Abstract
Recent transcription of GAL genes transiently leaves an H3K4 methylation mark at their promoters, providing an epigenetic memory for the recent transcriptional activity. However, the physiological significance of this mark is enigmatic. In our study, we show that the transient H3K4 di- and trimethylation at recently transcribed GAL1 inhibited the reinduction of GAL1. The H3K4 methylation functioned by recruiting the Isw1 ATPase onto GAL1 and thereby limiting the action of RNA polymerase II during GAL1 reactivation. Strikingly, the H3K4 methylation was also observed at the promoters of inositol- and fatty acid-responsive genes after recent transcription and played a negative role in their reinduction. Taken together, our data present a new mechanism by which H3K4 methylation regulates gene transcription.
Keywords: Cell Metabolism, Chromatin Remodeling, Gene Expression, Histone Methylation, Transcription
Introduction
Chromatin, the physiological template of all eukaryotic genetic information, is made of repeating nucleosomes. Each nucleosome consists of 147-bp DNA wrapping around a histone octamer, including two each of H2A, H2B, H3, and H4 (1). In the process of gene transcription, chromatin structure can be modulated at several levels, such as ATP-dependent chromatin remodeling (2), histone modifications (3), and nucleosome disassembly and reassembly (4).
Histone H3K4 methylation is one of the major histone modifications conserved in eukaryotes. Set1 is the catalytic subunit of a large complex named COMPASS (5), which is responsible for mono-, di-, and trimethylation observed in yeast (6). Set1-dependent methylation requires histone ubiquitination of lysine 123 of histone H2B via the ubiquitin-conjugating Rad6-Bre1 complex (7). Also, it is regulated by the COMPASS subunits (8). Set1-mediated H3K4 methylation positively regulates the activation of a subset of euchromatic genes, such as RAM2, HAS1, INO1, PPH3, and MET16 (9, 10), but negatively regulates the activation of PHO5 and GAL1 (11). Histone H3K4 methylation, especially trimethylation, is usually associated with active transcription (12). However, our recent study shows that H3K4 trimethylation also associates with the repressed PHO5 gene and remains essentially unchanged during PHO5 activation and inactivation (13).
In the case of GAL genes, Set1-mediated H3K4 methylation is absent from their promoters under repressed conditions. During galactose induction, Set1 is co-transcriptionally recruited by the elongating RNA polymerase II (RNAPII)2 and methylates the corresponding genes. Interestingly, hypermethylation of H3K4 persists after transcriptional inactivation for considerable time (∼5 h), constituting a molecular memory within the mRNA coding region for recent transcriptional activity (14). It seems unlikely that such a robust and highly specific phenomenon occurring in living wild-type cells has no biological meaning. In this study, we confirm the post-transcriptional histone H3K4 methylation at the promoters of GAL1 and GAL10. We further show that, after recent transcription, H3K4 was hypermethylated not only at the promoters but also across the ORF regions of GAL1/10. When analyzing the role of H3K4 methylation during the initial induction and reinduction of GAL1, we surprisingly found that elimination of H3K4 di- and trimethylation had a minor effect on the initial induction, especially within the first hour upon shifting cells from glucose to galactose culture, but greatly increased the reinduction kinetics of GAL1. Mechanistically, the Isw1 ATPase was absent from the GAL1 gene after long-term (overnight) glucose repression but was present in the GAL1 gene after a 1-h glucose repression. Isw1 functioned in the same genetic pathway as Set1-dependent H3K4 methylation to inhibit the reactivation of GAL1. Interestingly, we found that H3K4 methylation was also associated with many other inducible genes, such as INO1 and FOX2, after recent transcription and inhibited the reinduction of the corresponding genes.
EXPERIMENTAL PROCEDURES
Strains and Antibodies
All strains originated from EUROSCARF. The strains constructed and antibodies used in this study are listed in supplemental Tables 1 and 2, respectively.
Quantitative RT-PCR
Total RNA was isolated from yeast cells with an RNeasy mini kit (Qiagen). cDNA was synthesized using a Moloney murine leukemia virus reverse transcriptase system and oligo(dT) (Promega). One microliter of the reverse transcription reaction was used in the subsequent real-time fluorescent quantitative PCR (Eppendorf).
ChIP Assay
The ChIP assay was performed as described previously (15). DNA from immunoprecipitated fractions and whole cell extracts (input) was analyzed by real-time PCR. The relative enrichment value represents the ratio of immunoprecipitated fractions to input at the indicated loci relative to the corresponding internal control. All ChIP experiments were performed in triplicate on paired isogenic wild-type and mutant strains.
Preparation of Yeast Chromatin
Yeast chromatin was prepared as described previously (16). The chromatin pellets were resuspended in 1× SDS loading buffer, boiled, separated by 15% SDS-PAGE, and then subjected to Coomassie Blue staining or Western blotting.
RESULTS
Histone H3K4 Methylation Associates with Recently Repressed GAL1/10
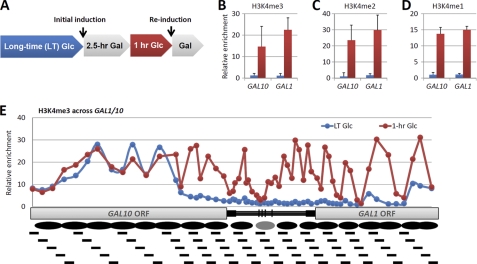
In a previous study, Struhl and co-workers (14) showed that histone H3K4 methylation persisted after transcriptional repression for at least 5 h. To study the function of the persisting H3K4 methylation, we set up a similar assay detecting the level of H3K4 methylation at GAL1. Briefly, long-term glucose-cultured yeast cells were shifted into galactose culture for 2.5 h (initial induction). The culture was then shifted back to glucose culture for an additional 1 h. Finally, the short-term repressed cells were rechallenged by galactose (reinduction) (Fig. 1A). We compared the level of H3K4 methylation at the promoters of GAL10 and GAL1 between the long- and short-term repressed states. ChIP combined with real-time PCR was performed to detect the relative enrichment of H3K4 trimethylation (H3K4me3), dimethylation (H3K4me2), and monomethylation (H3K4me1) at the gene promoters (3). The results showed that H3K4me3, H3K4me2, and H3K4me1 were almost absent from the long-term repressed promoters but were readily apparent at the 1-h repressed promoters (Fig. 1, B–D). These data are consistent with the previous report showing that H3K4 methylation persisted for >5 h after transcriptional repression of GAL10 (14). Distribution analysis of H3K4me3 with 60 overlapping primers throughout the promoter and gene coding regions of GAL1/10 (17) showed that, in 1-h repressed cells, H3K4me3 occupied both the promoter regions and the ORF regions of GAL1/10 genes (Fig. 1E). Together, these data suggest that repression-associated H3K4me3 covers the entire GAL1/10 genes.
FIGURE 1.
Persistence of H3K4 methylation at the GAL1/10 gene after transcriptional inactivation. A, schematic of galactose induction and reinduction time course used for the following data sets. B–D, ChIP of H3K4me3, H3K4me2, and H3K4me1, respectively, at the promoters of GAL1/10 under either long-term or 1-h glucose conditions. The y axis shows the abundance (immunoprecipitated fraction/input) at indicated loci relative to TEL-VIR (subtelomeric region of the right cerm of chromosome VI). Values that are >1 indicate more enrichment than the background. E, ChIP of H3K4me3 across GAL1/10 genes. The bottom black lines indicate the locations of the overlapping primers. The black ovals are predicted nucleosomal loci.
Elimination of Di- and Trimethylation of H3K4 Greatly Increases the Reactivation Rate of GAL1
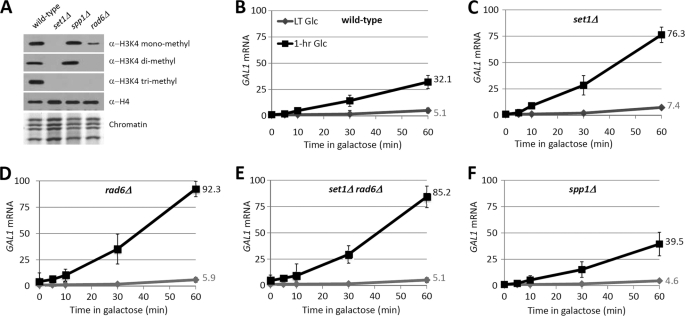
The transcriptional induction of GAL1 occurs with faster kinetics if it has been previously expressed, a phenomenon called transcriptional memory (18–20). Given that the H3K4 methylation was associated with recently expressed GAL1, we then wondered whether H3K4 methylation was involved in the formation of the transcriptional memory. To test this idea, we detected the effect of SET1 deletion, which abolished H3K4 methylation (Fig. 2A), on the initial induction and reinduction of GAL1. A time course mRNA analysis was performed under conditions of repeated glucose repression and galactose induction as described for Fig. 1A. We detected the mRNA level of GAL1 in the first hour upon galactose induction. From the results, we found that, in wild-type cells, the reactivation kinetics of GAL1 after a 1-h repression was much more robust than the initial activation kinetics (Fig. 2B), supporting the transcriptional memory of GAL1. Previous reports showed that Set1 repressed GAL1 transcription in galactose-containing medium plus 0.5% glucose (11) or GAL1 induction from raffinose to galactose culture (21). We found that SET1 deletion had a minor effect on the induction kinetics of GAL1 from long-term glucose to galactose culture, especially within the first 3 h in galactose culture (Fig. 2, B and C, and supplemental Fig. 1A). In striking contrast, 1-h repressed GAL1 was activated more rapidly in set1Δ cells than in wild-type cells (Fig. 2, compare B and C). We further checked if SET1 deletion also increased the peak level of GAL1 transcription during reactivation. The results demonstrated that the peak level in set1Δ cells during reactivation was comparable with that during initial activation (supplemental Fig. 1, compare A and B). Although Set1-dependent H3K4 methylation slightly inhibits the initial induction of GAL1 from glucose to galactose medium, it has a much stronger repressive effect on the reactivation of GAL1.
FIGURE 2.
Elimination of H3K4 di- and trimethylation accelerates the reactivation of GAL1. A, upper panel, Western blot of wild-type and mutant strains. Antibodies used are indicated on the right. Lower panel, Coomassie Blue-stained histones (chromatin). B–F, induction and reinduction kinetics of GAL1 in the genetic background as indicated. Cells were treated as depicted in Fig. 1A before galactose culturing. The mRNA level of GAL1 was normalized to that of ACT1. The relative mRNA level in long-term (LT) glucose was defined as 1.
Next, we wondered which type(s) of H3K4 methylation was involved in the regulation of GAL1 reactivation. We inactivated Spp1, one of the key subunits of COMPASS (8), and Rad6, the ubiquitin-conjugating enzyme responsible for H2BK123 ubiquitylation (supplemental Fig. 2A) (22). Western blot analysis showed that SPP1 deletion specifically eliminated the trimethylation of H3K4, whereas RAD6 deletion eliminated both di- and trimethylation of H3K4 (Fig. 2A). We then analyzed the roles of Rad6 and Spp1 in the initial induction and reinduction of GAL1. Notably, RAD6 deletion also increased the reactivation rates of GAL1 and RAD6; SET1 double deletion resembled this phenotype (Fig. 2, B–E). We also found that H2BK123 ubiquitylation was associated with GAL1/10 promoters 1 h after glucose repression (supplemental Fig. 2B). Therefore, H2B ubiquitylation functions in the same genetic pathway as H3K4 methylation. In contrast, SPP1 deletion did not affect the reactivation rate of GAL1 (Fig. 2, compare B and F), suggesting that either di- or trimethylation is sufficient for the function of H3K4. Therefore, we concluded that H2BK123 ubiquitylation-dependent H3K4 methylation after transcriptional inactivation inhibits the reactivation of GAL1.
Isw1 Negatively Regulates Galactose Memory in an H3K4 Methylation-dependent Manner
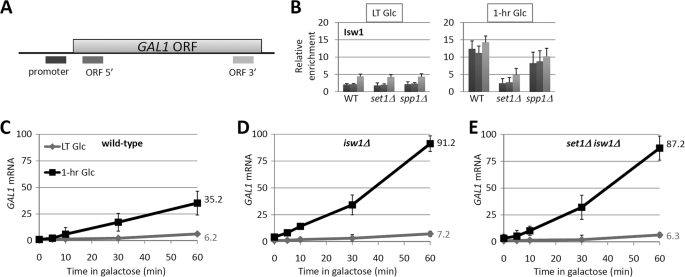
According to the literature, the repressive effect of H3K4me3 on transcription involves the Isw1 ATPase (23, 24). We hypothesized that the reduction of transcriptional competence of GAL genes during reactivation by H3K4 methylation requires Isw1. We then examined the binding of Isw1 protein at the promoter, 5′-region, and 3′-region of GAL1 (Fig. 3A) after long-term or 1-h glucose culturing. As shown in Fig. 3B, there was little Isw1 binding at GAL1 in the long-term repressed state. Interestingly, transcriptional inactivation recruited Isw1 to the GAL1 gene (Fig. 3B). Set1, but not Spp1, was essential for the association of Isw1 with GAL1 (Fig. 3B). Accordingly, ISW1 deletion led to a subtle increase in the kinetics of the initial induction of GAL1 but a substantial increase in mRNA levels during the reinduction of GAL1 (Fig. 3, compare C and D). isw1Δset1Δ double mutant cells resembled isw1Δ single mutant cells in the induction and reinduction kinetics of GAL1 (Fig. 3, C–E). Therefore, we concluded that the Isw1 ATPase occupies the recently repressed GAL1 gene in an H3K4 methylation-dependent fashion, and, like H3K4 methylation, the Isw1 ATPase particularly inhibits the reactivation of GAL1.
FIGURE 3.
Histone H3K4 methylation recruits the Isw1 ATPase to recently transcribed GAL1. A, schematic of the GAL1 gene and the location of the primers used in the following experiments. B, ChIP of Myc-tagged Isw1 in wild-type and mutant cells. Cells were treated as depicted in Fig. 1A. C–E, induction and reinduction kinetics of GAL1 in wild-type and mutant cells. mRNA analysis was performed as described in the legend to Fig. 2B. LT, long-term.
Histone H3K4 Methylation Suppresses the Action of RNAPII during GAL1 Reactivation by Isw1
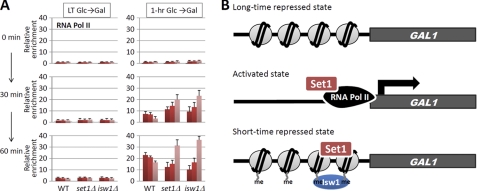
To address how the recruitment of Isw1 affects the induction of GAL1, we examined the occupancy of RNAPII at the promoter and 5′- and 3′-ends of the GAL1 ORF (Fig. 3A). ChIP of Rpb3, one of the subunits of RNAPII, showed that, in all strains, Rpb3 was absent from GAL1 before induction or reinduction (Fig. 4A), excluding the possibility that RNAPII pre-occupied GAL1 in the set1Δ or isw1Δ strain. As expected, when the GAL1 gene was initially induced, little Rpb3 was recruited in the wild-type and mutant strains (Fig. 4A). For the wild-type strain, we observed an obvious 5′-bias for RNAPII distribution during the first hour of the reinduction procedure. In striking contrast, in the isw1Δ strain, Rpb3 was localized predominantly to the 3′-region of GAL1 during GAL1 reactivation, suggesting that Isw1 inhibits transcription elongation during GAL1 reactivation. A similar 3′-bias for Rpb3 was also evident in the set1Δ strain (Fig. 4A), suggesting a related function.
FIGURE 4.
Histone H3K4 methylation reduces the processivity of RNAPII during GAL1 reactivation by Isw1. A, ChIP of Myc-tagged Rpb3 in wild-type and mutant cells cultured in glucose for various time as depicted in Fig. 1A. LT, long-term; RNA Pol II, RNAPII. B, model for the inhibition of GAL1 reactivation by short-term H3K4 methylation. In the long-term repressed state, histone H3K4 methylation is absent from GAL1. Upon gene activation, Set1 is recruited by the elongating RNAPII and persists for a short time after the shutoff of gene transcription. This short stay of Set1 is sufficient for methylating the newly incorporated histone at the corresponding gene upon transcriptional inactivation. The Isw1 ATPase is then recruited by di- and trimethylated H3K4 onto methylated GAL1 and suppresses the action of RNAPII during gene reactivation. Because Set1 is quickly released from the gene after repression, the induced H3K4 methylation is diluted and finally lost with cell division.
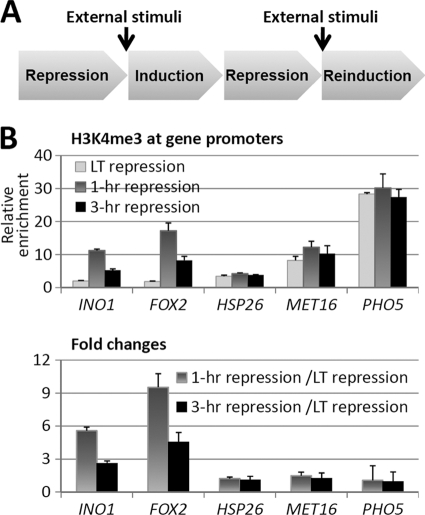
Epigenetic Memory of Inositol- and Fatty Acid-responsive Genes after Recent Transcription
We were curious whether the function of H3K4 methylation in galactose metabolism could be extended to other cellular processes. Therefore, we chose five other external stimulus-responsive genes, including INO1 (induced by inositol), FOX2 (induced by oleic acid), HSP26 (induced by high temperature), MET16 (induced by methionine starvation), and PHO5 (induced by phosphate starvation), and investigated whether these stimuli affect H3K4 methylation at the corresponding genes (Fig. 5A). In the long-term repressed state, H3K4me3 was absent from the promoters of INO1, FOX2, and HSP26 but present at the promoters of MET16 and PHO5 (Fig. 5B). Interestingly, in the 1- or 3-h repressed state, the H3K4me3 level was significantly increased at the promoters of INO1 and FOX2 but remained unchanged at the promoters of HSP26, MET16, and PHO5 (Fig. 5B), suggesting that post-nutrient stimuli are able to induce H3K4 methylation at the promoters of INO1 and FOX2.
FIGURE 5.
Recent transcription of inositol- or fatty acid-responsive genes establishes H3K4 methylation at the promoters of the corresponding genes. A, schematic of the induction and reinduction time course of the inducible genes used for the following data sets. B, ChIP of H3K4me3 at the promoter of inducible genes before and after recent induction as depicted in A. The upper panel shows the abundance of H3K4me3, and the lower panel shows the -fold changes after recent induction. LT, long-term.
SET1 Deletion Increases the Reactivation Rates of Inositol- and Fatty Acid-responsive Genes
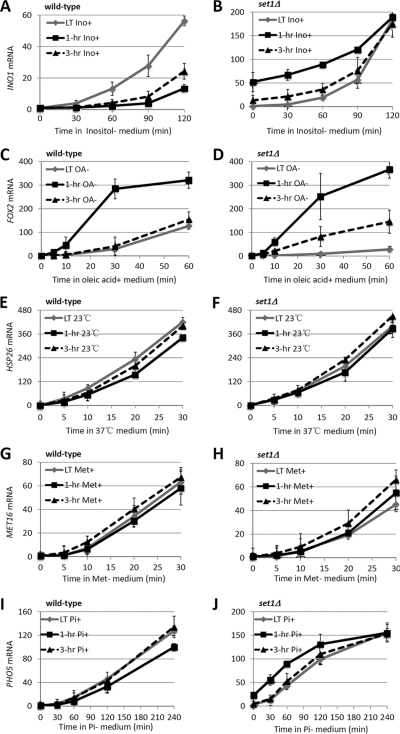
Next, we assessed the function of H3K4 methylation in the induction and reinduction of these genes. The mRNA analysis of INO1 showed that, in wild-type cells, the reactivation rate of INO1 was much slower than the initial one (Fig. 6A) (18, 25). In striking contrast, the reactivation kinetics of INO1 resembled the initial activation kinetics in set1Δ cells (Fig. 6, A and B), suggesting that, under physiological conditions, H3K4 methylation negatively regulates the reactivation of INO1. Because H3K4 methylation repressed the initial induction of INO1 (Fig. 6, A and B), we concluded that the repression-induced H3K4 methylation strengthens the repressive effect on INO1 reinduction. Additionally, we noticed that, in set1Δ cells, the mRNA level of INO1 after a 1-h repression was much higher than that in the long-term repressed state (Fig. 6, A and B), indicating that SET1 also delays the transcriptional repression of INO1.
FIGURE 6.
H3K4 methylation after transcriptional inactivation negatively regulates the reactivation of INO1 and FOX2. The Induction and reinduction kinetics are shown for INO1 (A and B), FOX2 (C and D), HSP26 (E and F), MET16 (G and H), and PHO5 (I and J) in wild-type and set1Δ cells. LT, long-term; Ino, inositol; OA, oleic acid.
By analyzing the induction and reinduction kinetics of FOX2, we were surprised to find that FOX2 also conferred transcriptional memory: FOX2 was activated much more rapidly when rechallenged with oleic acid (Fig. 6C). However, the transcriptional memory of FOX2 was lost within 3 h (Fig. 6C). This is likely a bona fide short-term memory because such a dramatic difference in mRNA levels is hard to be explained by dilution of the cells due to cell division. Strikingly, the Set1-mediated H3K4 methylation differentially affected the induction and reinduction of FOX2: in set1Δ cells, the initial induction rate of FOX2 was dramatically reduced, whereas the reinduction rate of FOX2 was restored to the wild-type level (Fig. 6, C and D). There are two possibilities for this phenomenon. First, Set1-mediated H3K4 methylation specifically regulates the initial induction of FOX2; second, Set1-mediated H3K4 methylation positively regulates the initial induction of FOX2, but, during reactivation of FOX2, the repression-associated H3K4 methylation overcomes the positive effect of co-transcriptional H3K4 methylation. We favor the later model because H3K4 methylation was absent in long-term repressed FOX2 but was abundant in recently repressed FOX2, providing a molecular basis for an additional function of Set1-mediated H3K4 methylation in FOX2 reactivation.
To investigate whether Set1-mediated H3K4 methylation regulates the reactivation of INO1 and FOX2 by the same mechanism as in GAL1, we detected the effect of RAD6 or SPP1 deletion on the induction and reinduction of INO1 and FOX2. The results showed that the induction and reinduction kinetics of both genes in either rad6Δ or spp1Δ cells resembled the wild-type levels (supplemental Fig. 3, A and B), implying that mono-, di-, and trimethylation of H3K4 are all required. Moreover, ISW1 deletion did not affect the transcription of INO1 and FOX2 (supplemental Fig. 3, A and B), suggesting that Set1 regulates the transcription of these genes independently of the Isw1 pathway. For the three other genes, whose H3K4 methylation was unaffected by recent stimuli, the reinduction kinetics of these genes were similar to the initial induction kinetics in both wild-type and set1Δ cells (Fig. 6, E–J), suggesting that they do not show transcriptional memory and that H3K4 methylation does not differentiate the reinduction rate from initial induction rate of these genes. Taken together, post-stimulus-dependent H3K4 methylation suppresses gene reactivation of genes.
DISCUSSION
In this study, we investigated the dynamic association of histone H3K4 methylation at GAL1 in the time course of repression-initial induction-repression-reinduction. Elimination of H3K4 methylation specifically affected the reactivation rate of GAL1 within the first hour of galactose induction. Furthermore, we showed that both di- and trimethylation of H3K4 were involved in the regulation of GAL1 reactivation. In contrast to that under long-term glucose conditions, the Isw1 ATPase was transiently localized to GAL1 and reduced the transcriptional competence of GAL1 under the short-term glucose conditions (Fig. 4B). Interestingly, the inhibition of reinduction of genes by Set1-mediated H3K4 methylation was also observed in many other inducible genes, such as inositol- and fatty acid-responsive genes. Therefore, our data suggest that the transient H3K4 methylation induced by recent transcription reduces the transcriptional competence of the corresponding genes for reactivation.
Repressive Role of Histone H3K4 Methylation during Gene Activation
Despite the general idea that Set1 is associated with active transcription (26–28), an increasing number of arguments suggest that Set1-mediated H3K4me2/3 is also involved in gene repression. It was shown that Set1 is required for silencing of Ty1 retrotransposons GAL1 and PHO84 (6, 11), subsequently found to be noncoding RNA-mediated (21, 29–31). Recently, scientists (21, 32) provided evidence that H3K4me2/3 recruits the Rpd3S complex to GAL1 as well as many other inducible genes that show cryptic transcription. In the case of PHO5, our recent study showed that H3K4 methylation recruits the Rpd3L complex, rather than the Rpd3S complex, to the PHO5 promoter and suppresses the aberrant chromatin remodeling at the PHO5 promoter (13). In this study, we have shown that H3K4 methylation recruited the Isw1 ATPase to GAL1 (Fig. 3). It has been documented that the chromatin association of Isw1 depends on Set1 in vivo and in vitro (24). Therefore, Isw1 is an important effector of Set1 in regulating gene transcription (23, 24). Our data showed that Isw1 mediated the repressive effect of Set1 on the reactivation of GAL1 (Figs. 3 and 4). Isw1 is also found to antagonize the positive role of the SWI/SNF complex in the reactivation of GAL genes (19). Therefore, it is likely that H3K4 suppresses GAL1 reactivation through the chromatin remodeling pathway. However, Isw1 seems dispensable for the function of Set1 in the transcription of INO1 and FOX2 (supplemental Fig. 3, A and B). This is not unexpected because the effectors of Set1 usually vary at different genes. It will be of interest to determine the effectors of Set1 in the transcription of INO1 and FOX2.
It was recently reported that Set1 does not preferentially affect GAL1 reactivation (19, 33), whereas our data argue that Set1 inhibits GAL1 reactivation. The discrepancy could be attributed to different time course settings for studying GAL1 induction and reinduction. Although SET1 deletion increased GAL1 reactivation in all three systems, the relative differences in previous systems were modest (supplemental Fig. 4). In addition, we noticed that either the PCR signal of GAL10 on the agarose gel (see Fig. 3B in Ref. 33) or the signal of GAL1 on the Northern blot (see Fig. 3A in Ref. 19) during reactivation in wild-type cells had already been saturated and was therefore hard to distinguish from that in set1Δ cells. In our study, we monitored the mRNA level of GAL1 in a more quantitative manner by using real-time PCR and thereby observed an obvious increase in the GAL1 reactivation rate by SET1 deletion.
Regulation of Gene Reactivation by Short-term H3K4 Methylation Represents a Distinct Function of H3K4 Methylation in Gene Transcription
The function of H3K4 methylation after transcriptional inactivation can be either in concert with or contrary to that of co-transcriptional H3K4 methylation. In the case of galactose- or inositol-responsive genes, co-transcriptional H3K4 methylation negatively regulated the expression of the corresponding genes during the activation processes (Fig. 2, B and C, and Fig. 6, A and B). Transient H3K4 methylation after transcriptional activation further reduced the transcriptional competence of these genes for reactivation. Hence, the function of H3K4 methylation in gene reactivation is a combined effect of both the recent transcription-induced one and the co-transcription one. In the case of fatty acid-responsive genes, the situation is different. Co-transcriptional H3K4 methylation positively regulated the initial induction of FOX2 (Fig. 6, C and D). However, SET1 deletion had little effect on the reinduction of FOX2 compared with the wild type (Fig. 6, C and D), suggesting that H3K4 methylation after transcriptional inactivation plays a negative role in FOX2 transcription, which counteracts the positive effect of co-transcriptional H3K4 methylation during FOX2 reinduction.
Possible Physiology of the Epigenetic Memory after Recent Transcription
The inhibition of stimulus-responsive genes by H3K4 methylation represents a mechanism for the yeast cells to delay the second-round transcriptional response to external stimuli. Why do the yeast cells do that? We propose that yeast cells undergo a series of cellular changes after recent stimuli, and, regarding the changes to a certain stimulus, some are beneficial, some are dispensable, whereas some are even harmful. Irreversible cellular changes may result in either evolution or diseases. Therefore, cells must possess an ability to quickly reset their inherited status after “adaptive” changes (34). Recent studies by others also suggest mechanisms for the yeast cells to accelerate the second-round transcriptional response to galactose stimuli (18–20). Therefore, we propose that there is a balance between different mechanisms when the yeast cells adapt to the environment.
Supplementary Material
Acknowledgment
We thank Dr. Brian A. Lenzmeier for critical reading of the manuscript.
This work was supported by National Natural Science Foundation of China Grant 90919027 and Ministry of Science and Technology Grant 2007CB914502.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables 1 and 2.
- RNAPII
- RNA polymerase II.
REFERENCES
- 1. Kornberg R. D. (1974) Science 184, 868–871 [DOI] [PubMed] [Google Scholar]
- 2. Cairns B. R. (2005) Curr. Opin. Genet. Dev. 15, 185–190 [DOI] [PubMed] [Google Scholar]
- 3. Li B., Carey M., Workman J. L. (2007) Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 4. Mellor J. (2006) Trends Genet. 22, 320–329 [DOI] [PubMed] [Google Scholar]
- 5. Miller T., Krogan N. J., Dover J., Erdjument-Bromage H., Tempst P., Johnston M., Greenblatt J. F., Shilatifard A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Briggs S. D., Bryk M., Strahl B. D., Cheung W. L., Davie J. K., Dent S. Y., Winston F., Allis C. D. (2001) Genes Dev. 15, 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun Z. W., Allis C. D. (2002) Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 8. Schneider J., Wood A., Lee J. S., Schuster R., Dueker J., Maguire C., Swanson S. K., Florens L., Washburn M. P., Shilatifard A. (2005) Mol. Cell 19, 849–856 [DOI] [PubMed] [Google Scholar]
- 9. Nislow C., Ray E., Pillus L. (1997) Mol. Biol. Cell 8, 2421–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. (2002) Nature 419, 407–411 [DOI] [PubMed] [Google Scholar]
- 11. Carvin C. D., Kladde M. P. (2004) J. Biol. Chem. 279, 33057–33062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ng H. H., Jeppesen P., Bird A. (2000) Mol. Cell. Biol. 20, 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang S. S., Zhou B. O., Zhou J. Q. (2011) Mol. Cell. Biol. 31, 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ng H. H., Robert F., Young R. A., Struhl K. (2003) Mol. Cell 11, 709–719 [DOI] [PubMed] [Google Scholar]
- 15. Zhou B. O., Wang S. S., Xu L. X., Meng F. L., Xuan Y. J., Duan Y. M., Wang J. Y., Hu H., Dong X., Ding J., Zhou J. Q. (2010) Mol. Cell. Biol. 30, 2391–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou B. O., Wang S. S., Zhang Y., Fu X. H., Dang W., Lenzmeier B. A., Zhou J. Q. (2011) PLoS Genet. 7, e1001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bryant G. O., Prabhu V., Floer M., Wang X., Spagna D., Schreiber D., Ptashne M. (2008) PLoS Biol. 6, 2928–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brickner D. G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P. C., Widom J., Brickner J. H. (2007) PLoS Biol. 5, e81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kundu S., Horn P. J., Peterson C. L. (2007) Genes Dev. 21, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zacharioudakis I., Gligoris T., Tzamarias D. (2007) Curr. Biol. 17, 2041–2046 [DOI] [PubMed] [Google Scholar]
- 21. Pinskaya M., Gourvennec S., Morillon A. (2009) EMBO J. 28, 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robzyk K., Recht J., Osley M. A. (2000) Science 287, 501–504 [DOI] [PubMed] [Google Scholar]
- 23. Morillon A., Karabetsou N., Nair A., Mellor J. (2005) Mol. Cell 18, 723–734 [DOI] [PubMed] [Google Scholar]
- 24. Santos-Rosa H., Schneider R., Bernstein B. E., Karabetsou N., Morillon A., Weise C., Schreiber S. L., Mellor J., Kouzarides T. (2003) Mol. Cell 12, 1325–1332 [DOI] [PubMed] [Google Scholar]
- 25. Tan-Wong S. M., Wijayatilake H. D., Proudfoot N. J. (2009) Genes Dev. 23, 2610–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 27. Shilatifard A. (2006) Annu. Rev. Biochem. 75, 243–269 [DOI] [PubMed] [Google Scholar]
- 28. Sims R. J., 3rd, Reinberg D. (2006) Genes Dev. 20, 2779–2786 [DOI] [PubMed] [Google Scholar]
- 29. Berretta J., Pinskaya M., Morillon A. (2008) Genes Dev. 22, 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Houseley J., Rubbi L., Grunstein M., Tollervey D., Vogelauer M. (2008) Mol. Cell 32, 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Camblong J., Beyrouthy N., Guffanti E., Schlaepfer G., Steinmetz L. M., Stutz F. (2009) Genes Dev. 23, 1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L. M. (2009) Nature 457, 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lainé J. P., Singh B. N., Krishnamurthy S., Hampsey M. (2009) Genes Dev. 23, 2604–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Margueron R., Reinberg D. (2010) Nat. Rev. Genet. 11, 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.