Abstract
Archaeal and eukaryotic tRNA (N2,N2-guanine)-dimethyltransferase (Trm1) produces N2,N2-dimethylguanine at position 26 in tRNA. In contrast, Trm1 from Aquifex aeolicus, a hyper-thermophilic eubacterium, modifies G27 as well as G26. Here, a gel mobility shift assay revealed that the T-arm in tRNA is the binding site of A. aeolicus Trm1. To address the multisite specificity, we performed an x-ray crystal structure study. The overall structure of A. aeolicus Trm1 is similar to that of archaeal Trm1, although there is a zinc-cysteine cluster in the C-terminal domain of A. aeolicus Trm1. The N-terminal domain is a typical catalytic domain of S-adenosyl-l-methionine-dependent methyltransferases. On the basis of the crystal structure and amino acid sequence alignment, we prepared 30 mutant Trm1 proteins. These mutant proteins clarified residues important for S-adenosyl-l-methionine binding and enabled us to propose a hypothetical reaction mechanism. Furthermore, the tRNA-binding site was also elucidated by methyl transfer assay and gel mobility shift assay. The electrostatic potential surface models of A. aeolicus and archaeal Trm1 proteins demonstrated that the distribution of positive charges differs between the two proteins. We constructed a tRNA-docking model, in which the T-arm structure was placed onto the large area of positive charge, which is the expected tRNA-binding site, of A. aeolicus Trm1. In this model, the target G26 base can be placed near the catalytic pocket; however, the nucleotide at position 27 gains closer access to the pocket. Thus, this docking model introduces a rational explanation of the multisite specificity of A. aeolicus Trm1.
Keywords: Crystal Structure, RNA Methylation, RNA Methyltransferase, RNA Modification, RNA-Protein Interaction, Transfer RNA (tRNA)
Introduction
Methylation is one of the most common chemical modifications that occurs in a broad range of biomolecules, including nucleic acids, proteins, lipids, and small compounds. It is implicated in a variety of cellular processes, such as translation, transcription, processing of RNA, epigenetics, development, carcinogenesis, neurotransmission, cellular transport, infection, and bacterial host defense. Among the RNA species, methylated nucleotides appear in most noncoding RNAs, constituting more than half of the post-transcriptional modifications identified so far. In particular, tRNA contains abundant methylated nucleotides, which stabilize the L-shaped tRNA structure and improve their molecular recognition (1–3).
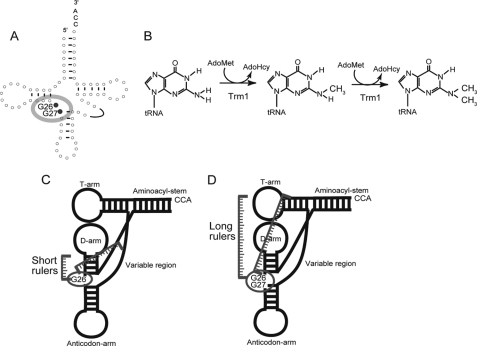
Among the methylated nucleotides in tRNA, N2,N2-dimethylguanine at position 26 (m22G26) is a common modification and is generated by tRNA (N2,N2-guanine)-dimethyltransferase (tRNA (m22G26) methyltransferase (m2, 2G26); EC 2.1.1.32) (Fig. 1, A and B) (2, 3). This enzyme catalyzes the methyl transfer reaction from S-adenosyl-l-methionine (AdoMet)3 to the N2 atom of G26, which is located at the junction between the D-arm and the anticodon-arm in tRNA. In the reaction, two molecules of AdoMet are consumed, and N2-methylguanine at position 26 (m2G26) is generated as an intermediate (4). Thus, in some cases, this enzyme catalyzes the transfer of only one methyl group and functions as a tRNA (m2G26) methyltransferase (5). The enzymatic activity was initially detected in rat liver (6, 7) and subsequently found in various organisms (8–12). The most highly purified enzyme from a native source was obtained from Tetrahymena pyriformis (4). The responsible gene was first determined to be trm1 from Saccharomyces cerevisiae (13, 14) and then experimentally identified from various eukaryotes (Schizosaccharomyces pombe (15), Caenorhabditis elegans (16), and human (17)) and archaea (Pyrococcus furiosus (18), Pyrococcus horikoshii (19), and Haloferax volcanii (20)), consistent with the distribution of the m22G26 (or m2G26) modification in tRNA. Furthermore, we have recently reported that Aquifex aeolicus, a hyper-thermophilic eubacterium, has a Trm1 protein that catalyzes methyl transfer not only to G26 but also to G27 (21).
FIGURE 1.
Methylation by Trm1 enzymes. A, methylation sites, G26 and G27, are highlighted in the cloverleaf structure of tRNA. Eukaryotic and archaeal Trm1 are single site-specific enzymes that modify only G26. In contrast, A. aeolicus Trm1 is a multisite-specific enzyme, which modifies both G26 and G27. B, Trm1 catalyzes methyl transfer to the target guanine from AdoMet, and m2G is produced as an intermediate. AdoMet is converted to AdoHcy by the reaction. C and D are slight modifications of Fig. 9 in our previous report (21). C, eukaryotic and archaeal Trm1 reported thus far recognize the D-arm and variable region in tRNA. D, in contrast, A. aeolicus Trm1 is expected to recognize the T-arm structure. Thus, the distance(s) from the recognition site(s) in tRNA to the modification site(s) may affect both single and multisite specificities.
In general, methylated nucleotides in tRNA are post-transcriptionally produced by site-specific methyltransferases (3). However, there are some cases in which one tRNA methyltransferase modifies multiple sites in substrate tRNA. Thus, some tRNA methyltransferases have a multisite specificity. For example, Pyrococcus abyssi TrmI catalyzes methyl transfer to both A57 and A58, forming m1A57 and m1A58, respectively (22). In addition, eukaryotic Trm4 is responsible for 5-methylcytidine formation at positions 34, 40, 48, and 49 in tRNA (23). Furthermore, it has been reported that P. abyssi PAB1947 protein (archaeal Trm4) changes its site specificity in the presence of PAB1946 protein (archaese) (24). Moreover, S. cerevisiae Trm7 protein catalyzes 2′-O-methylation of nucleotides at positions 32 and 34 (25). As described above, we previously reported that A. aeolicus Trm1 has a multisite specificity (21). Although these examples have been reported, there is little knowledge concerning the mechanism of multisite recognition. In this study, we focus on the multisite specificity of A. aeolicus Trm1.
EXPERIMENTAL PROCEDURES
Materials
[methyl-14C]AdoMet (1.95 GBq/mmol) and [methyl-3H]AdoMet (2.47 TBq/mmol) were purchased from ICN. Cold AdoMet was obtained from Sigma. DE52 is a product of Whatman. CM-Toyopearl 650 m was from Tosoh. DNA oligomers were bought from Invitrogen, and T7 RNA polymerase was from Toyobo. Other chemical reagents were of analytical grade.
Preparation of Recombinant A. aeolicus Trm1
The recombinant A. aeolicus Trm1 was purified according to our previous report (21). Site-directed mutagenesis was performed by a QuikChange® mutagenesis kit (Stratagene).
Measurement of Enzymatic Activity and Kinetic Parameters
The standard assay used during the purification process measured the incorporation of 14C-methyl groups from [methyl-14C]AdoMet into the A. aeolicus tRNAHis transcript by incubating 0.1 μm enzyme, 11 μm transcript, and 38 μm [methyl-14C]AdoMet in 45 μl of buffer A (50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 6 mm 2-mercaptoethanol, and 50 mm KCl) for 15 min at 55 °C. An aliquot (30 μl) was then used for the filter assay. RNA transcripts were prepared as reported previously (26). If discrimination between the m2G and m22G formation activities was necessary, we employed two-dimensional thin layer chromatography as described in our previous reports (21, 27). Prior to measuring the kinetic parameters, we performed time course experiments at 55 °C with 0.1 μm Trm1, 11 μm yeast tRNAPhe transcript, and 38 μm [methyl-14C]AdoMet in 210 μl of buffer A. Because the three-dimensional structure of yeast tRNAPhe has been well studied and this tRNA has a G26C27 sequence (28, 29), we used the yeast tRNAPhe transcript as substrate. Aliquots (30 μl each) were taken at appropriate times (0, 2, 5, 7.5, 10, and 15 min), and the formations of 14C-pm2G and 14C-pm22G were monitored by two-dimensional TLC. Under these conditions, only pm2G increased linearly for the first 10 min, and m22G formation was barely observable; the pm22G content was less than 5% of the pm2G content in the sample at 10 min. After this pilot experiment, we determined the kinetic parameters of the first methylation (m2G26 formation). For measurements of kinetic parameters for AdoMet, 0.1 μm Trm1, 11 μm tRNAPhe transcript, and various concentrations of [3H]AdoMet were incubated for 10 min at 55 °C.
Gel Mobility Shift Assay
A gel mobility shift assay was performed according to our previous reports (30, 31).
Crystallization and X-ray Crystal Structure Study
Purified protein (10 mg/ml) in buffer B (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm dithiothreitol) was mixed with an equal volume of buffer C (100 mm sodium citrate, pH 5.5, 2 m ammonium sulfate) in the presence of 1 mm AdoMet or sinefungin (AdoMet analog) and then crystallized at 4 °C by the sitting drop method. The Trm1 crystals grew within 14 days. The crystals were flash-cooled to 100 K using liquid paraffin oil as the cryoprotectant. X-ray diffraction data were collected at the Photon Factory NW12A and BL5A beamlines for the AdoMet- and sinefungin-bound crystals, respectively, and processed with the HKL2000 program suite (32). The structures were solved with the program MOLREP (33) by molecular replacement using the P. horikoshii Trm1 structure (PDB code 2EJT) as a search model. The models were built with the program Coot (34) and refined with the programs CNS (35) and Phenix (36). The information of crystal data collection is given in supplemental Table 1. The Rwork/Rfree factors of refined coordinates of Trm1-sinefungin complex crystal structure were 0.186/0.219.
Computer Programs for Figures
The structural figures were generated by using PyMOL (DeLano Scientific, Palo Alto, CA), and the electrostatic potential surface models were calculated by using APBS (37). The schematic diagram of interaction of sinefungin with the amino acid residues in Trm1 was generated by using LIGPLOT (38). The amino acid sequence alignment was generated by using ClustalW 1.83 (39) and ESPript (40) programs.
Inductivity Coupled Plasma Emission Spectrometry
The inductivity coupled plasma emission analysis was performed by the Chemical Analysis Laboratory, University of Georgia.
RESULTS
A. aeolicus Trm1 Recognizes the T-arm Structure in tRNA
The tRNA recognition mechanisms of eukaryotic and archaeal Trm1 proteins have been well studied (9–12, 16–18). Although there are several differences in the tRNA recognition mechanisms of eukaryotic and archaeal Trm1, both enzymes recognize the D-arm structure and variable region in tRNA (Fig. 1C). In our previous study (21), we reported that A. aeolicus Trm1, a eubacterial enzyme, has a completely different tRNA recognition mechanism from those of eukaryotic and archaeal Trm1. First, A. aeolicus Trm1 catalyzes methyl transfers not only to G26 but also to G27 in tRNA, whereas all eukaryotic and archaeal enzymes reported thus far catalyze methyl transfer to only G26. Second, A. aeolicus Trm1 catalyzes methyl transfers to class II tRNAs as well as all class I tRNAs tested, showing that the size and sequence of the variable region are not recognized by A. aeolicus Trm1. Third, tRNA transcripts containing G26 and/or G27, which were tested in our previous study, have different D-arm structures; however, all tRNA transcripts are methylated by A. aeolicus Trm1. Thus, the D-arm structure is not a positive determinant for A. aeolicus Trm1. Fourth, truncated tRNA experiments in our previous study showed that the T-arm structure is required for methylation by A. aeolicus Trm1. Taking these experimental results together, we proposed a long distance recognition mechanism of A. aeolicus Trm1 in our previous study (Fig. 1D).
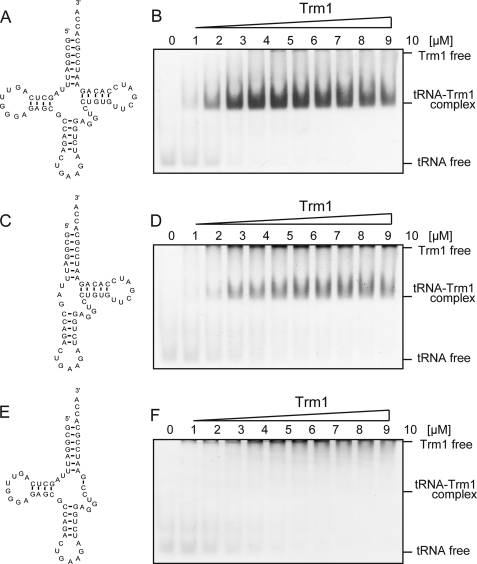
In our hypothetical mechanism, the T-arm structure is a key region in the substrate recognition mechanism of A. aeolicus Trm1. To confirm this idea, here we initially performed a gel mobility shift assay with A. aeolicus Trm1 (Fig. 2). This gel mobility assay system was originally devised for detection of tRNA elongation factor Tu-GTP ternary complex (41, 42). Because this system was used for measurement of affinity of nematoda mitochondrial elongation factor Tu for T-armless tRNAs (41), we selected this system. One of the virtues of this system is the usage of buffer containing 5 mm MgCl2 to maintain the L-shaped tRNA structure. Therefore, this gel shift system is applicable for various tRNA-related proteins. In the case of A. aeolicus Trm1, the enzyme does not migrate into the gel in the absence of RNA because the isoelectric point (pI) of A. aeolicus Trm1 is 9.06. When a full-length tRNAPhe transcript was used in the assay, A. aeolicus Trm1 clearly formed a tRNA-protein complex (Fig. 2, A and B), as visualized by double staining with methylene blue and Coomassie Brilliant Blue (Fig. 2B). When the D-arm structure was deleted, a similar band corresponding to the tRNA-Trm1 complex was observed (Fig. 2, C and D). In contrast, when the T-arm structure was deleted, the band corresponding to the tRNA-Trm1 complex disappeared (Fig. 2, E and F), demonstrating that the T-arm structure is indeed essential for substrate RNA binding by A. aeolicus Trm1. In these experiments, because the band corresponding to free tRNA was broadened in the presence of A. aeolicus Trm1, we avoided the calculation of Kd values.
FIGURE 2.
Gel mobility shift assay with A. aeolicus Trm1. A, cloverleaf structure of the full-length yeast tRNAPhe transcript. B, as the Trm1 concentration increased, full-length tRNAPhe and A. aeolicus Trm1 formed an RNA-protein complex. The gel was double-stained with Coomassie Brilliant Blue for the detection of protein and methylene blue for the detection of RNA. C, mutant tRNAPhe transcript, in which the D-arm was deleted. D, as the Trm1 concentration increased, the mutant tRNA transcript lacking the D-arm formed an RNA-protein complex although with decreased affinity. E, mutant tRNAPhe transcript, in which the T-arm was deleted. F, in contrast to B and D, bands corresponding to the RNA-protein complex were not observed with the mutant tRNA transcript lacking the D-arm, although bands corresponding to tRNA free were broadened with the increase in Trm1 concentration.
Overall Structure of A. aeolicus Trm1
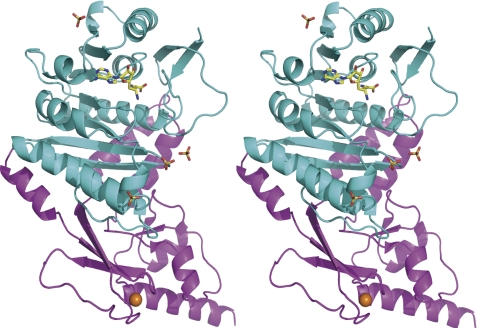
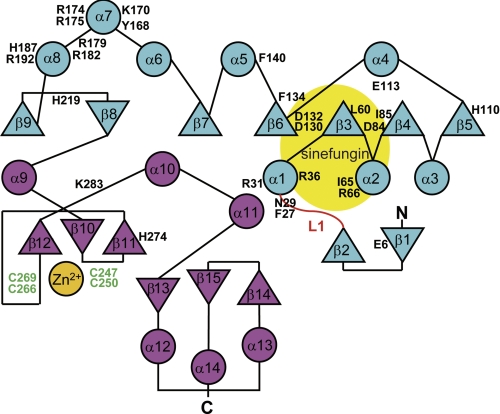
A. aeolicus Trm1 recognizes the T-arm structure in tRNA. This feature is considerably different from the mechanisms of archaeal and eukaryotic Trm1. To obtain structural information on A. aeolicus Trm1, we performed an x-ray crystal structure study. We have solved the structures of AdoMet-Trm1 (PDB code 3AXT) and sinefungin-Trm1 (PDB code 3AXS) complexes (supplemental Table 1). Sinefungin is an analog inhibitor, in which the activated methyl group of AdoMet is replaced by an amino group (see Fig. 5B) (43, 44). Because the structures of AdoMet-Trm1 and sinefungin-Trm1 complexes are almost identical and the resolution of sinefungin-Trm1 complex (2.15 Å) is better than that of the AdoMet-Trm1 complex, we performed this study based on the structure of sinefungin-Trm1 complex (Fig. 3). A. aeolicus Trm1 includes two domains as follows: an N-terminal Rossmann fold domain (Fig. 3, cyan), and a C-terminal novel structure domain (magenta). Although the C-terminal domain structure of A. aeolicus Trm1 is clearly different from that of P. horikoshii Trm1 (archaeal enzyme; PDB codes 2EJT and 2DUL) (19), the overall structure and topology of A. aeolicus Trm1 is similar to that of P. horikoshii Trm1 (Fig. 4). The N-terminal Rossmann fold domain is a typical catalytic domain of class I AdoMet-dependent methyltransferases. AdoMet-dependent methyltransferases are categorized into five classes based on the type of fold present in the catalytic domain (45). Here, a bound sinefungin (yellow) was found in the Rossmann fold domain (Figs. 3 and 4). The C-terminal domain was found to have a novel structure, in which one zinc atom (Fig. 4, orange) was bound to four cysteine residues (Cys-247, Cys-250, Cys-266, and Cys-269) (Figs. 3 and 4). Furthermore, four sulfate ions were bound to A. aeolicus Trm1; one sulfate ion was bound to the His-110 residue in the N-terminal domain, and three sulfate ions were bound to the border area between the N- and C-terminal domains (Fig. 3). These sulfate ions seemed to bind Trm1 instead of the phosphate groups in tRNA.
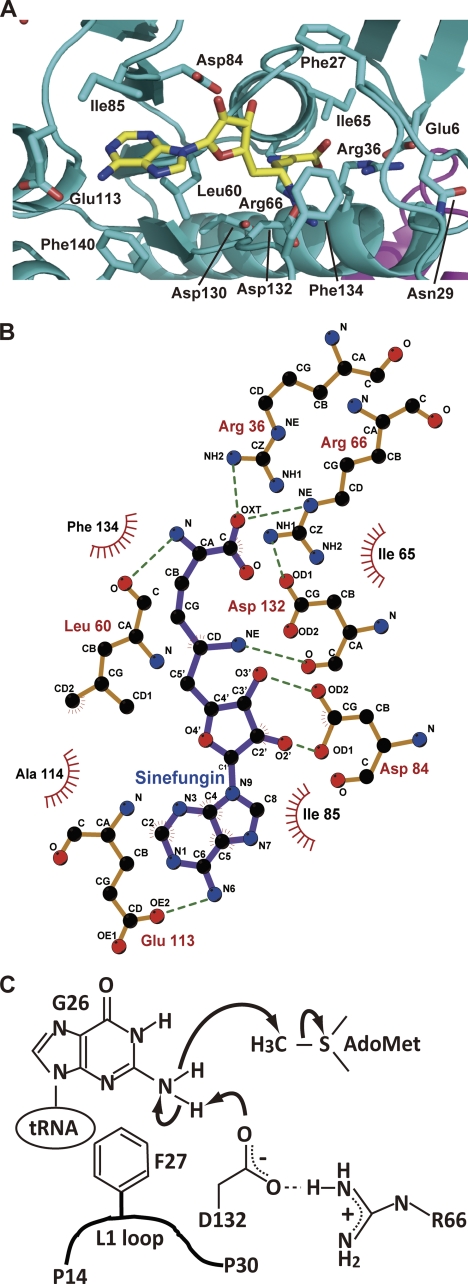
FIGURE 5.
AdoMet-binding site and hypothetical reaction mechanism. A, structure of AdoMet-binding site and bound sinefungin. Amino acid residues around the AdoMet-binding site and sinefungin are highlighted by stick models. B, interactions between amino acid residues and sinefungin. C, hypothetical catalytic mechanism. The catalytic center is the Asp-132 residue. As described in the text, the Arg-66 residue contributes to the electron relay pathway. The activated amino group of G26 in tRNA attacks the methyl group of AdoMet. Phe-27 in the L1-loop probably contributes to fixation of the target guanine base by a hydrophobic interaction.
FIGURE 3.
Stereo view of the overall structure of A. aeolicus Trm1. A. aeolicus Trm1 includes an N-terminal catalytic domain (cyan) and a C-terminal domain (magenta). Bound sinefungin (yellow) is highlighted by a stick model. Four sulfate ions are also shown by a stick model. An orange ball represents a zinc atom.
FIGURE 4.
Topology of A. aeolicus Trm1 and mutation sites. Topology of A. aeolicus Trm1 is schematically drawn. The α-helices and β-strands are shown by circles and triangles, respectively. The L1-loop is highlighted in red. An orange circle shows the bound zinc atom. The N- and C-terminal domains are indicated in cyan and magenta, respectively. Amino acid residues are the mutation sites.
Structure-based Amino Acid Sequence Alignment and Target Residues of Site-directed Mutagenesis
On the basis of the structural information, we aligned the amino acid sequences of the Trm1 proteins reported thus far (supplemental Fig. 1). Although the tRNA recognition mechanism of A. aeolicus Trm1 differs from those of eukaryotic and archaeal Trm1, the amino acid sequence of A. aeolicus Trm1 seems to be an intermediate of those of eukaryotic and archaeal Trm1. For example, A. aeolicus Trm1 lacks a long stretch between the α1 helix and β3 strand in the N-terminal region of eukaryotic enzymes; this is a feature of archaeal enzymes. However, A. aeolicus Trm1 has a cysteine cluster (green in Fig. 4 and supplemental Fig. 1), which is observed only in eukaryotic enzymes. Furthermore, the intervening sequence between β11 and β12 of A. aeolicus Trm1 is shorter than that of eukaryotic enzymes.
To predict the reaction mechanism and substrate-binding sites, we introduced single alanine and serine substitutions into A. aeolicus Trm1. The target sites (Fig. 4 and triangles and stars in supplemental Fig. 1) were selected on the basis of the crystal structure and amino acid sequence alignment. We performed site-directed mutagenesis and prepared 30 mutant Trm1 proteins. All mutant Trm1 proteins were purified to near-homogeneity, as judged by 15% SDS-PAGE (supplemental Fig. 2).
AdoMet-binding Site and Hypothetical Reaction Mechanism
Fig. 5A shows the AdoMet-binding site in the N-terminal catalytic domain. A conserved aspartic acid (Asp-132) residue is located near the amino group of sinefungin, which corresponds to the methyl group of AdoMet. This residue was expected to be the catalytic center of A. aeolicus Trm1 on the basis of the amino acid sequence alignment and crystal structure (Fig. 4) (19, 21). Indeed, alanine substitution of Asp-132 caused a complete loss of methyl transfer activity (Table 1), consistent with the idea that Asp-132 is the catalytic center. In the crystal structure, the carboxyl group of Asp-132 forms a hydrogen bond with the Arg-66 residue (Fig. 5B). The alanine substitution mutant of Arg-66 showed a severe loss of methyl transfer activity (1:1000 of the wild-type enzyme) (Table 1). This loss was probably due to two reasons. One is that the guanidino group of Arg-66 functions as a part of the electron relay system. The other is that the hydrogen bond between Arg-66 and Asp-132 determines the direction of the carboxyl group of Asp-132 (predicted catalytic center). Arg-66 forms a hydrogen bond with another aspartic acid (Asp-130) (Fig. 5A). Asp-130 is highly conserved (Fig. 4) and does not contact AdoMet directly. The alanine substitution of this residue caused an increase in Km values for AdoMet. Therefore, Asp-130 seems to contribute to the formation of AdoMet binding pocket to contact Arg-66. There are two conserved phenylalanine residues (Phe-27 and Phe-134) located near the amino group of sinefungin (Fig. 5, A and B). The alanine substitution mutant of Phe-27 showed a severe loss of methyl transfer activity (1:400 of the wild-type enzyme) through an increase in the Km value and a decrease in the Vmax value for AdoMet (Table 1). Phe-27 may have two functions because it is located in the flexible loop region (L1-loop). First, the Phe-27 is a part of the AdoMet-binding site through a hydrophobic interaction between Phe-27 and the methionine of AdoMet because the Km value for AdoMet was increased more than 100-fold as compared with the wild-type enzyme. Second, Phe-27 may fix the target guanine (G26 or G27) by a hydrophobic interaction in the presence of tRNA. However, we could not measure the kinetic parameters of the F27A mutant for tRNA (data not shown); the methyl transfer activity was too low to calculate the values, and two-dimensional TLC analysis showed that second methylation (m22G26 formation) occurred in the low concentration samples (data not shown). Therefore, we could not conclude whether Phe-27 is responsible for tRNA (guanine) binding, although it is the most likely candidate for guanine binding. The importance of Phe-27 residue in the L1-loop was also previously reported for P. horikoshii Trm1 (19). In contrast, alanine substitution of Phe-134 did not cause a significant effect; the methyl transfer activity was decreased to a third of the level of the wild-type enzyme through an increase in Km and a decrease in Vmax values (Table 1), suggesting that Phe-134 is a structural element of the AdoMet-binding site. This result is different from previous results with the P. horikoshii Trm1 mutant protein (19). In the case of P. horikoshii Trm1, the substitution of Phe-140, which corresponds to Phe-134 in A. aeolicus Trm1, by alanine caused a severe loss of methyl transfer activity (19). Thus, structural contribution of this phenylalanine (Phe-134 in A. aeolicus Trm1) residue to the architecture of catalytic pocket seems to differ in Trm1 proteins. Taken together, these results enable us to predict a hypothetical reaction mechanism of A. aeolicus Trm1 (Fig. 5C). In this mechanism, the role of Phe-27 is not clear, although its location suggests that this residue fixes the guanine base in tRNA by a hydrophobic interaction. Furthermore, the result of alanine substitution did not contradict the idea that the Asp-132 is a catalytic center.
TABLE 1.
Kinetic parameters for AdoMet
ND means that methyl transfer activity is not detectable. The relative Km/Vmax value is expressed that of the wild type enzyme as 1.00.
| Alanine substitution, mutant name | Km | Vmax | Relative Vmax/Km |
|---|---|---|---|
| nm | nmol mg−1h−1 | ||
| Wild type | 170 | 90 | 1.00 |
| Glu-6 | 300 | 130 | 0.84 |
| Phe-27a | >23,000 | 30–100 | <0.0025 |
| Asn-29 | 300 | 90 | 0.57 |
| Arg-36 | ND | ND | ND |
| Leu-60 | 670 | 170 | 0.49 |
| Ile-65 | 200 | 170 | 1.70 |
| Arg-66a | >45,000 | 30–100 | <0.0011 |
| Asp-84a | >330,000 | 100–200 | <0.0012 |
| Ile-85 | 170 | 50 | 0.51 |
| Glu-113 | 670 | 170 | 0.49 |
| Asp-130 | 1300 | 170 | 0.25 |
| Asp-132 | ND | ND | ND |
| Phe-134 | 800 | 260 | 0.65 |
| Phe-140 | 670 | 430 | 1.30 |
a Kinetic parameters for these mutants could not be measured correctly because of low methyl transfer activities.
As shown in Fig. 5, A and B, sinefungin forms hydrogen bonds with many amino acid residues in the catalytic domain. These interactions were also observed in the AdoMet-P. horikoshii Trm1 complex (19). The amino group of adenine of sinefungin forms a hydrogen bond with one glutamic acid (Glu-113). In the case of P. horikoshii Trm1, this glutamic acid is replaced by an aspartic acid (Asp-120). Thus, this interaction between an acidic amino acid and amino group of adenine is conserved. However, substitution of Glu-113 by alanine did not have a significant effect, showing that the Glu-113 is a weak AdoMet-binding site. One isoleucine residue (Ile-85) hydrophobically interacts with the adenine of sinefungin; however, alanine substitution of Ile-85 did not cause a severe effect, showing that Ile-85 is also a weak AdoMet-binding site. The ribose of sinefungin contacts one conserved aspartic acid residue (Asp-84), and alanine substitution of Asp-84 caused a severe decrease of methyl transfer activity (1:1000 of the wild-type enzyme) through an increase in the Km value for AdoMet. Thus, Asp-84 is very important for AdoMet binding and, in fact, is highly conserved in Trm1 proteins (supplemental Fig. 1). The carboxyl group of methionine of sinefungin formed a hydrogen bond with the Arg-36 residue. Disruption of this interaction by alanine substitution brought a severe loss of methyl transfer activity (1:1000) through an increase in Km values for AdoMet, demonstrating that Arg-36 is also very important for AdoMet binding. Arg-36 residue forms a hydrogen bond with a semi-conserved glutamic acid (Glu-6), which constitutes half of a distinctive amino acid sequence motif, the so-called “EG motif” (19, 46). Because the EG motif forms a sharp β-turn and is located near the AdoMet-binding site, it has been predicted to be involved in the catalytic mechanism (19, 46). Furthermore, it has been reported that the deletion of the EG motif in S. cerevisiae Trm1 causes complete loss of the enzymatic activity (47). However, alanine substitution of Glu-6 did not have an effect on methyl transfer activity. Thus, this result revealed that the Glu-6 residue itself is not involved in the catalytic mechanism. Taking these results together, we conclude that AdoMet mainly binds to Trm1 by hydrogen bonds. In general, a hydrogen bond is weakened at high temperatures. Even though A. aeolicus is a hyper-thermophile, this AdoMet-binding mode, which is a typical feature of class I AdoMet-dependent methyltransferases (43), is conserved. This AdoMet-binding mode is completely different from that of class IV enzymes such as TrmH; in the case of TrmH, for example, AdoMet binds to the enzyme hydrophobically (30, 48).
Use of Alanine Substitution Mutants to Search for the tRNA-binding Site
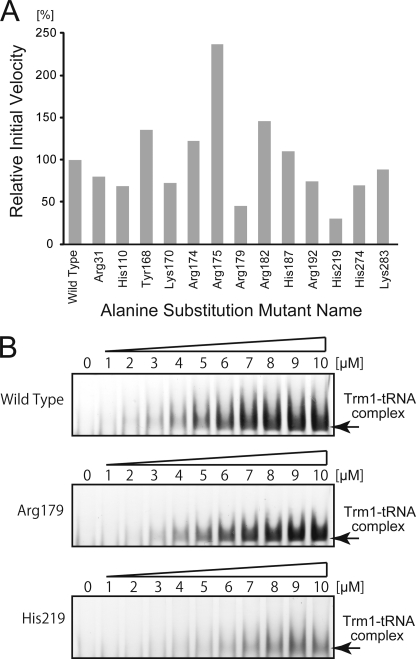
As described above, four sulfate ions were found on the surface of A. aeolicus Trm1 because we used ammonium sulfate for crystallization. We assumed that some of these sulfate ions occupied the phosphate (tRNA)-binding sites. We therefore selected 13 amino acid residues (Arg-31, His-110, Tyr-168, Lys-170, Arg-174, Arg-175, Arg-179, Arg-182, His-187, Arg-192, His-219, His-274, and Lys-283; marked by stars in supplemental Fig. 1) to test tRNA binding on the basis of the sulfate ion positions and amino acid sequence alignment. In general, searching for a tRNA-binding site by alanine substitution is not so easy because a single mutation does not bring the severe loss of methyl transfer activity observed in the search for the catalytic center and AdoMet-binding site (49). Nevertheless, our alanine substitution experiments identified residues important for tRNA binding as follows: alanine substitutions of eight residues (Arg-31, His-110, Lys-170, Arg-179, Arg-192, His-219, His-274, and Lys-283) led to a clear decrease in methyl transfer activity and, in particular, the alanine substitution mutants of Arg-179 and His-219 showed a significant decrease in the methyl transfer activity (Fig. 6A). To confirm whether Arg-179 and His-219 residues are part of the tRNA-binding site or not, we performed a gel mobility shift assay (Fig. 6B). Both alanine substitution mutants (R179A and H219A) showed a decrease in affinity for the tRNA transcript. Thus, we confirmed that Arg-179 and His-219 are important for tRNA binding and that Arg-31, His-110, Lys-170, Arg-192, His-274, and Lys-283 probably form part of the tRNA-binding site.
FIGURE 6.
Determination of tRNA-binding site by site-directed mutagenesis. A, methyl transfer activity of the alanine-substituted mutant proteins. The mutant name indicates the site of alanine substitution. The initial velocity of wild-type Trm1 is expressed as 100%. Data are the average of four independent experiments. B, gel mobility shift assay of wild-type Trm1 (upper panel), the alanine substitution mutant of Arg-179 (middle panel), and the alanine substitution mutant of His-219 (lower panel). As the protein concentration increased, the protein-tRNA complex was formed. However, the Arg-179 and His-219 mutants have weaker affinity for the yeast tRNAPhe transcript as compared with the wild-type enzyme.
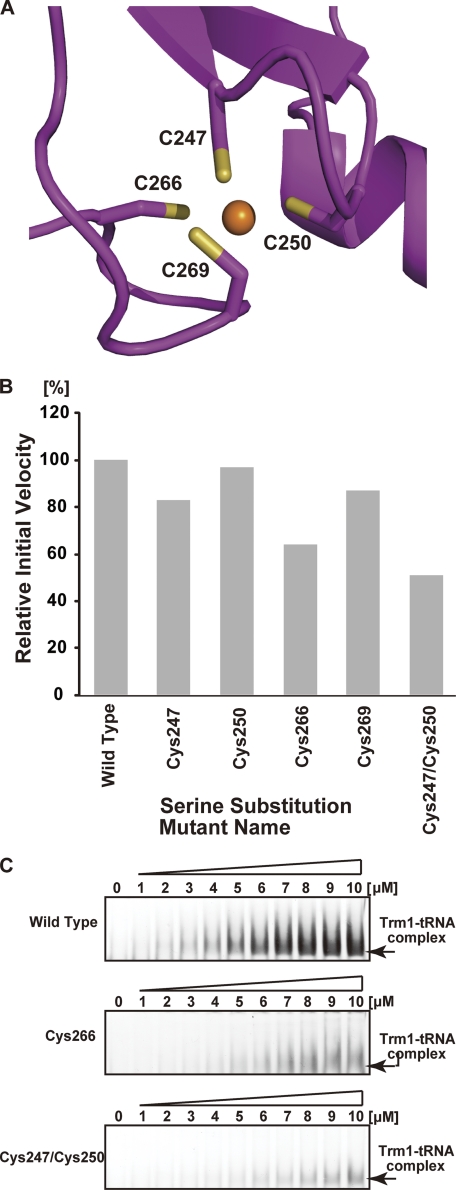
Zinc Atom and Cysteine Cluster
As described above, one zinc atom was bound to four cysteine residues (Cys-247, Cys-250, Cys-266, and Cys-269). The bound metal was identified as zinc by x-ray absorption fluorescence spectroscopic analysis and inductivity coupled plasma emission analysis (data not shown), and its position was consistent with the cysteine cluster motif (50, 51). It should be mentioned that zinc binding by this cysteine cluster was previously predicted by a bioinformatics study by Bujnicki et al. (46). Thus, this study is in good agreement with their bioinformatics study (46). Because this zinc atom is located in the C-terminal domain far from the catalytic center, we considered that the zinc atom and cysteine cluster comprise a structural element of the C-terminal domain. To confirm this idea, we performed serine substitution of four cysteine residues individually (green in Figs. 4 and 7A). As shown in Fig. 7B, serine substitution of each cysteine residue caused a decrease in methyl transfer activity. However, considerable activity remained; for example, the C250S mutant had more than 90% of the activity of the wild-type enzyme. Therefore, we considered that single mutations might not disrupt the zinc-bound cysteine cluster completely. To test the stability of the cluster, we dialyzed the wild-type enzyme against buffer A containing 10 mm EDTA at 4 °C overnight; however, no loss of activity was observed (data not shown). Furthermore, after incubation of the wild-type enzyme with 50 mm EDTA at 90 °C for 30 min, more than 90% of the methyl transfer activity remained (data not shown). Thus, the zinc-cysteine cluster is considerably stable. We therefore prepared a double-mutated Trm1 (C247S and C250S) protein to disrupt the cluster. As expected, this mutant protein showed a significant decrease in activity (Fig. 7A). Furthermore, the gel mobility shift assay showed a decrease in affinity of the double-mutated protein for tRNA (Fig. 7B). On the basis of these results, we concluded that the cysteine cluster is a structural element that maintains the tRNA-binding site between the N- and C-terminal domains.
FIGURE 7.
Cysteine cluster mutants. A, structure of the cysteine cluster chelating a zinc atom. Sulfur atoms and the zinc atom are indicated in yellow and orange, respectively. B, comparison of methyl transfer activities of the serine substitution mutants with that of the wild-type Trm1. The initial velocity of wild-type Trm1 is expressed as 100%. Data are the average of four independent experiments. C, gel mobility shift assay of the wild-type Trm1 (upper panel), the serine substitution mutant of Cys-266 (middle panel), and the double serine substitution mutant of Cys-247 and Cys-250 (lower panel). Disruption of the zinc-cysteine cluster causes a severe decrease in affinity for tRNA.
Comparison of A. aeolicus Trm1 with Archaeal Trm1 and tRNA Docking Model
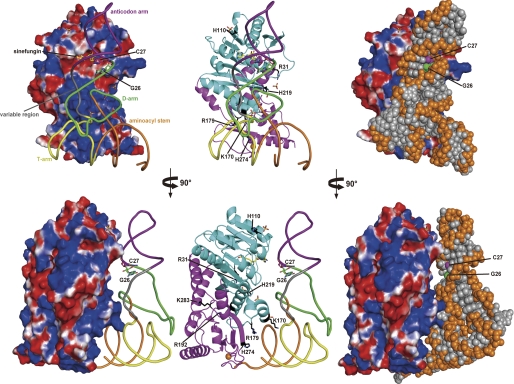
Finally, we compared A. aeolicus Trm1 with P. horikoshii Trm1 (archaeal enzyme). As shown in Fig. 8A, the apparent overall structures of both Trm1 proteins resemble each other. However, the electrostatic potential surface models of the two proteins show that the electric charges on the protein surfaces are considerably different (Fig. 8B). In the case of A. aeolicus Trm1 (Fig. 8B, left), there is a broad positive charge area at the border between the N- and C-terminal domains. The amino acid residues (Lys-170, Arg-179, Arg-182, His-187, and His-219) involved in tRNA binding are present in this area. In fact, three sulfate ions were found in this region. In the case of P. horikoshii Trm1 (Fig. 8B, right), by contrast, the area of positive charge is observed only around the catalytic pocket; instead, a lot of negative charge exists around the border area between the N- and C-terminal domains. These differences in the distribution of positive charge suggest that the tRNA-binding mode differs between the A. aeolicus and P. horikoshii Trm1 proteins.
FIGURE 8.
Comparison of A. aeolicus Trm1 with archaeal Trm1 (P. horikoshii Trm1). A, comparison of the structures of the two proteins. As described in the text, A. aeolicus Trm1 (left panel) has a multisite specificity and modifies both G26 and G27 in tRNA. In contrast, P. horikoshii Trm1 (right panel) has single site specificity and modifies only G26 in tRNA. Although A. aeolicus Trm1 has a zinc-cysteine cluster in the C-terminal domain, the overall structures of both proteins are similar. The catalytic domains (cyan in A. aeolicus Trm1 and green in P. horikoshii Trm1) are almost identical. B, in contrast, comparison of the electrostatic potential surface models of both proteins shows that the electric charges of the protein surfaces are considerably different. A. aeolicus Trm1 has a large area of positive charge at the border between the N- and C-terminal domains (left panel). In contrast, the area of positive charge in P. horikoshii Trm1 is relatively small (right panel).
To address the tRNA-binding mode of A. aeolicus Trm1, we constructed a tRNA-docking model, in which the T-arm region of yeast tRNAPhe was manually placed at the border between the N- and C-terminal domains, i.e. at the expected tRNA-binding site (Fig. 9). This model explains why A. aeolicus Trm1 catalyzes methyl transfers to G27, as well as G26. In the model, the catalytic pocket of A. aeolicus Trm1 can be placed near G26 in tRNA. However, another target guanine, G27 (C27 in the model), is positioned closer to the catalytic pocket than G26. If the anticodon stem structure is disrupted according to the formation of a tRNA-Trm1 complex, then G27 will be more accessible to the catalytic center than G26. Thus, the T-arm recognition and G27 methylation of A. aeolicus Trm1 are explainable by the distributions of positive charge determined from our structural study.
FIGURE 9.
Model of tRNA docking on A. aeolicus Trm1. Yeast tRNAPhe (phosphate-ribose backbone) was docked onto the expected tRNA-binding site in the A. aeolicus Trm1. The G26 and C27 bases are highlighted by stick models. In the docking model, we placed the T-arm at the border region between the N- and C-terminal domains. The right panel shows the space-filling model of tRNA. The target G26 base can be placed near the catalytic pocket; however, the C27 base gains closer access to the pocket in the model. If the anticodon stem structure (i.e. the base pair between nucleotides at positions 27 and 43) is disrupted in the Trm1-tRNA complex, the docking model suggests that tRNA species with G27 could be modified by A. aeolicus Trm1. Lower panels show a side view of the model after a 90° anticlockwise rotation.
DISCUSSION
In this study, we have focused on the multisite specificity of A. aeolicus Trm1, a eubacterial enzyme, which recognizes the T-arm structure of tRNA. Our x-ray crystal structural study has revealed that the apparent overall structure of A. aeolicus Trm1 is similar to that of P. horikoshii Trm1 (archaeal enzyme), although the A. aeolicus Trm1 has a novel C-terminal domain with a zinc atom. On the basis of the crystal structure and amino acid sequence alignment, we have performed a site-directed mutagenesis study, enabling us to propose a hypothetical reaction mechanism of A. aeolicus Trm1 and to determine the amino acid residues responsible for AdoMet binding. Our hypothetical reaction mechanism seems to be reasonable for an amino group methyltransferase (19, 52). The AdoMet-binding site is typical of class I AdoMet-dependent methyltransferases (45).
We have also predicted the tRNA-binding site by the site-directed mutagenesis study based on the sulfate ion binding in the crystal structure and amino acid sequence alignment. The amino acid residues involved in tRNA binding mainly map to the border area between the N- and C-terminal domains. The electrostatic potential surface model of A. aeolicus Trm1 has demonstrated that the border area between the N- and C-terminal domains forms a large area of positive charge, which is not observed in the model of P. horikoshii Trm1. The tRNA docking model of A. aeolicus Trm1, in which the T-arm was placed at the border area between the N- and C-terminal domains, has elucidated the multisite recognition mechanism of A. aeolicus Trm1 by showing that G27 can be placed closer to the catalytic pocket than G26. Among multisite-specific tRNA modification enzymes, Escherichia coli TruA modifies U38, U39, and U40 to Ψ38, Ψ39, and Ψ40, respectively (53, 54), and is the only enzyme whose multisite recognition mechanism has been studied by the tRNA-enzyme complex (55). In this study, we have been able to propose a hypothetical mechanism for the multisite specificity of A. aeolicus Trm1; however, a structural study of tRNA-Trm1 complex will be necessary, like that of TruA, for a discussion of the detailed mechanism.
The multisite specificity and tRNA recognition mode of A. aeolicus Trm1 is completely different from those of eukaryotic and archaeal Trm1, although A. aeolicus Trm1 seems to be an intermediate of eukaryotic and archaeal Trm1 proteins from the amino acid sequence alignment. These facts suggest that the RNA recognition mechanism of RNA modification enzymes may have changed during molecular evolution process, even though the enzymes bring about the same modification(s) at the same position(s) in RNA. In another case of single- and multisite specific enzymes, eubacterial TrmI modifies only A58 in the T-loop of tRNA (56, 57), whereas archaeal TrmI modifies both A57 and A58 (22). The RNA recognition mechanisms of eubacterial and archaeal TrmI may differ from each other, although both enzymes share structural relationships (57, 58). In fact, it has been recently reported that the P. abyssi (archaeal) TrmI requires the A59 sequence for A58 methylation in tRNA, whereas Thermus thermophilus (eubacterial) TrmI does not require the A59 (58). Furthermore, in their report, the A59 requirement of archaeal TrmI is structurally revealed; the His-78 residue is involved in the A59 recognition (58). It should be mentioned that the eukaryotic counterpart of m1A58 methylation in tRNA is a hetero-subunit enzyme, Trm6 (gcd10-gcd14 complex in yeast) (59, 60). In the case of m1A58 modification, the eukaryotic enzyme probably has a different RNA recognition mechanism from those of the eubacterial and archaeal TrmI enzymes. To verify this idea, further studies will be necessary. In addition, this study has revealed that the RNA recognition mechanism cannot be simply predicted by an amino acid sequence alignment. Combined use of biochemical approaches and structural studies is required for full understanding of the RNA recognition mechanism.
Supplementary Material
Acknowledgment
We thank Madoka Nishimoto (RIKEN Yokohama, Systems and Structural Biology Center) for technical support with crystallization.
This work was supported in part by Grant-in-aid for Science Research on Priority Areas 20034041 (to H. H.), Grants-in-aid for Science Research 19350087 and 2350081 (to H. H.) and 21603018 (to Y. B.), and the National Project on Targeted Proteins Research Program for Science Research from the Ministry of Education, Science, Sports, and Culture of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
The atomic coordinates and structure factors (codes 3AXT and 3AXS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- AdoMet
- S-adenosyl-l-methionine.
REFERENCES
- 1. Rozenski J., Crain P. F., McCloskey J. A. (1999) Nucleic Acids Res. 27, 196–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dunin-Horkawicz S., Czerwoniec A., Gajda M. J., Feder M., Grosjean H., Bujnicki J. M. (2006) Nucleic Acids Res. 34, D145–D149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garcia G. A., Goodenough-Lashhua D. M. (1998) in Modification and Editing of RNA (Grosjean H., Benne R. eds) pp. 555–560, American Society for Microbiology, Washington, D. C [Google Scholar]
- 4. Reinhart M. P., Lewis J. M., Leboy P. S. (1986) Nucleic Acids Res. 14, 1131–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Constantinesco F., Motorin Y., Grosjean H. (1999) J. Mol. Biol. 291, 375–392 [DOI] [PubMed] [Google Scholar]
- 6. Kuchino Y., Nishimura S. (1970) Biochem. Biophys. Res. Commun. 40, 306–313 [DOI] [PubMed] [Google Scholar]
- 7. Glick J. M., Averyhart V. M., Leboy P. S. (1978) Biochim. Biophys. Acta 518, 158–171 [DOI] [PubMed] [Google Scholar]
- 8. Stange N., Beier H. (1987) EMBO J. 6, 2811–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edqvist J., Grosjean H., Stråby K. B. (1992) Nucleic Acids Res. 20, 6575–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edqvist J., Blomqvist K., Stråby K. B. (1994) Biochemistry 33, 9546–9551 [DOI] [PubMed] [Google Scholar]
- 11. Grosjean H., Edqvist J., Stråby K. B., Giegé R. (1996) J. Mol. Biol. 255, 67–85 [DOI] [PubMed] [Google Scholar]
- 12. Constantinesco F., Motorin Y., Grosjean H. (1999) Nucleic Acids Res. 27, 1308–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Phillips J. H., Kjellin-Stråby K. (1967) J. Mol. Biol. 26, 509–518 [DOI] [PubMed] [Google Scholar]
- 14. Ellis S. R., Morales M. J., Li J. M., Hopper A. K., Martin N. C. (1986) J. Biol. Chem. 261, 9703–9709 [PubMed] [Google Scholar]
- 15. Niederberger C., Gräub R., Costa A., Desgrès J., Schweingruber M. E. (1999) FEBS Lett. 464, 67–70 [DOI] [PubMed] [Google Scholar]
- 16. Liu J., Zhou G. Q., Stråby K. B. (1999) Gene 226, 73–81 [DOI] [PubMed] [Google Scholar]
- 17. Liu J., Strâby K. B. (2000) Nucleic Acids Res. 28, 3445–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Constantinesco F., Benachenhou N., Motorin Y., Grosjean H. (1998) Nucleic Acids Res. 26, 3753–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ihsanawati, Nishimoto M., Higashijima K., Shirouzu M., Grosjean H., Bessho Y., Yokoyama S. (2008) J. Mol. Biol. 383, 871–884 [DOI] [PubMed] [Google Scholar]
- 20. Grosjean H., Gaspin C., Marck C., Decatur W. A., de Crécy-Lagard V. (2008) BMC Genomics 9, 470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Awai T., Kimura S., Tomikawa C., Ochi A., Ihsanawati, Bessho Y., Yokoyama S., Ohno S., Nishikawa K., Yokogawa T., Suzuki T., Hori H. (2009) J. Biol. Chem. 284, 20467–20478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roovers M., Wouters J., Bujnicki J. M., Tricot C., Stalon V., Grosjean H., Droogmans L. (2004) Nucleic Acids Res. 32, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Motorin Y., Grosjean H. (1999) RNA 5, 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Auxilien S., El Khadali F., Rasmussen A., Douthwaite S., Grosjean H. (2007) J. Biol. Chem. 282, 18711–18721 [DOI] [PubMed] [Google Scholar]
- 25. Pintard L., Lecointe F., Bujnicki J. M., Bonnerot C., Grosjean H., Lapeyre B. (2002) EMBO J. 21, 1811–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takeda H., Toyooka T., Ikeuchi Y., Yokobori S., Okadome K., Takano F., Oshima T., Suzuki T., Endo Y., Hori H. (2006) Genes Cells 11, 1353–1365 [DOI] [PubMed] [Google Scholar]
- 27. Keith G. (1995) Biochimie 77, 142–144 [DOI] [PubMed] [Google Scholar]
- 28. Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. (1974) Nature 250, 546–551 [DOI] [PubMed] [Google Scholar]
- 29. Kim S. H., Sussman J. L., Suddath F. L., Quigley G. J., McPherson A., Wang A. H., Seeman N. C., Rich A. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 4970–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watanabe K., Nureki O., Fukai S., Ishii R., Okamoto H., Yokoyama S., Endo Y., Hori H. (2005) J. Biol. Chem. 280, 10368–10377 [DOI] [PubMed] [Google Scholar]
- 31. Ochi A., Makabe K., Kuwajima K., Hori H. (2010) J. Biol. Chem. 285, 9018–9029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 33. Vagin A., Teplyakov A. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 36. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 39. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 41. Ohtsuki T., Sakurai M., Sato A., Watanabe K. (2002) Nucleic Acids Res. 30, 5444–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Doi Y., Ohtsuki T., Shimizu Y., Ueda T., Sisido M. (2007) J. Am. Chem. Soc. 129, 14458–14462 [DOI] [PubMed] [Google Scholar]
- 43. Pugh C. S., Borchardt R. T., Stone H. O. (1978) J. Biol. Chem. 253, 4075–4077 [PubMed] [Google Scholar]
- 44. Vedel M., Robert-Géro M. (1981) FEBS Lett. 128, 87–89 [DOI] [PubMed] [Google Scholar]
- 45. Schubert H. L., Blumenthal R. M., Cheng X. (2003) Trends Biochem. Sci. 28, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bujnicki J. M., Leach R. A., Debski J., Rychlewski L. (2002) J. Mol. Microbiol. Biotechnol. 4, 405–415 [PubMed] [Google Scholar]
- 47. Liu J., Liu J., Stråby K. B. (1998) Nucleic Acids Res. 26, 5102–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nureki O., Watanabe K., Fukai S., Ishii R., Endo Y., Hori H., Yokoyama S. (2004) Structure 12, 593–602 [DOI] [PubMed] [Google Scholar]
- 49. Watanabe K., Nureki O., Fukai S., Endo Y., Hori H. (2006) J. Biol. Chem. 281, 34630–34639 [DOI] [PubMed] [Google Scholar]
- 50. Michel S. L., Berg J. M. (2002) Chem. Biol. 9, 667–668 [DOI] [PubMed] [Google Scholar]
- 51. Sharpe B. K., Matthews J. M., Kwan A. H., Newton A., Gell D. A., Crossley M., Mackay J. P. (2002) Structure 10, 639–648 [DOI] [PubMed] [Google Scholar]
- 52. Garcia G. A., Goodenough-Lashhua D. M. (1998) in Modification and Editing of RNA (Grosjean H., Benne R. eds) pp. 135–168, American Society for Microbiology, Washington, D. C [Google Scholar]
- 53. Cortese R., Kammen H. O., Spengler S. J., Ames B. N. (1974) J. Biol. Chem. 249, 1103–1108 [PubMed] [Google Scholar]
- 54. Arena F., Ciliberto G., Ciampi S., Cortese R. (1978) Nucleic Acids Res. 5, 4523–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hur S., Stroud R. M. (2007) Mol. Cell 26, 189–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Droogmans L., Roovers M., Bujnicki J. M., Tricot C., Hartsch T., Stalon V., Grosjean H. (2003) Nucleic Acids Res. 31, 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barraud P., Golinelli-Pimpaneau B., Atmanene C., Sanglier S., Van Dorsselaer A., Droogmans L., Dardel F., Tisné C. (2008) J. Mol. Biol. 377, 535–550 [DOI] [PubMed] [Google Scholar]
- 58. Guelorget A., Roovers M., Guérineau V., Barbey C., Li X., Golinelli-Pimpaneau B. (2010) Nucleic Acids Res. 38, 6206–6218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Anderson J., Phan L., Cuesta R., Carlson B. A., Pak M., Asano K., Björk G. R., Tamame M., Hinnebusch A. G. (1998) Genes Dev. 12, 3650–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Anderson J., Phan L., Hinnebusch A. G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5173–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.