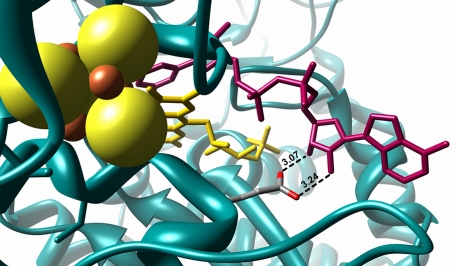
NADPH is known to be a very poor substrate for the complex (22). It was established that submitochondrial particles exhibit an NADPH oxidase activity (23). The optimum for this activity is around pH 6, whereas the NADH oxidase activity shows a broad optimum around pH 7. The NADPH oxidase activity was about one-fifth of the NADH oxidase activity at pH 6 (24). The activity was shown to be an intrinsic property of the mitochondrial complex I and not related to the activity of the NADPH/NAD transhydrogenase (23, 24). The structure of the peripheral arm of complex I with NADH bound to subunit NuoF revealed that the carboxylate group of Glu-183F (nomenclature according to the Escherichia coli sequence; the superscript denotes the name of the corresponding Nuo subunit) is hydrogen-bonded to the hydroxyl groups of the adenosine ribose (13). The ribose is shielded by the protein in such a way that there is some but not sufficient space below the O2B atom to provide space for the phosphate group of NADPH (Fig. 1), thus explaining its low reactivity with complex I. In principle, NADPH could be a substrate for the complex, but most likely due to the steric hindrance and electrostatic repulsion with Glu-183F, its reaction rate with NADPH is at least five times slower than with NADH as a substrate (24, 25). Here, we report on engineering complex I to an energy-converting NADPH:ubiquinone oxidoreductase by a single mutation of the conserved Glu-183F.
FIGURE 1.
Structure of the nucleotide-binding site of complex I with bound NADH. The protein is shown in blue, iron in red, sulfur in yellow, the FMN in yellow, and the bound NADH in magenta. The radius of the sulfur ion was set to 1.7 Å and that of the iron ions to 0.7 Å (48). The length of the hydrogen bonds between Glu-183F and the adenosine ribose hydroxyl groups are given in Å. The picture was drawn with Chimera (49) using the Protein Data Bank code 3IAM (13).
EXPERIMENTAL PROCEDURES
Materials and Strains
A derivative from E. coli strain BW25113 (26) was used, in which the nuo-operon was deleted by genomic replacement methods (27).3 In addition, E. coli strain DH5αΔnuo (28) and the plasmids pBADnuoHis (29), pCA24NnuoF (30), and pKD46 (28) were used. Chloramphenicol (170 μg/ml), ampicillin (100 μg/ml), and kanamycin (50 μg/ml) were supplemented where necessary. All enzymes used for recombinant DNA techniques were from Fermentas (St. Leon-Roth, Germany). DNA oligonucleotides were from MWG Operon (Ebersberg, Germany).
Site-directed Mutagenesis
The plasmid pCA24NnuoF (30) was used to introduce mutations in nuoF. The primer pairs nuoF E183D, nuoF E183H, nuoF E183N, and nuoF E183Q (supplemental Table S1) were used to create the plasmids pCA24NnuoF E183D/E183H/E183N and E183Q.
λ-Red-mediated Recombineering
Electrocompetent DH5αΔnuo/pKD46 cells were prepared and electroporated (28). The nptI-sacRB cartridge was amplified from pVO1100 by PCR with the primer pair nuoF::nptI-sacRB (supplemental Table S2). To integrate the cartridge into pBADnuoHis by λ-Red-mediated recombineering (27), electrocompetent DH5αΔnuo/pKD46 was mixed with 50 ng of pBADnuoHis and 350 ng of the PCR product. Recombinants were selected on LB-agar supplemented with kanamycin. Plasmids were isolated from KmR clones and purified by transformation of DH5α and growth on LB-agar supplemented with kanamycin. The nptI-sacRB cartridge on pBADnuoHis was replaced by the PCR product containing the mutation by recombineering. A linear dsDNA was amplified from pCA24NnuoF carrying the point mutation by PCR with the primer pair nuoF2_fwd and nuoF_rev (supplemental Table S2). Electrocompetent DH5αΔnuo/pKD46 was co-transformed with 50 ng pBADnuoHis nuoF::nptI-sacRB and 400–500 ng of PCR product. Recombinants were selected on YP-agar supplemented with chloramphenicol and 10% (w/v) sucrose at 30 °C. Plasmids from CamR and SucR clones were isolated. All mutations were confirmed by DNA sequencing.
Isolation of Complex I and the Variants
The complex and its variants were isolated as described (29). Typical course of the preparations is shown in supplemental Tables S3–S6.
Determination of Complex I Activity
E. coli cytoplasmic membranes were isolated as described (31). The NAD(P)H oxidase activity of the cytoplasmic membranes was measured with a Clark-type oxygen electrode at 30 °C. The assay contained ∼200 μg of membrane protein in 50 mm MES/NaOH, 50 mm NaCl, 5 mm MgCl2, pH 6.0. The reaction was started by adding 2.5 mm of either NADH or NADPH (both from Sigma), respectively. After 1 min, 10 μm piericidin A was added to the assay. The NAD(P)H:decyl-ubiquinone oxidoreductase activity of the isolated complex was measured either as a decrease of the NAD(P)H concentration at 340–400 nm using an ϵ of 6.3 mm−1 cm−1 or as a decrease in the decyl-ubiquinone concentration at 248–268 nm using an ϵ of 7.8 mm−1 cm−1 (32). The very high NADPH concentrations were measured at 378–400 nm using an ϵ of 1.59 mm−1 cm−1. The enzyme was reconstituted in E. coli polar lipids (Avanti) by mixing the preparation (10 mg/ml) in 1:1 (w/w) ratio with the lipids (10 mg/ml) and by incubating for 20 min on ice. One μl of the protein/lipid mixture (or 3 μl with NADPH as substrate for the complex from the parental strain) was added to the assay buffer (50 mm MES/NaOH, 50 mm NaCl, pH 6.0) at 30 °C. The assay for the determination of the KM to NAD(P)H contained 60 μm decyl-ubiquinone, and the reaction was started by an addition of the corresponding nucleotide in various concentrations. The pH dependence of the NAD(P)H:decyl-ubiquinone oxidoreductase activity was assayed in the range from pH 5.0 to 8.0 using 50 mm MES, MOPS, or Tris as buffers. At lower pH values, the Fe/S clusters fall apart, and at higher values, the E. coli complex I disintegrates (8).
Determination of a Proton Gradient
The generation of a proton gradient was determined by the fluorescence quench of 9-amino-6-chloro-2-methoxyacridine (ACMA, Sigma). Proteoliposomes containing complex I were prepared as described (32). Five μl of proteoliposomes, 0.2 μm ACMA, and 50 μm decyl-ubiquinone were incubated for 3 min at 25 °C in 5 mm MES/NaOH, 50 mm KCl, 2 mm MgCl2, pH 6.0. The fluorescence was detected with an LS 45 luminescence spectrometer (PerkinElmer Life Sciences), using an excitation wavelength of 430 nm and an emission wavelength of 480 nm. The reaction was started by an addition of 150 μm nucleotide. In control experiments either 20 μm CCCP or 20 μm piericidin A was added to the reaction mixture.
EPR Spectroscopy
EPR measurements were conducted with a Bruker EMX 1/6 spectrometer operating at X-band (9.2 GHz). The sample temperature was controlled with an Oxford instrument ESR-9 helium flow cryostat. The magnetic field was calibrated using a strong pitch standard. The isolated proteins (5 mg/ml) were reduced with a 1,000-fold molar excess NAD(P)H and frozen after a 30 s incubation on ice. The reaction tubes and the EPR tubes were flushed with nitrogen before filling to prevent oxidation by air. The oxygen concentration of the buffers was reduced by flushing with argon.
Superoxide radical formation was detected by EPR spectroscopy at room temperature. One mm NAD(P)H was added to 1 ml of 50 mm MES/NaOH, 50 mm NaCl, pH 6.0, containing 45 μg of complex I reconstituted in phospholipids, 100 μm decyl-ubiquinone, and 100 mm of the spin trap DEPMPO. 10 μm piericidin A was added where indicated. For control, superoxide dismutase (100 units/ml; Sigma) was added to the assay buffer. 20 μl aliquots were withdrawn from the solutions and placed in small quartz tubes in the cavity.
Amplex Red Assay
Superoxide dismutates rapidly to H2O2. Thus, the total rate of superoxide and H2O2 production was additionally measured by the horseradish peroxidase-dependent oxidation of Amplex Red (46). The 1 ml assay contained 5 μg of complex I in polar lipids, 2 units of horseradish peroxidase, and 10 μm Amplex Red. The reaction was started by the addition of 30 μm NAD(P)H. When indicated, 60 μm decyl-ubiquinone or 10 μm piericidin A was added. The oxidation of the nucleotide was followed at 340 nm; the production of resorufin was monitored at 557–620 nm (ϵ = 23.1 mm−1 cm−1, pH 6.0) using an UV-visible diode array photometer (J&M Analytik AG, Aalen, Germany).
Other Analytical Assays
The purity of the nucleotides used in this study was determined by HPLC as described (33). Analysis was performed with a reverse phase C-18 column (5 μm Hypersil ODS, 250 × 10 mm, Knauer Wissenschaftlicher Gerätebau, Berlin, Germany) coupled with a multiwavelength detector (GE Healthcare). The mobile phase was 200 mm NaCl, 1 mm Tris/HCl, pH 8.2. NADH/ferricyanide oxidoreductase activity was measured at room temperature with an Ultrospec 1000 (GE Healthcare) spectrophotometer at 410 nm in 50 mm MES/NaOH, 50 mm NaCl, pH 6.0, using an ϵ of 1 mm−1 cm−1. The reaction was started with 0.2 mm NADH. Protein concentrations were determined according to the biuret method.
RESULTS
Reaction of Complex I with NAD(P)H
So far, a kinetic analysis with NADPH as substrate was only reported for the mitochondrial complex I (22, 34) in submitochondrial particles. To determine the activity of a bacterial complex I with NADPH, the E. coli enzyme was purified by affinity chromatography from an overproducing strain (29) and reconstituted in E. coli polar lipids to regain full enzymatic activity (32). The kinetic parameters were calculated from the dependence of the reaction rate on the nucleotide concentration in Hanes plots (Table 1) (35). By measuring the rate of NADH oxidation at various NADH concentrations, a KmNADH of 13 μm and a Vmax of 2.9 μmol/(min·mg) were determined. Measuring the rate of the decyl-ubiquinone reduction at various NADH concentrations led to the same KmNADH and a Vmax of 2.8 μmol/(min·mg). The reaction was inhibited by more than 95% by an addition of 10 μm piericidin A, a specific complex I inhibitor. These data demonstrate that the physiological electron transfer from NADH to quinone was measured in the reconstituted system. The KmNADPH was determined to be 1.9 mm (Table 1). The Vmax with NADPH as substrate was 8-fold lower than with NADH (Table 1). These data are in agreement with those reported for the mitochondrial complex (22–25).
TABLE 1.
Kinetic parameters of the NAD(P)H:decyl-ubiquinone oxidoreductase activity of E. coli complex I and variants of the complex after reconstitution in phospholipids
Activity was determined by measuring the rate of NAD(P)H oxidation.
| Variant |
KM |
Vmax |
kcat |
kcat/KM |
||||
|---|---|---|---|---|---|---|---|---|
| NADH | NADPH | NADH | NADPH | NADH | NADPH | NADH | NADPH | |
| μm | μmol·min−1·mg−1 | s−1 | s−1·μm−1 | |||||
| Parental | 13.0 ± 1 | 1,870 ± 30 | 2.9 ± 0.2 | 0.4 ± 0.1 | 26 ± 2 | 3 ± 1 | 2.0 ± 0.3 | 0.0018 ± 0.001 |
| E183DF | 5.8 ± 1 | 390 ± 30 | 4.2 ± 0.2 | 3.8 ± 0.2 | 37 ± 2 | 34 ± 2 | 6.4 ± 0.4 | 0.09 ± 0.006 |
| E183QF | 12.0 ± 2 | 45 ± 4 | 3.1 ± 0.2 | 1.6 ± 0.1 | 28 ± 2 | 14 ± 1 | 2.3 ± 0.5 | 0.32 ± 0.003 |
| E183NF | 14.0 ± 1 | 480 ± 50 | 1.2 ± 0.2 | 1.2 ± 0.3 | 11 ± 2 | 11 ± 3 | 0.8 ± 0.1 | 0.02 ± 0.006 |
| E183HF | 5.7 ± 1 | 25 ± 2 | 4.1 ± 0.2 | 1.1 ± 0.1 | 37 ± 2 | 10 ± 1 | 6.5 ± 0.3 | 0.40 ± 0.003 |
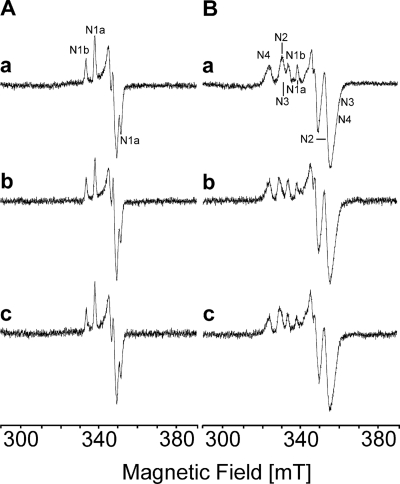
To determine whether the oxidation of NADPH by complex I led to a reduction of the Fe/S clusters, aliquots of the preparation were treated either with NADH or NADPH and was characterized by helium temperature EPR spectroscopy. The aim was not to determine the absolute amount of the reduced clusters but to detect the difference in the degree of reduction. Therefore, the amount of the individual clusters was estimated from the amplitude of the individual signals at the g values indicated below. Redox mediators were omitted because they give a strong signal at g = 2.00 overlapping with the gz of N1a (31). The spectra of the reconstituted complex reduced with either NADH or NADPH were very similar (Fig. 2). A comparison of both spectra revealed that NADPH reduced N1a (at g = 2.00; 40 K) by 100%, N1b (at g = 2.03, 40 K), N3 (at g = 2.04; 13 K), and N4 (at g = 2.09; 13 K) by 90% and N2 (at g = 2.05; 13 K) by 85% compared with the reduction of the Fe/S clusters by NADH, which was attributed to 100% (Fig. 2).
FIGURE 2.
Reduction of the Fe/S clusters of complex I and the E183HF variant by nucleotides. Complex I was reduced with a 1,000-fold molar excess of both NADH (trace a) and NADPH (trace b). The E183HF variant was reduced with a 1,000-fold molar excess NADPH (trace c). The spectra in A were recorded at 40 K and 2 milliwatts of microwave power, and the spectra in B were recorded at 13 K and 5 milliwatts of microwave power. Other EPR conditions were as follows: microwave frequency, 9.44 GHz; modulation amplitude, 0.6 mT; time constant: 0.124 s; scan rate: 17.9 mT/min.
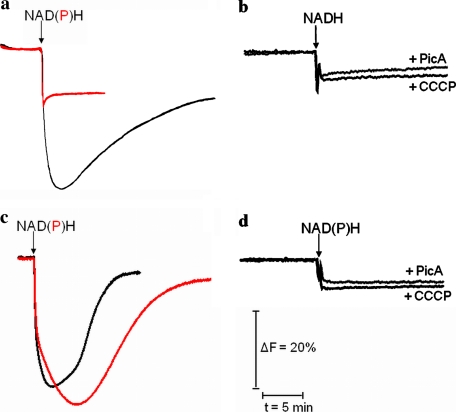
To investigate whether the NAD(P)H-driven redox reaction led to proton translocation, the complex was reconstituted in proteoliposomes (32), and the generation of a proton gradient was determined by the fluorescence of the pH-sensitive dye ACMA (Fig. 3). The addition of NADH gave rise to a proton gradient that was completely sensitive to the proton uncoupler CCCP and the specific inhibitor piericidin A (Fig. 3, a and b). Thus, the signal reflects the presence of a proton gradient generated by the redox reaction of the complex. No signal was obtained after an addition of 5 mm NADPH to the proteoliposomes. This could be due to the slow reaction rate of the complex with NADPH as a substrate compared with the proton leakiness of the proteoliposomes. However, it is also possible that the reaction of complex I with NADPH is not coupled with proton translocation.
FIGURE 3.
Generation of a proton gradient by complex I and the E183HF variant. The proteins were reconstituted in proteoliposomes, and the ACMA fluorescence was monitored (28). The reaction of complex I (a) and the E183HF variant (c) was started by adding either NADH (black curve) or NADPH (red curve) as indicated. The addition of NADPH to proteoliposomes containing complex I did not lead to the generation of a detectable proton gradient. As a control, either CCCP or piericidin A was added to the complex (b) before the reaction was started with the addition of NADH and to the variant (d) before the reaction was started with either NADH or NADPH.
Role of Glu-183F
From the mode of NADH binding (Fig. 1), we expected Glu-183F to be essential for the specificity of complex I for NADH. We inspected the homologues of NuoF in the databases to check whether the residue is conserved (supplemental Fig. S1). All sequences that were unambiguously identified as coding for the complex I subunit NuoF contained a glutamic acid at the homologous position.
Structural comparisons between other NAD- and NADP-binding proteins revealed that the space needed for binding the additional phosphate group of NADP is occupied by a hydrogen bond between a conserved aspartate and the adenosine ribose as described for NAD-dependent dehydrogenases (36). To accommodate the binding of NADP, an adjustment of the nucleotide binding region is necessary (37). It has been described that either a conformational change takes place leading to a hydrogen bond between a nitrogen of the backbone and a phosphate oxygen (38) or that the aspartic acid residue is replaced by an asparagine residue forming hydrogen bonds to the adenosine ribose and the phosphate group (39). For the NADP-dependent glutathione reductase, it was shown that the high affinity of NADP binding correlates with the presence of two basic amino acid residues around NADPH's additional phosphate group (40). Because of the data reported in the literature, we replaced the conserved Glu-183F of complex I by Asp, Gln, Asn, and His.
Generation and Characterization of Variants
The conserved Glu-183F was mutated using an E. coli strain lacking all 13 genes coding for subunits of complex I on the chromosome. This strain was transformed with the 21-kb expression plasmid pBADnuoHis containing the 13 complex I genes under the control of an inducible promoter (29). Because the plasmid is too large for mutagenesis, mutations were introduced in the 6-kb plasmid pCA24NnuoF containing only nuoF. The gene nuoF on the plasmid pBADnuoHis was inactivated by the insertion of an nptI-sacRB selection cartridge via λ-Red-mediated recombination. In a second recombineering step, the inactivated gene nuoF was individually replaced by the mutated versions of nuoF, again using λ-Red-mediated recombination. All mutations were confirmed by DNA sequencing.
Cytoplasmic membranes were prepared from the mutants, and the NAD(P)H conferred activities were measured (29). Compared with the parental strain, the mutant strains E183DF, E183NF, and E183HF exhibited a 2-fold increase in NADH/ferricyanide oxidoreductase activity, although that of the mutant strain E183QF was not significantly changed (Table 2). This indicates either an enhanced amount of complex I in the corresponding mutant membranes or an enhanced enzymatic turnover due to the mutation in the active site. The NADH oxidase activity of the mutants E183QF, E183NF, and E183HF was only slightly enhanced, although that of the mutant E183DF was increased more than 2-fold. In addition, the conservative substitution E183DF led to a more than 5-fold enhanced NADPH oxidase activity, although the NADPH oxidase activity of the other mutants was increased ∼2-fold (Table 2). The NAD(P)H oxidase activities were sensitive to piericidin A.
TABLE 2.
NADH/ferricyanide oxidoreductase activity (measured as ferricyanide reduction) and NAD(P)H oxidase activity (measured as oxygen consumption) of cytoplasmic membranes of various E. coli strains used in this study
| Strain | NADH/ferricyanide oxidoreductase activity | NADH oxidase activity | NADPH oxidase activity |
|---|---|---|---|
| μmol·min−1·mg−1 | |||
| Parental | 7.0 | 0.8 | 0.2 |
| E183DF | 15.4 | 1.8 | 1.1 |
| E183QF | 7.9 | 0.8 | 0.6 |
| E183NF | 15.4 | 1.1 | 0.5 |
| E183HF | 11.2 | 1.1 | 0.5 |
Purification and Characterization of Variants
The wild type complex and the variants were isolated from the corresponding strains by affinity chromatography (29). From 20 g of cells, ∼3–4 mg of protein were obtained from all strains (supplemental Tables S3–S6), indicating that the enhanced NADH/ferricyanide oxidoreductase activity of the mutant membranes (Table 2) reflects an enhanced catalytic activity and not an increased amount of the complex I variants in the membrane. SDS-PAGE of the preparations demonstrates the presence of all complex I subunits without any significant traces of impurities (supplemental Fig. S2).
After reconstitution in lipids, the kinetic parameters of the variants for the NAD(P)H:decyl-ubiquinone oxidoreductase activity were determined (Table 1). The E183NF and the E183QF variants showed a reactivity toward NADH like the complex from the parental strain. In contrast, the variants E183HF and E183DF exhibited a significantly higher reaction rate with NADH and higher affinity to NADH than the complex from the parental strain. All variants showed a better reactivity with NADPH than the complex from the parental strain, although to varying extents. The E183HF variant exhibited a 3-fold higher activity with NADPH and an 75-fold higher affinity. Most strikingly, the E183DF variant showed a 10-fold higher reaction rate with NADPH, which is even higher than the reaction rate of the complex from the parental strain with NADH. However, the affinity of the E183DF variant to NADPH was only 5-fold greater than that of the parental strain. Although its reactivity with NADH was not significantly changed, the E183QF variant showed a more than 40-fold higher affinity to NADPH and a 4-fold enhanced reaction rate. The NAD(P)H:decyl-ubiquinone oxidoreductase activity of the preparations of all variants was inhibited more than 95% by piericidin A with an IC50 of 5 μm, which is identical to that obtained for complex I from the parental strain. Thus, the variants exhibited NADPH:ubiquinone oxidoreductase activity. The catalytic efficiency with NADH was not enhanced in the E183NF and E183QF variants but was 3-fold higher in the E183DF and E183HF variants. Most strikingly, the catalytic efficiency with NADPH was enhanced 220-fold in the E183HF variant, 178-fold in the E183QF variant, 50-fold in the E183DF variant, and 12-fold in the E183NF variant (Table 1).
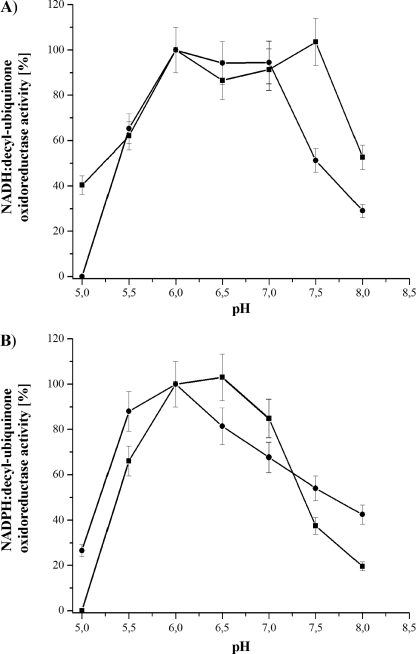
The pH dependence of the NAD(P)H oxidase activity of bovine mitochondria has been reported in the literature (22–25). To determine the pH dependence of the E. coli complex I NAD(P)H:decyl-ubiquinone oxidoreductase activity, the preparation was measured in the range from pH 5 to 8 (Fig. 4). The pH activity profile of the NADH:decyl-ubiquinone oxidoreductase activity showed a broad maximum between 6.0 and 7.5 with an abrupt decrease to more acidic and more basic pH values. The NADPH:decyl-ubiquinone oxidoreductase activity showed a similar dependence with a more narrow plateau between pH 6.0 and 6.5. The contribution of the conserved Glu-183F to the pH dependence of the activity was examined by measuring the activity of the variants at different pH values (Fig. 4). The activity profiles were very similar to that of the complex from the parental strain. The NADH oxidation profile of the E183HF variant showed a plateau of maximum activity between pH 6.0 and 7.0 and a greater decrease to more extreme pH values. The NADPH oxidation showed a distinct activity maximum at pH 6.0 and a marked decrease to more acidic pH values but a less marked decrease to more basic pH values. As all variants showed a maximum activity at pH 6.0 to 7.0, Glu-183F does not significantly contribute to the pH dependence of the NAD(P)H:decyl-ubiquinone oxidoreductase activity.
FIGURE 4.
pH dependence of the NADH:decyl-ubiquinone (A) and NADPH:decyl-ubiquinone (B) oxidoreductase activity of complex I (squares) and the E183HF variant (circles). The activity was measured between pH 5.0 and 8.0. All activities were more than 95% sensitive to the addition of 10 μm piericidin A. The maximum activity of the NADH and NADPH oxidation was 2.8 and 0.6 μm·min−1·mg−1 for complex I, respectively, and 4.0 and 1.3 μm·min−1·mg−1 for the E183HF variant. The other variants showed a similar pH activity profile with a maximum NADH oxidation rate between pH 6.0. and 7.0 and a maximum NADPH oxidation rate between 6.0 and 6.5.
To determine whether the Fe/S clusters of the variants were reduced by NADPH, EPR spectra at helium temperature were recorded as described above. The signals of all Fe/S clusters of the E183HF variant were detectable after an addition of NADPH to the preparation (Fig. 2, trace c). A comparison with the spectra of the NADH-reduced sample of complex I from the parental strain revealed that NADPH completely reduced the EPR-detectable Fe/S clusters in this variant.
The ability of the E183HF variant to translocate protons was examined after reconstitution in proteoliposomes (32). A pH-gradient generated by the redox reaction of complex I was detected by the ACMA fluorescence. Addition of NADH led to the generation of a pH gradient as with the wild type enzyme (Fig. 3c). In contrast to the wild type complex, the variant E183HF generated a pH gradient with NADPH as a substrate. Because of the higher turnover rate with NADH compared with that of NADPH (Table 1), the rise of the gradient is faster after an addition of NADH. The ACMA signals correspond to a proton gradient established by the redox reaction of the variant as it is sensitive to piericidin A and CCCP (Fig. 3d). The different rate of the decay of the NADH- and NADPH-induced ACMA quench is due to the different tightness of the proteoliposomes.
Generation of Reactive Oxygen Species
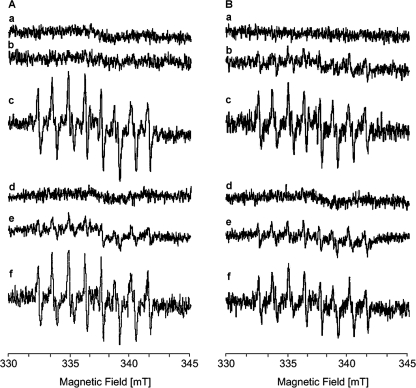
Studies on mitochondria have shown that complex I-mediated lipid peroxidation due to the generation of ROS is enhanced in the presence of NADPH (41). Therefore, we examined whether an addition of either NADH or NADPH to complex I and the E183HF variant gives rise to the formation of superoxide or hydrogen peroxide radicals. ROS were directly detected by scavenging radicals with the phosphorylated nitrone-type spin trap compound DEPMPO. The DEPMPO-OOH and DEPMPO-OH adduct radicals are easily detected by EPR spectroscopy due to their long half-life (t½ of ∼8 min) and its distinct superoxide trapping rate (42). The sample containing either complex I or the variant in phospholipids showed a similar behavior; no EPR signals were detectable in the presence of a 10-fold molar excess of decyl-ubiquinone and 100 mm DEPMPO or in the absence of any nucleotides (Fig. 5). An addition of 1 mm NADH led to a small background signal that could hardly be detected in the complex from the parental strain but was clearly present in the variant. However, the addition of 1 mm NADPH gave rise to the characteristic EPR signals of the DEPMPO-OOH adduct radical. These signals increased with time concomitant with the appearance of signals deriving from the DEPMPO-OH adduct radical. The amplitude of the signals was higher with the complex from the parental strain than with the variant. The formation of the EPR signals of the spin trap was completely suppressed by the addition of superoxide dismutase. The generation of superoxide depended on the presence of decyl-ubiquinone in the assay because the signals of the spin trap were considerably diminished in the samples lacking decyl-ubiquinone. No signals were detected after an addition of NADPH to phospholipids lacking the complex in the presence of decyl-ubiquinone. In addition, the appearance of the signals was not inhibited by an addition of piericidin A (Fig. 5).
FIGURE 5.
NAD(P)H-induced superoxide production by complex I (A) and the E183HF variant (B). The spectra were recorded with aliquots of the proteins reconstituted with phospholipids in the presence of 100 mm DEPMPO and 100 μm decyl-ubiquinone (trace a), plus 1 mm NADH (trace b), plus 1 mm NADPH (trace c), plus 1 mm NADPH and superoxide dismutase (100 units/ml) (trace d), plus 1 mm NADPH but without decyl-ubiquinone (trace e), and plus 1 mm NADPH and 20 μm piericidin A (trace f). Other EPR conditions were as follows: microwave frequency, 9.65 GHz; modulation amplitude, 0.1 mT; time constant: 0.164 s; scan rate: 5.4 mT/min.
To obtain information about the rate and the total amount of superoxide and H2O2 formation under various experimental conditions, the ROS production was determined by measuring the horseradish peroxidase-dependent oxidation of Amplex Red (Table 3). Neither H2O2 nor superoxide was produced in the absence of the nucleotides. The addition of NADH to complex I led to a limited ROS production, although the amount was enhanced more than 2-fold by the addition of NADPH. The variant showed an increased ROS production in the presence of NADH but only a slight increase after the addition of NADPH. The specific activity of complex I with NADPH is lower than that of the E183HF variant (Table 1). However, complex I produced more ROS than the variant in the presence of NADPH (Fig. 5; Table 3). Simultaneous measurement of the NADPH oxidation rate revealed that the ROS production added up to 14% of the NADPH oxidation in complex I, whereas only 4% was used in the side reaction in the variant. Addition of piericidin A in the presence of NADPH had just a small effect on the production of superoxide and H2O2 by the variant but it further enhanced the production by complex I. Significantly less ROS were formed in the absence of decyl-ubiquinone in both samples. Thus, the data obtained by the Amplex Red assay are in agreement with those obtained by the spin trap method.
TABLE 3.
Superoxide and H2O2 production by complex I and the E183HF variant measured as Amplex Red oxidation
Piericidin A is abbreviated as PicA; decyl-ubiquinone is abbreviated as Dec-Q.
| Sample | NAD(P)H-induced superoxide and H2O2 production |
||||
|---|---|---|---|---|---|
| None | +NADH | +NADPH | +NADPH, +PicA | +NADPH, −Dec-Q | |
| nmol·min−1·mg−1 | |||||
| Complex I | 0 | 17 ± 0.3 | 40 ± 0.9 | 75 ± 1.2 | 26 ± 0.7 |
| E183HF | 0 | 25 ± 0.4 | 29 ± 0.6 | 23 ± 0.6 | 11 ± 0.3 |
From these data, we propose that under these experimental conditions the ROS evolved at the NADH-binding site were mediated to a certain extent by decyl-ubiquinone, which has a distinct water solubility. Although the E183HF variant shows a strongly enhanced catalytic efficiency with NADPH, this reactivity is coupled with the generation of ROS. This might be due either to the low KmNADPH, which is twice the KmNADH of the complex from the parental strain, or to the slow reaction rate with NADPH, which is one-third that of the wild type complex with NADH as a substrate (Table 1). The E183DF variant is most helpful in discriminating between these two possibilities because it exhibits a very low KmNADPH of 390 μm but has a faster reaction rate with NADPH than the wild type complex with NADH (Table 1). This variant also produces ROS under these experimental conditions (supplemental Fig. S3), demonstrating that the binding of the nucleotide determines whether ROS are produced or not.
DISCUSSION
By changing a single amino acid residue in a complex consisting of nearly 5,000 amino acid residues, we have created a novel enzymatic function, an energy-converting NADPH:ubiquinone oxidoreductase, which has so far not been described in natural systems. By replacing Glu-183F with a histidine residue, the catalytic efficiency of the respiratory complex I with NADPH as substrate was more than 200-fold enhanced (Table 1). This is experimental proof of the assumption that Glu-183F is a key residue in substrate recognition as derived from structural analysis. The strictly conserved Glu-183F (supplemental Fig. S1) ensures that complex I only uses NADH in vivo and not NADPH. Although the catalytic efficiency of the E183HF variant with NADH is 3-fold higher than with the wild type complex (Table 1), the mutation has not evolved in the sequenced species, most likely because the higher efficiency is at the cost of the loss of substrate selectivity. In addition, the oxidation of NADPH by the variants produced in this study is always coupled with the generation of ROS. Thus, a conserved glutamic acid residue at this position in the nucleotide binding domain of NAD-dependent dehydrogenases is indicative of a selective use of NADH (36, 37, 40, 43). Some examples in the literature discuss that it is not a trivial undertaking to change the nature of the nucleotide cofactor for dehydrogenases by site-directed mutagenesis, even in more simply structured enzymes (44).
The various mutations had different effects on the NAD(P)H oxidation kinetics (Table 1). Changing the acidic amino acids to the corresponding amides had no effect on NADH binding. The substitution to an asparagine residue increased the catalytic efficiency with NADPH 10-fold, whereas the substitution to a glutamine residue led to a 177-fold enhanced catalytic efficiency with NADPH indicating a specific interaction most likely between the amide oxygen atom of the side chain and NADPH. The data indicate that the electrostatic repulsion between the side chain carboxylate of Glu-183F and the phosphate group bound to the adenosine ribose is the key interaction preventing a rapid oxidation of NADPH by complex I. The conservative replacement of Glu-183F by an aspartic acid residue led to a slightly higher catalytic efficiency with NADH but to a 50-fold higher efficiency with NADPH, which can be explained by the less steric hindrance for binding NADPH. The low affinity of this variant to NADPH (Table 1) reflects the contribution of the electrostatic repulsion in this variant. The introduction of a histidine residue at this position led to a 3-fold enhanced catalytic efficiency with NADH but a drastic 220-fold increase of the efficiency with NADPH (Table 1). This might derive from a less steric hindrance at this position and an electrostatic stabilization of the negative charge of the phosphate group (40).
The single mutation neither changed the other activities of complex I such as the reduction of the Fe/S clusters (Fig. 2), the reduction of the quinone (Tables 1 and 2), and the translocation of protons (Fig. 3) nor did it influence the coupling of the processes. Using the proteoliposomes, we were not able to detect proton translocation by the complex from the parental strain with NADPH. This is most likely due to the low reaction rate of this enzyme with NADPH in combination with the proton leakiness of the proteoliposomes. The assumption that the mutations introduced resulted in NADPH-driven proton translocation is very unlikely, because the sites of nucleotide binding and proton translocation are separated by a distance of more than 100 Å (20, 21).
The oxidation of NADPH by complex I and its variants is always accompanied by ROS production, although the NADPH-induced ROS production is slightly diminished in the variants (Fig. 5; Table 3). However, they also show, in contrast to the wild type complex, an NADH-induced ROS production. The source of ROS evolving during the redox reaction of complex I is still under discussion (45). Most investigators agree on the flavin site, whereas the ubiquinone-binding site and the Fe/S clusters N1a and N2 have also been proposed to be responsible for the generation of ROS (46). Recently, it was proposed that the site where ROS are generated is variable and determined by the reduction state of the individual cofactors (47). Thus, the various sites of ROS production reported under different experimental conditions might derive from different states of the complex (47). Under our experimental conditions the ROS are produced at the flavin site because piericidin A did not inhibit ROS production (Fig. 5 and Table 3).
The E183DF variant, exhibiting a higher maximal reaction rate with NADPH than complex I with NADH and a very low affinity to NADPH (Table 1), showed an NADPH-induced ROS production. In addition, the NADPH-induced ROS production is enhanced in complex I compared with the E183HF variant (Fig. 3; Table 3) because the portion of electrons involved in the side reaction leading to ROS production is more than 4-fold enhanced. These data indicate that the mode of nucleotide binding contributes to the possibility of ROS being generated at this site. To understand the molecular details of this process, a high resolution structure of the nucleotide-binding site of complex I with bound NADP(H) is needed.
Supplementary Material
Acknowledgments
We kindly thank Helga Lay for an excellent HPLC analysis of the nucleotides and Lothar Kussmaul and Karoline Aierstock, Boehringer Ingelheim, for their help in establishing the Amplex Red assay. We are grateful to Linda Williams for help in preparing the manuscript.
This work was supported by the Volkswagen Stiftung and the Deutsche Forschungsgemeinschaft.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S6 and Figs. S1–S3.
M. Vranas and T. Friedrich, unpublished results.
- complex I
- proton-pumping NADH:ubiquinone oxidoreductase
- ACMA
- 9-amino-6-chloro-2-methoxyacridine
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- Fe/S
- iron-sulfur
- decyl-ubiquinone
- 2,3-dimethoxy-5-methyl-6-decylbenzoquinone
- ROS
- reactive oxygen species
- mT
- millitesla
- DEPMPO
- 5-(diethoxyphosphoryl)-5-methyl-1-pyrroline-N-oxide, (2-methyl-3,4-dihydro-1-oxide-2H-pyrrol-2-yl)diethylphosphonate.
REFERENCES
- 1. Weiss H., Friedrich T., Hofhaus G., Preis D. (1991) Eur. J. Biochem. 197, 563–576 [DOI] [PubMed] [Google Scholar]
- 2. Walker J. E. (1992) Q. Rev. Biophys. 25, 253–324 [DOI] [PubMed] [Google Scholar]
- 3. Ohnishi T. (1998) Biochim. Biophys. Acta 1364, 186–206 [DOI] [PubMed] [Google Scholar]
- 4. Friedrich T. (2001) J. Bioenerg. Biomembr. 33, 169–177 [DOI] [PubMed] [Google Scholar]
- 5. Yagi T., Matsuno-Yagi A. (2003) Biochemistry 42, 2266–2274 [DOI] [PubMed] [Google Scholar]
- 6. Brandt U. (2006) Annu. Rev. Biochem. 75, 69–92 [DOI] [PubMed] [Google Scholar]
- 7. Carroll J., Fearnley I. M., Skehel J. M., Shannon R. J., Hirst J., Walker J. E. (2006) J. Biol. Chem. 281, 32724–32727 [DOI] [PubMed] [Google Scholar]
- 8. Friedrich T. (1998) Biochim. Biophys. Acta 1364, 134–146 [DOI] [PubMed] [Google Scholar]
- 9. Friedrich T., Scheide D. (2000) FEBS Lett. 479, 1–5 [DOI] [PubMed] [Google Scholar]
- 10. Friedrich T., Böttcher B. (2004) Biochim. Biophys. Acta 1608, 1–9 [DOI] [PubMed] [Google Scholar]
- 11. Baranova E. A., Holt P. J., Sazanov L. A. (2007) J. Mol. Biol. 366, 140–154 [DOI] [PubMed] [Google Scholar]
- 12. Sazanov L. A., Hinchliffe P. (2006) Science 311, 1430–1436 [DOI] [PubMed] [Google Scholar]
- 13. Berrisford J. M., Sazanov L. A. (2009) J. Biol. Chem. 284, 29773–29783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dupuis A., Prieur I., Lunardi J. (2001) J. Bioenerg. Biomembr. 33, 159–168 [DOI] [PubMed] [Google Scholar]
- 15. Fendel U., Tocilescu M. A., Kerscher S., Brandt U. (2008) Biochim. Biophys. Acta 1777, 660–665 [DOI] [PubMed] [Google Scholar]
- 16. Friedrich T., Weiss H. (1997) J. Theor. Biol. 187, 529–540 [DOI] [PubMed] [Google Scholar]
- 17. Mathiesen C., Hägerhäll C. (2003) FEBS Lett. 549, 7–13 [DOI] [PubMed] [Google Scholar]
- 18. Brandt U., Kerscher S., Dröse S., Zwicker K., Zickermann V. (2003) FEBS Lett. 545, 9–17 [DOI] [PubMed] [Google Scholar]
- 19. Mamedova A. A., Holt P. J., Carroll J., Sazanov L. A. (2004) J. Biol. Chem. 279, 23830–23836 [DOI] [PubMed] [Google Scholar]
- 20. Efremov R. G., Baradaran R., Sazanov L. A. (2010) Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 21. Hunte C., Zickermann V., Brandt U. (2010) Science 329, 448–451 [DOI] [PubMed] [Google Scholar]
- 22. Vinogradov A. D. (1998) Biochim. Biophys. Acta 1364, 169–185 [DOI] [PubMed] [Google Scholar]
- 23. Hatefi Y., Hanstein W. G. (1973) Biochemistry 12, 3515–3522 [DOI] [PubMed] [Google Scholar]
- 24. Djavadi-Ohaniance L., Hatefi H. (1975) J. Biol. Chem. 250, 9397–9403 [PubMed] [Google Scholar]
- 25. Rydström J., Montelius J., Bäckström D., Ernster L. (1978) Biochim. Biophys. Acta 501, 370–380 [DOI] [PubMed] [Google Scholar]
- 26. Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Court D. L., Sawitzke J. A., Thomason L. C. (2002) Annu. Rev. Genet. 36, 361–388 [DOI] [PubMed] [Google Scholar]
- 28. Pohl T., Bauer T., Dörner K., Stolpe S., Sell P., Zocher G., Friedrich T. (2007) Biochemistry 46, 6588–6596 [DOI] [PubMed] [Google Scholar]
- 29. Pohl T., Uhlmann M., Kaufenstein M., Friedrich T. (2007) Biochemistry 46, 10694–10702 [DOI] [PubMed] [Google Scholar]
- 30. Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H. (2005) DNA Res. 12, 291–299 [DOI] [PubMed] [Google Scholar]
- 31. Leif H., Sled V. D., Ohnishi T., Weiss H., Friedrich T. (1995) Eur. J. Biochem. 230, 538–548 [DOI] [PubMed] [Google Scholar]
- 32. Stolpe S., Friedrich T. (2004) J. Biol. Chem. 279, 18377–18383 [DOI] [PubMed] [Google Scholar]
- 33. Markham K. A., Sikorski R. S., Kohen A. (2004) Anal. Biochem. 325, 62–67 [DOI] [PubMed] [Google Scholar]
- 34. Albracht S. P., Mariette A., de Jong P. (1997) Biochim. Biophys. Acta 1318, 92–106 [DOI] [PubMed] [Google Scholar]
- 35. Hanes C. S. (1932) Biochem. J. 26, 1406–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lesk A. M. (1995) Curr. Opin. Struct. Biol. 5, 775–783 [DOI] [PubMed] [Google Scholar]
- 37. Mittl P. R., Berry A., Scrutton N. S., Perham R. N., Schulz G. E. (1994) Protein Sci. 3, 1504–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thorn J. M., Barton J. D., Dixon N. E., Ollis D. L., Edwards K. J. (1995) J. Mol. Biol. 249, 785–799 [DOI] [PubMed] [Google Scholar]
- 39. Phillips C., Gover S., Adams M. J. (1995) Acta Crystallogr. D Biol. Crystallogr. 51, 290–304 [DOI] [PubMed] [Google Scholar]
- 40. Scrutton N. S., Berry A., Perham R. N. (1990) Nature 343, 38–43 [DOI] [PubMed] [Google Scholar]
- 41. Glinn M. A., Lee C. P., Ernster L. (1997) Biochim. Biophys. Acta 1318, 246–254 [DOI] [PubMed] [Google Scholar]
- 42. Vasquez-Vivar J., Kalyanaraman B., Kennedy M. C. (2000) J. Biol. Chem. 275, 14064–14069 [DOI] [PubMed] [Google Scholar]
- 43. Schulz G. E. (1992) Curr. Opin. Struct. Biol. 2, 61–67 [Google Scholar]
- 44. Dudek H. M., Torres Pazmiño D. E., Rodríguez C., de Gonzalo G., Gotor V., Fraaije M. W. (2010) Appl. Microbiol. Biotechnol. 88, 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hirst J., King M. S., Pryde K. R. (2008) Biochem. Soc. Trans. 36, 976–980 [DOI] [PubMed] [Google Scholar]
- 46. Kussmaul L., Hirst J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ohnishi S. T., Shinzawa-Itoh K., Ohta K., Yoshikawa S., Ohnishi T. (2010) Biochim. Biophys. Acta 1797, 1901–1909 [DOI] [PubMed] [Google Scholar]
- 48. Bondi A. (1964) J. Phys. Chem. 68, 441–451 [Google Scholar]
- 49. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.