Abstract
N-Linked glycosylation is a frequent protein modification that occurs in all three domains of life. This process involves the transfer of a preassembled oligosaccharide from a lipid donor to asparagine side chains of polypeptides and is catalyzed by the membrane-bound oligosaccharyltransferase (OST). We characterized an alternative bacterial pathway wherein a cytoplasmic N-glycosyltransferase uses nucleotide-activated monosaccharides as donors to modify asparagine residues of peptides and proteins. N-Glycosyltransferase is an inverting glycosyltransferase and recognizes the NX(S/T) consensus sequence. It therefore exhibits similar acceptor site specificity as eukaryotic OST, despite the unrelated predicted structural architecture and the apparently different catalytic mechanism. The identification of an enzyme that integrates some of the features of OST in a cytoplasmic pathway defines a novel class of N-linked protein glycosylation found in pathogenic bacteria.
Keywords: Bacteria, Glucose, Glycosylation, Glycosyltransferases, Post-translational Modification, Consensus Sequence
Introduction
N-Linked glycosylation is characterized by an N-glycosidic linkage between the side chain amide of an asparagine residue of proteins and an oligosaccharide. This type of glycosylation occurs in both prokaryotes and eukaryotes and requires the assembly of an oligosaccharide on a polyisoprenoid lipid by sequential addition of monosaccharides, catalyzed by cytosolic glycosyltransferase. The resulting lipid-linked oligosaccharide is translocated to the luminal side of the endoplasmic reticulum membrane or the plasma membrane of prokaryotes, where it may be further elongated. The glycan is then transferred to the δ-amino group of asparagine residues within the consensus sequence NX(S/T) of polypeptides. This reaction is catalyzed by oligosaccharyltransferase (OST),4 a membrane-bound enzyme that can be composed of several different subunits (1). As oligosaccharide transfer takes place in the endoplasmic reticulum or in the periplasm, N-glycosylation affects proteins trafficking along the secretory pathway.
N-Glycosylation exhibits important physiological functions. In the early secretory pathway of eukaryotes, N-glycans present on newly synthesized proteins orchestrate the folding of glycoproteins and act as a signal for directing misfolded polypeptides to degradation (2). After being processed in the Golgi organelle, N-linked glycans are relevant, among other processes, for the modulation of the immune system and for the control of immune cell homeostasis and inflammation (3, 4). The importance of this protein modification is supported by its incidence: more than half of all eukaryotic proteins are glycosylated (5).
About 8 years ago, a study uncovered that the extracellular HMW1A adhesin of the Gram-negative bacterium Haemophilus influenzae is modified with hexose monosaccharides on asparagine residues (6). The pioneering work of St Geme and co-workers (7, 8) subsequently showed that the modified asparagine residues are found within the same consensus sequence recognized by OST (i.e. NX(S/T)) and that the enzyme responsible for this modification is the HMW1C protein, capable of transferring Glc or Gal from nucleotide-activated substrates. Moreover, it has been proposed that HMW1C also forms hexose-hexose bonds. Modification of HMW1A adhesin prevents its premature degradation and promotes HMW1A to be displayed at the cell surface, a prerequisite for HMW1A-mediated adherence to human epithelial cells (8). Additionally, a recent report demonstrated that an HMW1C homolog from Actinobacillus pleuropneumoniae mediates N-linked glycosylation of the H. influenzae HMW1A protein (9).
To characterize accurately the mechanisms of this alternative N-glycosylation pathway and to gain insight into the specificity of the key enzyme N-glycosyltransferase (NGT), we established a platform suitable for in vitro glycosylation and undertook a detailed analysis of the reaction products. We adopted MS and gel electrophoresis for detection of glycosyltransferase activity using peptides and proteins as substrates and performed NMR studies to characterize the reaction adducts. We show that the HMW1C homolog from A. pleuropneumoniae is an inverting NGT that transfers a glucose or galactose moiety to asparagine, but it does not further elongate the N-linked monosaccharide. Instead, we found that another glucosyltransferase was able to elaborate the N-linked glucose. We compared the acceptor substrate range of NGT and OST and observed a highly similar specificity of the two different enzymes. We concluded that NGT integrates polypeptide recognition common to the OST-based modification into a novel framework, resulting in a general N-glycosylation system that operates in the cytoplasm.
EXPERIMENTAL PROCEDURES
Materials
Restriction enzymes were purchased from Fermentas. T4 DNA ligase was from PerkinElmer Life Sciences. UDP-Glc, UDP-GlcNAc, and UDP-GalNAc were from Sigma. UDP-Gal was obtained from VWR International. Synthetic peptides were purchased from JPT Peptide Technologies.
Construction of Plasmids
Escherichia coli DH5α was chosen as host for cloning. The NGT and α1,6-glucosyltransferase (α6GlcT) genes were amplified by PCR using genomic DNA from Yersinia enterocolitica strain 8081, A. pleuropneumoniae strain L20, or A. pleuropneumoniae strain AP76 as a template. Fragments containing the NGT gene were cut with XhoI and ligated into pEC(AcrA-cyt), previously digested with NdeI, blunted by treatment with Klenow fragment, and digested with XhoI. The α6GlcT gene was cloned into the pEXT21 plasmid. All open reading frames are in-frame with a hexahistidine tag at the C terminus. All plasmid constructs were verified by sequencing of relevant fragments (Microsynth AG).
Protein Expression, Purification, and Analysis
E. coli DH5α cells harboring a plasmid for expression of a relevant protein were grown in volumes of 1 liter at 37 °C in LB medium. Ampicillin (100 mg/liter) or chloramphenicol (25 mg/liter) was added to the medium as needed. When cultures reached A600 = 0.5, 0.2% arabinose or 1 mm isopropyl β-d-thiogalactopyranoside was added for induction of protein expression. After 4 h of incubation at 37 °C, cells were harvested by centrifugation, resuspended in 30 mm Tris (pH 8) and 300 mm NaCl supplemented with 1 mm EDTA and 1 g/liter lysozyme, and incubated for 1 h at 4 °C. MgCl2 and DNase I (Roche Applied Science) were added to final concentrations of 5 mm and 0.1 mg/ml, respectively. Cells were broken with a French press. Extracts were spun at 150,000 × g for 30 min at 4 °C. Supernatants were supplemented with 20 mm imidazole and loaded on a HisTrap column (GE Healthcare). Purification of proteins was done as recommended by the provider. Purification of Xanthomonas campestris O-GlcNAc-transferase (OGT) was performed according to a published procedure (10). Buffer exchange to 25 mm Tris (pH 7.2) and 150 mm NaCl was performed by gel filtration chromatography using HiTrap desalting columns (GE Healthcare). Proteins were analyzed by SDS-PAGE and quantified by measuring absorbance at 280 nm.
Glycosylation Analysis of Synthetic Peptides
Enzymatic activity using different sugar donors or peptide acceptors was assessed with 1.4 μg (0.46 μm) of NGT and/or α6GlcT in a 50-μl final volume of 25 mm Tris buffer (pH 7.2). Acceptor peptides and sugar donors were mixed at a 1:100 molar ratio. Glycosylation reactions were incubated for 16 h at 30 °C. Analysis of the reaction products was performed by MALDI-TOF/TOF-MS, NMR, or gel electrophoresis. For electrophoretic analysis, carboxytetramethylrhodamine (TAMRA)-labeled peptides were supplemented with reducing sample buffer (0.0625 m Tris-HCl (pH 6.8), 2% (v/w) SDS, 5% (v/v) β-mercaptoethanol, 10% (v/v) glycerol, and 0.01% (w/v) bromphenol blue), boiled at 95 °C for 5 min, and separated by Tricine/SDS-PAGE (11). Fluorescence was acquired with a Bio-Rad RX imager. For removal of salts and enzyme, peptides were bound to a Sep-Pak C18 cartridge (Waters) or to a ZipTipC18 pipette tip (Millipore), washed with 0.1% formic acid, and eluted with a solution of 70% acetonitrile and 0.1% formic acid.
Glycosylation Analysis of Proteins
50 μg of AcrA was incubated with 1 mm UDP-Glc and 1.4 μg of NGT in 25 mm Tris (pH 7.2) and 150 mm NaCl for 16 h at 30 °C. Samples were digested with trypsin (Promega) overnight at 37 °C. Peptides were bound to a C18 cartridge for removal of salts, eluted with a solution of 70% acetonitrile and 0.1% formic acid, and subjected to MS analysis.
MS Analysis
MALDI-MS and MS/MS analyses of synthetic peptides were performed on a model 4800 proteomics analyzer (Applied Biosystems) operated in the positive reflectron mode. The peptides eluted from the C18 cartridge or the ZipTipC18 pipette tip were mixed 1:1 with α-cyano-4-hydroxycinnamic acid (5 mg/ml in 70% acetonitrile and 0.1% trifluoroacetic acid) as a matrix for spotting onto the target plate. Peptide mixtures from the AcrA glycosylation reaction were analyzed with an Eksigent nano-HPLC system using an autosampler with a self-made reverse-phase tip column (75 μm × 80 mm) packed with C18 material (AQ, 3 μm, 200 Å; Bischoff GmbH, Germany). The gradient consisted of 10–30% acetonitrile in 0.2% formic acid at a flow rate of 250 nl/min for 60 min and 30–50% acetonitrile in 0.2% formic acid for 5 min. High accuracy mass spectra were acquired with an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) in the mass range of m/z 300–1700 and a target value of 1 × 106 ions. Up to 20 data-dependent MS/MS spectra of the most intense ions with a charge state of 2+ or higher were recorded in parallel at the ion trap using collision-induced dissociation.
NMR Analysis
Unless indicated otherwise, samples were lyophilized and dissolved in D2O at 1 mm. All samples were measured at 303 K on a 500-MHz AVANCE III spectrometer equipped with an inverse triple-resonance cryogenic probe (Bruker). Standard two-dimensional 13C-1H heteronuclear single-quantum coherence (HSQC), 13C-1H heteronuclear multiple-bond correlation, and 1H-1H total correlation spectroscopy (TOCSY) (τmix = 13 and 120 ms) spectra were recorded to assign the peptide and glycan chemical shifts. In addition, a two-dimensional 13C-1H relayed HMQC-COSY spectrum (12, 38) was measured to assist the assignment process. To detect through-bond correlations between glycan protons and the asparagine Hδ21, a two-dimensional 1H-1H TOCSY spectrum was recorded in 95% H2O and 5% D2O with a mixing time of 80 ms. All spectra are referenced to 2,2-dimethyl-2-silapentanesulfonic acid. 13C chemical shifts are indirectly referenced using a scaling factor (Ξ) of 0.251449530 (13). All spectra were processed with Topspin 2.1 (Bruker) and analyzed by Sparky (39).
RESULTS
A. pleuropneumoniae and Y. enterocolitica HMW1C Homologs Modify a DANYTK Peptide
To gain a comprehensive view of potential NGT proteins, we searched the genome data base for bacteria encoding HMW1C homologs and identified a restricted group of candidate proteins (supplemental Fig. 1). Interestingly, many of the microorganisms that encode a putative NGT are pathogenic and cluster in the γ-proteobacteria group, but only some strains within the same species appear to carry an hmw1c-like gene. When we extended the search to retrieve low score hits, we found that HMW1C of H. influenzae displays low similarity to the C-terminal region of the product of the XCC0866 gene from X. campestris (24% identity within a 236-amino acids sequence). Although there is no exhaustive evidence of the activity of XCC0866 protein, it is considered to be a bona fide OGT due to the high sequence identity to its eukaryotic counterpart (10, 14). OGT is an inverting glycosyltransferase that utilizes UDP-GlcNAc as a donor to modify hydroxyl groups of serine or threonine residues of nuclear, mitochondrial, and cytoplasmic proteins (15, 16). Notably, HMW1C homologs are classified in the same family (GT41) as OGT in the CAZy Database, probably due to their low sequence identity.
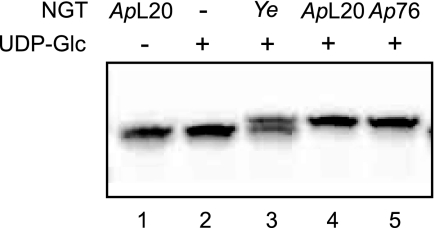
We expressed the HMW1C homologs from Y. enterocolitica strain 8081, A. pleuropneumoniae strain L20, A. pleuropneumoniae strain AP76, and X. campestris ATCC 33913 in E. coli and purified them. To test the proteins for glycosyltransferase activity, we adapted an in vitro assay developed for analysis of OST activity (17). We incubated the purified proteins with UDP-Glc and the hexapeptide DANYTK labeled at the N terminus with the fluorescent dye TAMRA. After separation of the reaction products by Tricine/SDS-PAGE and detection of fluorescence signals, we observed that Y. enterocolitica and the two A. pleuropneumoniae homologs modified the TAMRA-labeled peptide, visualized by a shift in electrophoretic mobility (Fig. 1). By contrast, X. campestris OGT did not exhibit glycosyltransferase activity for this acceptor peptide in the presence of UDP-Glc, UDP-Gal, UDP-GlcNAc, or UDP-GalNAc under the experimental conditions tested (supplemental Fig. 2). In the following, we focused our study on the A. pleuropneumoniae strain AP76 enzyme.
FIGURE 1.
Modification of TAMRA-labeled peptides by putative glycosyltransferases. Reaction products were separated by Tricine/SDS-PAGE, and fluorescence signals were acquired by an image analyzer. Lanes 1 and 2 are negative controls, where no UDP-Glc (lane 1) or no enzyme (lane 2) was added to the reaction. ApL20, A. pleuropneumoniae strain L20; Ye, Y. enterocolitica strain 8081; Ap76, A. pleuropneumoniae strain AP76.
A. pleuropneumoniae HMW1C Homolog Is an Inverting N-Glucosyltransferase
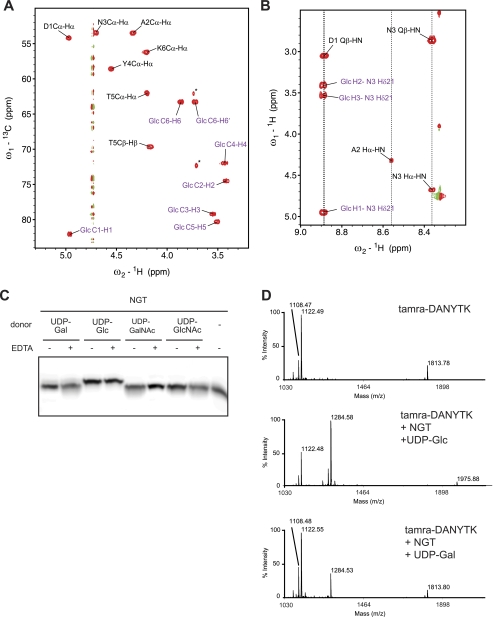
To prove glycosylation of the peptide directly and to characterize the site and chemical structure of the modification, we analyzed the glycosylation products using NMR spectroscopy. The assignment of the peptide and glycan was achieved using through-bond short- and long-range J couplings. The 13C-1H HSQC spectrum, a fingerprint of the glycopeptide, showed 13C-1H correlations, with the signals of the glycan being separated from those of the peptide (Fig. 2A). As is typical for a hexose, seven carbohydrate-specific signals were observed. The anomeric C1 13C chemical shift of 82 ppm was a strong indication of a linkage to nitrogen. Similar anomeric C1 shifts have been found in bacterial and eukaryotic N-glycans (18–20), in sharp contrast to chemical shifts around 100 ppm typical for O-glycosidic linkages. A comparison of all observed 1H and 13C chemical shifts of the glycan with those obtained for published model compounds revealed a striking coincidence with those of Glc-βAsn (supplemental Table 1) (21). With a perfect match of all six 13C chemical shift values and the single reported 1H chemical shift (22), we identified the Glc-βAsn structure. To confirm the linkage, we measured a JH1,H2 coupling constant of 9.9 Hz typical for a β-configuration and recorded a two-dimensional TOCSY spectrum in H2O (Fig. 2B). Through-bond correlations between the asparagine side chain NH and glucose protons were clearly visible, with the asparagine side chain NH chemical shift at 8.88 ppm being similar to those reported for bacterial and eukaryotic N-glycans of 8.62–8.73 ppm (18, 23). Thus, we established that the A. pleuropneumoniae HMW1C homolog is an inverting NGT.
FIGURE 2.
A. pleuropneumoniae NGT is an inverting glycosyltransferase with specificity for UDP-glucose. A, 13C-1H correlations in an HSQC spectrum of TAMRA-DANYTK-Glc. Signals originating from the covalently linked glucose are shown in purple. The asterisk denotes signals from an impurity of the original peptide. B, 1H-1H TOCSY spectrum recorded with a mixing time of 80 ms and four transients. C, Tricine/SDS-PAGE analysis of glycosylation products. TAMRA-labeled peptides were incubated with NGT and different UDP-monosaccharide donors in the presence or absence of EDTA. D, MALDI-MS analysis of the TAMRA-labeled peptide (upper panel) in the presence of NGT and UDP-glucose (middle panel) or UDP-galactose (lower panel).
Next, we analyzed the donor specificity of this NGT in vitro. The enzyme transferred glucose or galactose, but not GlcNAc or GalNAc, to the DANYTK peptide (Fig. 2C). In the presence of a 100-fold molar ratio of donor to acceptor, the conversion to glycopeptide was quantitative in the presence of UDP-Glc, whereas it was marginal in the presence of UDP-Gal. Importantly, NGT glycosylated the peptide in the presence of EDTA, proving that glycosyl transfer does not require metal ions. We also monitored the products of the reaction by MS (Fig. 2D). Analysis of unmodified TAMRA-DANYTK resulted in two major species, matching with TAMRA-DANYTK (calculated molecular mass, 1122.19 Da; observed, 1122.49 Da) and a by-product, TAMRA-(DANYTK)2 (calculated molecular mass, 1814.50 Da; observed, 1813.78 Da). After incubation with NGT and UDP-Glc, we detected species corresponding to TAMRA-DANYTK-Glc (calculated molecular mass, 1284.35 Da; observed, 1284.58 Da) and TAMRA-(DANYTK)2-Glc (calculated molecular mass, 1975.66 Da; observed, 1975.88 Da). A similar result was obtained after incubation with UDP-Gal. In all cases, we observed addition of a single hexose moiety to the asparagine residue.
A. pleuropneumoniae Encodes a Polymerizing Glucosyltransferase That Elongates N-Linked Glucose
We inspected the NGT-encoding genomic region of A. pleuropneumoniae strain AP76 (supplemental Fig. 3). This region contains genes encoding putative proteins involved in the uptake of mannitol and its conversion to glucosamine 6-phosphate, two isomerases, a nucleosidase, and a methylthiotransferase. Interestingly, the ORF next to the NGT gene encodes a putative glycosyltransferase (APP7_1696). We expressed a C-terminally tagged protein in E. coli. When we incubated the purified protein with the TAMRA-labeled product of the NGT reaction, we detected a mobility shift upon Tricine/SDS-PAGE indicative of elaboration of the glycopeptide (Fig. 3A). This modification occurred in the presence of UDP-Glc, but not in the presence of UDP-Gal, UDP-GlcNAc, or UDP-GalNAc. The glucosyltransferase activity appeared to be cation-independent (supplemental Fig. 4).
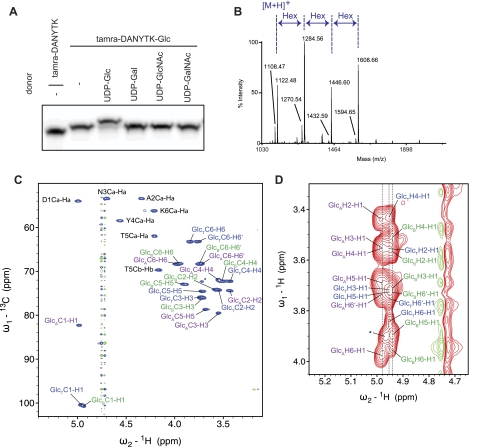
FIGURE 3.
An α6GlcT elongates N-linked glucose. A, glucosylated TAMRA-labeled peptides were incubated with α6GlcT and different UDP-monosaccharide donors and then separated by Tricine/SDS-PAGE. B, MALDI-MS analysis of glucosylated products showed that the glucosyltransferase added two units of glucose to the N-glucose. C, HSQC spectrum showing 13C-1H correlations of TAMRA-DANYTK-Glc3. Signals of three different glucose species are visible. GlcT, terminal glucose; GlcB, bridging glucose; GlcA, N-linked glucose. D, 1H-1H TOCSY spectrum (120-ms mixing time, D2O) allows identification of the different glucose species within the sample. The signal labeled with an asterisk might come from a second signal set for a bridging glucose different from GlcB.
We analyzed the reaction product by MS and found an addition of two glucose moieties in the presence of a 1:100 acceptor/donor ratio (Fig. 3B). Notably, we observed addition of up to six glucose units in the presence of an excess of the donor and with increasing amounts of glucosyltransferase (supplemental Fig. 5). MS/MS analysis of the precursor ions corresponding to glucosylated peptides (m/z = 1446.61, 1608.68, 1770.74, 1932.80, 2094.86, and 2256.93) gave fragmentation compatible to modified TAMRA-DANYTK (data not shown).
To determine the chemical structure and stereochemistry of the reaction product, we analyzed the glycopeptide by NMR spectroscopy. The 13C-1H HSQC spectrum displayed signals of three different glucose units (Fig. 3C). Two new signals appeared in the anomeric region at ∼100 ppm, in addition to the previously observed signal at ∼82 ppm originating from the N-linked glucose (supplemental Fig. 6). The signals were assigned with a two-dimensional TOCSY spectrum (Fig. 3D) and 13C-1H long-range correlations via J couplings. The first set of signals belonging to the N-linked glucose displayed a C6 chemical shift of 68.3 ppm that differed from the initial glycopeptide harboring a single glucose unit (C6, 63.3 ppm). This was indicative of a carbohydrate attachment at O6. The signals of a second glucose unit originated from a terminal glucose (C6, 63.2 ppm), whose chemical shifts coincided with those of Glc-α1,6Glc. The third set of signals displayed chemical shifts of a bridging glucose unit that is α1,6-linked on either side. Chemical shifts of Glc-α1,6Glc-α1,6Glc reported previously (24, 25) fitted perfectly the experimental data of the terminal and bridging glucoses, providing strong evidence for α1,6-linked glucose residues (supplemental Table 1). Chemical shifts calculated with the algorithm CASPER (26) further supported the assignment. We concluded that the APP7_1697 gene encodes a polymerizing α6GlcT that elaborates the product of the NGT reaction.
NGT Exhibits Acceptor Site Specificity Overlapping OST
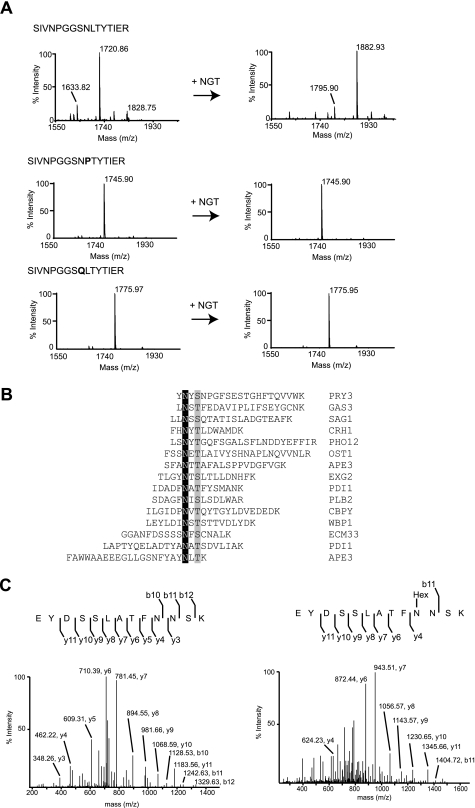
We analyzed glycosylation of the SIVNPGGSNLTYIER peptide present in the yeast glycoprotein lysophospholipase 2 (Plb2). After incubation with NGT and UDP-Glc, analysis of the reaction products by MS indicated that the peptide was modified (Fig. 4A, upper panels). The fragmentation spectrum of the ion at m/z 1882.93 was consistent with glucosylation of the asparagine within the NLT site (supplemental Fig. 7). Importantly, alteration of the NLT sequon to QLT or NPT abolished glycosylation (Fig. 4A), demonstrating a similar property of NGT as for OST with respect to acceptor sequon specificity (27).
FIGURE 4.
Overlapping substrate specificity of NGT and OST. A, MALDI-MS analysis of the glycosylation products using model peptides in the presence of UDP-glucose without (left panels) or with (right panels) NGT. Upper panels, spectrum of the SIVNPGGSNLTYTIER peptide (calculated molecular mass, 1720.89 Da); middle panels, spectrum of the acetyl-SIVNPGGSNPTYTIER-amide peptide (calculated molecular mass, 1745.87 Da); lower panels, spectrum of the acetyl-SIVNPGGSQLTYTIER-amide peptide (calculated molecular mass, 1775.97 Da). B, list of synthetic peptides that are glycosylated by NGT. The modified asparagine residues are highlighted in black. The amino acids in position +2 are shown in gray. The peptide sequences derive from the yeast glycoproteins identified by the UniProt entry names indicated on the right. C, LC-electrospray ionization-MS/MS analysis of the tryptic products of AcrA. Left panel, the spectrum from fragmentation of the doubly charged precursor ion at m/z 738.34 corresponds to the peptide EYDSSLATFNNSK. Right panel, the spectrum from fragmentation of the doubly charged ion at m/z 819.36 matches the glycopeptide EYDSSLATFN(Glc)NSK.
We extended our analysis and examined glycosylation of a group of model peptides with the NX(S/T) consensus sequence in different positions (N terminus, central, C terminus). We chose sequences found to be glycosylated in yeast glycoproteins (Fig. 4B). Glycosylation at the NX(S/T) site was observed for all peptides (supplemental Fig. 8), indicating that the NX(S/T) consensus sequence was recognized by NGT. Although our analysis by MS/MS was not quantitative, the position of the consensus sequence within the peptide did not seem to affect glycosylation, and we did not observe a sequence preference for Ser or Thr at position +2. Importantly, we did not detect glycosylation of short peptides such as DQNAT and DFNVT (data not shown), known substrates identified in vitro for bacterial OST from Campylobacter jejuni (28).
To further probe the acceptor site specificity of NGT, we tested glycosylation of the AcrA protein from C. jejuni, a substrate of bacterial and eukaryotic OSTs (29). MS analysis revealed that NGT was able to modify all four glycosites present in AcrA with a glucose moiety (Fig. 4C and supplemental Fig. 9).
Altogether, these experiments proved that NGT and OST share the basis for recognition of acceptor substrates. Moreover, glycosylation of AcrA demonstrated that NGT operates on folded proteins.
DISCUSSION
In this work, we have provided direct proof that the soluble enzyme NGT glycosylates asparagine side chains of peptides and proteins. The modified asparagines are found within the NX(S/T) sequence. Proline is not tolerated in the X position, and asparagine cannot be replaced by glutamine. Any amino acid is allowed in positions next to the sequon. NGT shows preference for long polypeptides, indicating that binding of the substrate to the enzyme might require an extended contact surface. At the same time, modification of different NX(S/T) sequences on one protein suggested that NGT does not recognize a specific target but acts as a key component of a general protein glycosylation system.
NGT transfers one glucose unit from UDP-glucose to acceptor sites, forming β-glycosidic linkages in a metal ion-independent manner. UDP-galactose appears to be accepted as a substrate donor, albeit with low efficiency.
These features mark a clear distinction from the conventional N-glycosylation, wherein OST is localized in a membrane, uses a complex lipid-bound oligosaccharide as a donor, requires a metal ion for catalysis, and exclusively modifies proteins trafficking along the secretory pathway. Moreover, the two enzymes are structurally unrelated, as NGT is predicted to have a GT-B fold, whereas OST clusters in the GT-C group (30). Given these premises, it is remarkable that NGT and OST exhibit the same NX(S/T) acceptor sequence specificity. The amino acid at position +2 appears not be essential for catalysis by OST, but is rather involved in binding of the acceptor substrate by the enzyme (31). Indeed, both NGT and OST can glycosylate sites that do not contain Ser or Thr at position +2 (7, 32–34).5 It will be important to determine the substrate recognition mode of NGT, the biophysical properties of the consensus sequence, and the mechanisms that selected this sequon for general glycosylation systems in two convergent evolutionary processes. In this context, it is interesting to note that the glycoproteins produced by the cytoplasmic glycosylation system of H. influenzae are secreted proteins.
Cytosolic N-glycosylation does not appear to be a single-monosaccharide modification, as the N-glucose can be extended with α1,6-linked glucose in A. pleuropneumoniae. The enzyme responsible for glucose transfer is considered a distant relative of family GT4 glycosyltransferases based on sequence similarity. Remarkably, whereas we confirmed that the enzyme exhibits a retaining mechanism, its polymerase activity appears to be a novel feature of a member of this class of glycosyltransferases.
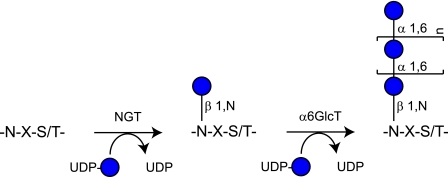
In summary, this study illustrates a cytoplasmic pathway of N-linked protein glycosylation (Fig. 5) with overlapping properties with the conventional N-glycosylation process. Future research will have to address the mechanism, extent, and physiological roles of this protein modification. Furthermore, as NGTs are encoded on the genome of relevant pathogens, it will be interesting to study how the immune system of the hosts reacts to this type of N-glycosylation. Finally, NGTs represent a promising tool that complements established platforms for production of glycoconjugates and for site-specific modification of proteins (35–37).
FIGURE 5.
Model of the cytoplasmic pathway of N-linked protein glycosylation in A. pleuropneumoniae. NGT recognizes NX(S/T) sequences within polypeptides and glycosylates asparagine residues. NGT is an inverting glycosyltransferase with specificity for UDP-glucose as a donor substrate. The N-linked glucose can be elongated by a polymerizing α6GlcT, which is sensitive to the UDP-glucose concentration.
Supplementary Material
Acknowledgments
We thank the members of the Aebi laboratory and Fred Allain for fruitful discussions and support. We thank Vincent Perreten and Guy Cornelis for providing genomic DNA and strains. We are grateful to Daan van Aalten for providing the pGEX-6P-1-XcOGT plasmid. Magnus Lundborg and Göran Widmalm are acknowledged for help with the chemical shift prediction server CASPER and immediate implementation of suggestions.
This work was supported by Swiss National Science Foundation Grant 3100AQ-105541 (to M. A.), Sinergia Grant CRSII3_127333, and ETH Zürich.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–9, Table 1, and additional references.
A. Nägeli, F. Schwarz, Y.-Y. Fan, and M. Aebi, unpublished data.
- OST
- oligosaccharyltransferase
- NGT
- N-glycosyltransferase
- α6GlcT
- α1,6-glucosyltransferase
- OGT
- O-GlcNAc-transferase
- TAMRA
- carboxytetramethylrhodamine
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- HSQC
- heteronuclear single-quantum coherence
- TOCSY
- total correlation spectroscopy.
REFERENCES
- 1. Kelleher D. J., Gilmore R. (2006) Glycobiology 16, 47R–62R [DOI] [PubMed] [Google Scholar]
- 2. Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) Trends Biochem. Sci. 35, 74–82 [DOI] [PubMed] [Google Scholar]
- 3. Anthony R. M., Nimmerjahn F., Ashline D. J., Reinhold V. N., Paulson J. C., Ravetch J. V. (2008) Science 320, 373–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Kooyk Y., Rabinovich G. A. (2008) Nat. Immunol. 9, 593–601 [DOI] [PubMed] [Google Scholar]
- 5. Apweiler R., Hermjakob H., Sharon N. (1999) Biochim. Biophys. Acta 1473, 4–8 [DOI] [PubMed] [Google Scholar]
- 6. Grass S., Buscher A. Z., Swords W. E., Apicella M. A., Barenkamp S. J., Ozchlewski N., St Geme J. W., 3rd. (2003) Mol. Microbiol. 48, 737–751 [DOI] [PubMed] [Google Scholar]
- 7. Gross J., Grass S., Davis A. E., Gilmore-Erdmann P., Townsend R. R., St Geme J. W., 3rd. (2008) J. Biol. Chem. 283, 26010–26015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grass S., Lichti C. F., Townsend R. R., Gross J., St Geme J. W., 3rd. (2010) PLoS Pathog. 6, e1000919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi K. J., Grass S., Paek S., St Geme J. W., 3rd, Yeo H. J. (2010) PLoS ONE 5, e15888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clarke A. J., Hurtado-Guerrero R., Pathak S., Schüttelkopf A. W., Borodkin V., Shepherd S. M., Ibrahim A. F., van Aalten D. M. (2008) EMBO J. 27, 2780–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schägger H. (2006) Nat. Protoc. 1, 16–22 [DOI] [PubMed] [Google Scholar]
- 12. Clore G. M., Bax A., Wingfield P., Gronenborn A. M. (1988) FEBS Lett. 238, 17–21 [DOI] [PubMed] [Google Scholar]
- 13. Markley J. L., Bax A., Arata Y., Hilbers C. W., Kaptein R., Sykes B. D., Wright P. E., Wüthrich K. (1998) J. Biomol. NMR 12, 1–23 [DOI] [PubMed] [Google Scholar]
- 14. Martinez-Fleites C., Macauley M. S., He Y., Shen D. L., Vocadlo D. J., Davies G. J. (2008) Nat. Struct. Mol. Biol. 15, 764–765 [DOI] [PubMed] [Google Scholar]
- 15. Hart G. W., Housley M. P., Slawson C. (2007) Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 16. Hanover J. A., Krause M. W., Love D. C. (2010) Biochim. Biophys. Acta 1800, 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohda D., Yamada M., Igura M., Kamishikiryo J., Maenaka K. (2007) Glycobiology 17, 1175–1182 [DOI] [PubMed] [Google Scholar]
- 18. Slynko V., Schubert M., Numao S., Kowarik M., Aebi M., Allain F. H. (2009) J. Am. Chem. Soc. 131, 1274–1281 [DOI] [PubMed] [Google Scholar]
- 19. van Zuylen C. W., de Beer T., Leeflang B. R., Boelens R., Kaptein R., Kamerling J. P., Vliegenthart J. F. (1998) Biochemistry 37, 1933–1940 [DOI] [PubMed] [Google Scholar]
- 20. Yamaguchi Y., Takizawa T., Kato K., Arata Y., Shimada I. (2000) J. Biomol. NMR 18, 357–360 [DOI] [PubMed] [Google Scholar]
- 21. Takeda T., Sugiura Y., Ogihara Y., Shibata S. (1980) Can. J. Chem. 58, 2600–2603 [Google Scholar]
- 22. Ogawa T., Nakabayashi S., Shibata S. (1983) Agric. Biol. Chem. 47, 1213–1218 [Google Scholar]
- 23. Wood M. J., Sampoli Benitez B. A., Komives E. A. (2000) Nat. Struct. Biol. 7, 200–204 [DOI] [PubMed] [Google Scholar]
- 24. Hansen P. I., Larsen F. H., Motawia S. M., Blennow A., Spraul M., Dvortsak P., Engelsen S. B. (2008) Biopolymers 89, 1179–1193 [DOI] [PubMed] [Google Scholar]
- 25. Williamson M. P., Trevitt C., Noble J. M. (1995) Carbohydr. Res. 266, 229–235 [Google Scholar]
- 26. Lundborg M., Widmalm G. (2011) Anal. Chem. 83, 1514–1517 [DOI] [PubMed] [Google Scholar]
- 27. Marshall R. D. (1974) Biochem. Soc. Symp. 40, 17–26 [PubMed] [Google Scholar]
- 28. Chen M. M., Glover K. J., Imperiali B. (2007) Biochemistry 46, 5579–5585 [DOI] [PubMed] [Google Scholar]
- 29. Kowarik M., Young N. M., Numao S., Schulz B. L., Hug I., Callewaert N., Mills D. C., Watson D. C., Hernandez M., Kelly J. F., Wacker M., Aebi M. (2006) EMBO J. 25, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 31. Lizak C., Gerber S., Numao S., Aebi M., Locher K. P. (2011) Nature 474, 350–355 [DOI] [PubMed] [Google Scholar]
- 32. Zielinska D. F., Gnad F., Wiśniewski J. R., Mann M. (2010) Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 33. Schwarz F., Lizak C., Fan Y. Y., Fleurkens S., Kowarik M., Aebi M. (2011) Glycobiology 21, 45–54 [DOI] [PubMed] [Google Scholar]
- 34. Valliere-Douglass J. F., Kodama P., Mujacic M., Brady L. J., Wang W., Wallace A., Yan B., Reddy P., Treuheit M. J., Balland A. (2009) J. Biol. Chem. 284, 32493–32506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang L. X. (2008) Carbohydr. Res. 343, 1509–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Antoine T., Priem B., Heyraud A., Greffe L., Gilbert M., Wakarchuk W. W., Lam J. S., Samain E. (2003) ChemBioChem 4, 406–412 [DOI] [PubMed] [Google Scholar]
- 37. Schwarz F., Huang W., Li C., Schulz B. L., Lizak C., Palumbo A., Numao S., Neri D., Aebi M., Wang L. X. (2010) Nat. Chem. Biol. 6, 264–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simon B. (2000) Structure Determination of the Active Center in the Integral Membrane Protein Bacteriorhodopsin and Development of Methods for Calculating the Effect of Pulsed Field Gradients on High-resolution NMR Spectra. PhD thesis, Free University of Berlin [Google Scholar]
- 39. Goddard T. D., Kneller D. G. (2008) Sparky, Version 3, University of California, San Francisco [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.