Background: Endoplasmic reticulum (ER) stress initiates apoptosis by an unknown mechanism.
Results: ATF6, a component of the cytoprotective mechanism triggered by ER stress, or a newly identified effector of ATF6 mediates apoptosis with reduction of an anti-apoptotic protein.
Conclusion: There is a potential link between ATF6 and an apoptotic regulator.
Significance: The ATF6 pathway may control switching the cell fate from self-defense to self-destruction.
Keywords: Apoptosis, Bcl-2 Family Proteins, Caspase, Cell Death, Cell Differentiation, ER Stress
Abstract
Endoplasmic reticulum (ER) stress is involved in both physiological and pathological apoptosis. ER stress triggers the unfolded protein response (UPR), which can then initiate apoptosis, when the cell fails to restore ER homeostasis. However, the mechanism employed by the UPR to lead cells into apoptosis is unknown. Among the three proximal sensors of ER stress, activating transcription factor-6 (ATF6) is specifically activated in apoptotic myoblasts during myoblast differentiation. This implies that active ATF6 has the ability to mediate apoptosis. Here, we demonstrate that overexpression of active ATF6 induced apoptosis in myoblast cells. Moreover, coexpression of a dominant negative form of ATF6 suppressed apoptosis. This suggested that apoptosis-related pathways depended on ATF6-mediated transcription activation. ATF6 caused up-regulation of the WBP1 (WW domain binding protein 1), probably via an indirect mechanism. Furthermore, WBP1 was also found to be proapoptotic. The silencing of WBP1 with small hairpin RNAs caused partial, but significant suppression of ATF6-induced apoptosis. Overexpression of active ATF6 or WBP1 caused a specific reduction in an anti-apoptotic protein, Mcl-1 (myeloid cell leukemia sequence 1). This suggested a molecular link between the UPR and an apoptosis regulator. Neither Bcl-2 nor Bcl-xL were reduced upon apoptosis induction in C2C12 cells that overexpressed ATF6 or WBP1. Cells treated with ER stressors underwent apoptosis concomitant with an up-regulation of WBP1 and suppression of Mcl-1. These results suggested that Mcl-1 is a determinant of cell fate, and ATF6 mediates apoptosis via specific suppression of Mcl-1 through up-regulation of WBP1.
Introduction
Endoplasmic reticulum (ER)2 stress occurs when ER homeostasis is lost due to an overload of protein folding in the ER (1). ER stress triggers an evolutionarily conserved response termed the unfolded protein response (UPR) (2). The UPR alters transcriptional and translational programs to cope with the accumulation of unfolded or misfolded proteins. Failure to resolve a protein-folding defect and restore ER homeostasis induces the UPR to initiate apoptosis. This protects the organism by removing the stressed cell (2, 3). ER stress-induced apoptosis is involved in many diseases (1), but it is also observed during normal development (4).
The UPR is mediated by three ER-resident transmembrane proteins that sense ER stress and signal downstream pathways. These proximal sensors include the kinase and ribonuclease IRE1, the eIF2α kinase PERK, and activating transcription factor-6 (ATF6) (5). ER stress induces the autophosphorylation and activation of IRE1 and PERK, which results in the activation of downstream transcription factors. In addition, ER stress leads to ATF6 transit through the Golgi complex, where it is sequentially cleaved for activation by the proteases S1P and S2P (6). The cleaved N-terminal ATF6 cytoplasmic domain is released from the Golgi membrane, and it translocates to the nucleus to regulate transcription. These three sensor proteins comprise parallel pathways connected by signaling cross-talk through gene expression (7).
Several mechanisms have been proposed that link the distressed ER to apoptosis, including the activation of transcription factors and expression of Bcl-2 family proteins. However, the mechanism triggered by the UPR that eventually leads to apoptosis after prolonged ER stress is unknown (8). For example, in some experimental settings, overexpression of a protein homologous to CHOP (CCAAT enhancer-binding protein, also known as GADD153) may be involved. CHOP is an ER stress-inducible member of the C/EBP family of bZIP transcription factors that induces apoptosis through a Bcl-2 inhibitable mechanism (9, 10). However, CHOP−/− cells are only partially resistant to ER stress-induced apoptosis. This observation suggested that a CHOP-independent mechanism can also induce apoptosis in response to ER stress. In a previous study, a genome-wide screen of an RNAi library did not identify any genes associated with ER stress-induced apoptosis in HeLa cell derivatives (11). This may reflect multiplicity in apoptotic signal transduction during ER stress-induced apoptosis. Multiplicity may occur because ER stressors used in typical experiments activate all three arms of the UPR.
We previously showed that ER stress signaling was involved in the induction of developmental apoptosis in muscle tissues through the activation of caspase-12 (4). Myoblast cells exhibit considerable morphological changes during myogenesis, and they fuse into multinucleated myotubes. Myoblast fusion is associated with apoptosis in a subpopulation of cells (12), a phenomenon that has long been considered an example of cell degeneration during normal vertebrate ontogeny (13). Our previous study suggested that, during myogenesis, ER stress signaling is mediated mainly by ATF6. When C2C12 mouse myoblast cells were cultured under differentiation conditions, we observed specific activation of the ATF6 pathway in dying cells, but not the IRE1 or PERK pathways (4). When a specific inhibitor of S1P was added, these cells were resistant to apoptosis; moreover, concomitant with the reduction in cell death, we found no detectable activation of ATF6 (4). These results prompted us to explore the possibility that active ATF6 has the ability to mediate apoptosis on its own. In the present study, we examined the effect of forced expression of active ATF6 in C2C12 cells without simultaneously activating all three proximal arms of the UPR.
EXPERIMENTAL PROCEDURES
Plasmid DNAs
FANTOM3 cDNA clones of ATF6, WBP1, CDKap2, Creld2, Sel1, Tmem50b, PDCD4, and arginine rich, mutated in early stage of tumors (ARMET) were obtained from the Genome Exploration Research Group, Genomic Sciences Center of RIKEN (14). The names of these genes are listed in supplemental Table 1. CHOP, Mcl (myeloid cell leukemia sequence)-1, and Aloxe3 cDNAs were amplified by PCR from mouse pancreas cDNA pools (Zyagen). Human ATF6 cDNA was kindly provided by R. Prywes (Columbia University, Biological Sciences). cDNAs were cloned into a variety of vectors, including pcDNA3.1(−) (Invitrogen), pEGFPC1, pEGFPN3, and pDsRed-Monomer-C1 (Clontech).
Cell Culture
C2C12 cells (RIKEN Cell Bank, Tsukuba, Japan) were grown in DMEM medium (Invitrogen) with 20% FBS (Invitrogen) at 37 °C and 5% CO2 as described previously (4). NIH-3T3 and COS-1 cells were grown in DMEM medium with 10% FBS. mIMCD-3 cells (American Type Culture Collection) were grown in DMEM:F12 medium (Invitrogen) with 10% FBS. MCF-7 cells (Cell Resource Center for Biomedical Research, Tohoku University) were cultured in RPMI1640 medium (Invitrogen) with 10% FBS. Cell treatment with ER stressors was performed as described previously (15). Cell transfection was performed with Superfect Transfection Reagent (Qiagen) (16). C2C12 cells (5 × 106 cells/ml) were electroporated with 10 μg of DNA with a Microporator MP-100 (Invitrogen) and 100-μl microporation tips, according to the manufacturer's protocol. Six to eight h after electroporation, dead cells (floating cells) were removed by suction, and electroporated cells were incubated in fresh medium.
Apoptosis Assay by Transfection
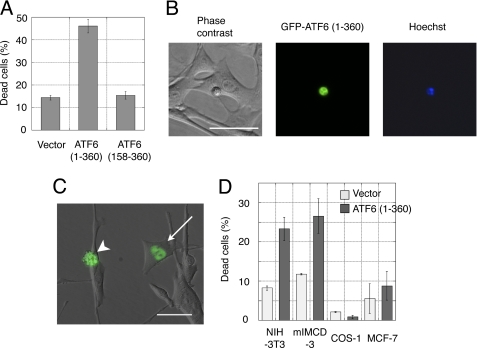
At 24 h post-transfection, cells were observed under a fluorescence microscope (16). More than 200 GFP-positive cells were randomly selected from each transfection experiment, and dead cells were identified by their morphology (small, round cells). Dead cells also showed nuclear condensation, a hallmark of apoptosis, detected by staining with 1 μg/ml Hoechst dye 33342 (Fig. 1B). Some dead cells underwent membrane blebbing (Fig. 1C).
FIGURE 1.
Overexpression of active ATF6 induced apoptosis in myoblast cells. A, apoptosis induced by GFP-ATF6(1–360) was assessed by cell morphology. Bars represent an average of three independent experiments. B, cell morphology 24 h after cell transfection with GFP-ATF6(1–360), shows nuclei stained with Hoechst 33342 dye. Scale bar, 50 μm. C, merged image of GFP-ATF6(1–360) and corresponding phase contrast images. Arrow and arrowhead indicate live and dead cells, respectively. Scale bar, 50 μm. D, active ATF6 induced apoptosis in different cell lines.
Statistical Analysis
Data are presented as the mean ± S.D. All bar graphs represent n = 3 for each experimental group. Significant differences among groups were determined by analysis of variance followed by the Student's t test.
Microarray Analysis
Twenty-four h after electroporation, total RNA was isolated with an RNeasy mini kit (Qiagen). First- and second-strand cDNAs were synthesized from 1 μg of total RNA with the One-cycle cDNA Synthesis Kit (Affymetrix) according to the manufacturer's instructions. cRNA was synthesized and labeled with biotinylated UTP by in vitro transcription with the in vitro transcription labeling kit (Affymetrix) and the T7 promoter-coupled double-stranded cDNA as template. The labeled cRNA was separated from unincorporated ribonucleotides by filtering through an in vitro transcription cRNA cleanup spin column (Affymetrix). Biotin-labeled cRNAs were hybridized to GeneChip Mouse Genome 430a 2.0 Array chips (Affymetrix) and analyzed with the GeneChip Scanner 3000 7G (Affymetrix). Raw expression data were generated with GeneSpring software (Silicon Genetics).
Real-time Quantitative PCR Analysis
cDNA was synthesized from 1 μg of total RNA with the High Capacity cDNA reverse transcription kit (Applied Biosystems). Real-time PCR was performed on an Applied Biosystems 7900HT with TaqMan probes (Applied Biosystems). Relative gene expression levels were calculated with standard curves generated by serial dilution of cDNA isolated from C2C12 cells. Each cDNA sample was diluted with EASY Dilution (Takara Bio) and analyzed in triplicate. To determine relative gene expression, the expression of GAPDH was used as an internal standard. The expression of each gene was assessed by three independent PCR analyses.
Prediction of Transmembrane Regions and Orientation
cDNA sequences were analyzed by TMpred software.
Western Blot Analysis
Cells were lysed in radioimmune precipitation assay buffer that contained COMPLETE protease inhibitor mixture (Roche Applied Science). Protein concentration was quantified with a protein assay (Bio-Rad), and BSA was used as a standard. Western blot analysis was performed as described previously (4). For several experiments, dead cells were separated from live cells for sample preparation (16). Dead cells (floating) were isolated from the culture medium after centrifugation at 1000 × g for 10 min. After several washes, live cells were scraped from culture dishes.
Microscopy
Images were captured with an ORCA-ER cooled charge-coupled camera (Hamamatsu Photonics) mounted on an IX70 microscope (Olympus Optical Co.). All images were captured at either 20-fold or 40-fold magnification with Plan-SemiApochromat objective lenses (20×, 0.40 numerical aperture; 40×, 0.60 numerical aperture). Images were acquired and processed with IPLab software (Scanalytics, Inc.).
Immunocytochemistry
C2C12 cells were grown in four-chamber slides (Nalge-Nunc), fixed in 4% paraformaldehyde/PBS, and permeabilized in 0.1% Triton X-100 (4). Fixed, permeabilized cells were blocked in PBS that contained 3% BSA (Jackson ImmunoResearch Laboratories) and incubated overnight at 4 °C with anti-Sar1 antibody in blocking solution. Immunoreactivity was detected with a biotin-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) and Alexa Fluor 594-streptavidin (Molecular Probes). Immunostained images were captured with an FV1000D confocal microscope (Olympus). All images were captured at 60-fold magnification with a PLAPON 60× oil objective lens (1.42 numerical aperture). Images were acquired and processed with FV10-ASW software (Olympus). Selected images were pseudo-colored for presentation in ImageJ software.
Antibodies
The primary antibodies for immunostaining were as follows: anti-Mcl-1 (Epitomics), anti-Bcl-xL (Sigma-Aldrich), anti-Bcl-2 (Medical & Biological Laboratories), anti-caspase-12 (17), anti-active caspase-9 and anti-caspase-3 (Cell Signaling), anti-CHOP and anti-α-tubulin (Santa Cruz Biotechnology), anti-GAPDH (Chemicon), anti-GFP (Molecular Probes), anti-BiP (BD Transduction Laboratories), anti-WBP1 (ProteinTech), and anti-Sar1 (Abcam).
shRNA Plasmid
The following pairs of synthetic DNAs were annealed and cloned into the pGeneClip hMGFP vector (Promega) according to the manufacturer's protocol. The MGFP coding region was deleted to make an shRNA plasmid for co-transfection with GFP-ATF6(1–360). The following primers were used: WBP1 version A (sense, TCTCGGACTGTCCTCATCCTCTTTACTTCCTGTCATAAAGAGGATGAGGACAGTCCCT; and antisense, CTGCAGGGACTGTCCTCATCCTCTTTATGACAGGAAGTAAAGAGGATGAGGACAGTCC); WBP1 version C (sense, TCTCGGCTAAACTCAGGCTGCAACACTTCCTGTCATGTTGCAGCCTGAGTTTAGCCCT; and antisense, CTGCAGGGCTAAACTCAGGCTGCAACATGACAGGAAGTGTTGCAGCCTGAGTTTAGCC); Mcl-1 version A (sense, TCTCGCGTAAACCAAGAAAGCTTCACTTCCTGTCATGAAGCTTTCTTGGTTTACGCCT; and antisense, CTGCAGGCGTAAACCAAGAAAGCTTCATGACAGGAAGTGAAGCTTTCTTGGTTTACGC); Mcl-1 version B (sense, TCTCGCTGGTCTGGCATATCTAATACTTCCTGTCATATTAGATATGCCAGACCAGCCT; and antisense, CTGCAGGCTGGTCTGGCATATCTAATATGACAGGAAGTATTAGATATGCCAGACCAGC); and Mcl-1 version C (sense, TCTCGGACTGGCTTGTCAAACAAAGCTTCCTGTCACTTTGTTTGACAAGCCAGTCCCT; and antisense, CTGCAGGGACTGGCTTGTCAAACAAAGTGACAGGAAGCTTTGTTTGACAAGCCAGTCC).
RESULTS
Apoptosis Induced by Active ATF6
The mature form of human ATF6 comprises amino acids 1–373 (18); this corresponds to amino acids 1–360 of mouse ATF6 (supplemental Fig. 1). When the mature form of mouse ATF6 was N-terminally tagged with green fluorescent protein (GFP-ATF6(1–360)) and transiently transfected into C2C12 cells, it induced apoptosis (Fig. 1A); ∼45% of cells died. The dead cells exhibited the typical morphological features of apoptosis: round shape, condensed nuclei, very small size compared with healthy cells (Fig. 1B), and some membrane blebbing (Fig. 1C). Visualization of GFP-ATF6(1–360) in live cells indicated that the GFP tag did not impair ATF6(1–360) nuclear localization (Fig. 1C). When the full-length ATF6(1–656) was fused to GFP and transiently transfected into C2C12 cells, it also induced apoptosis in ∼40% of cells (data not shown). This may have been due to spontaneous processing of the full-length ATF6 to the mature form, as previously reported (19). In contrast, a dominant negative form of ATF6 (19, 20), which contained the DNA binding domain but lacked the activation domain (ATF6 (158–360)), did not induce apoptosis (Fig. 1A). Similarly, the mature form of human ATF6 also induced apoptosis but not the dominant negative form (data not shown).
ATF6 has been overexpressed in several other cell lines (e.g. COS (21)) without remarkable induction of apoptosis. This suggests that sensitivity to ATF6 overexpression is cell line dependent. Fig. 1D shows that GFP-ATF6(1–360) induced apoptosis in mammalian cells with different efficiencies. A mouse fibroblast cell line and a kidney cell line (NIH-3T3 and mIMCD-3, respectively) underwent apoptosis with 20–30% efficiency, but little apoptosis was induced in COS-1 (a monkey kidney cell line) and MCF-7 (a human breast cancer cell line) cells. These results indicated that resistance to active ATF6 varies among cell lines.
Activation of Caspases during ATF6-induced Apoptosis
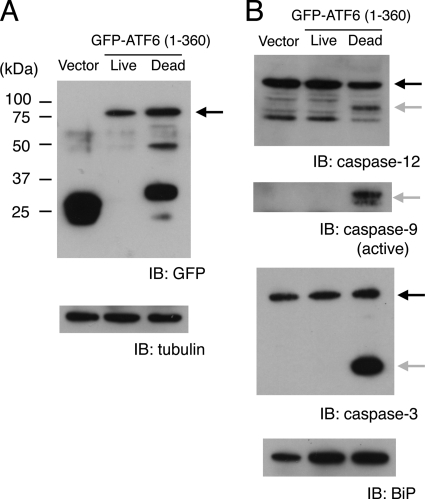
To confirm that the observed apoptosis depended on the amount of active ATF6 in C2C12 cells, we examined GFP-ATF6(1–360) expression in separate lysates of dead and live cells (see “Experimental Procedures”). Western blot analysis (Fig. 2A) showed that GFP-ATF6(1–360) expression in apoptotic cells was higher, on average, than in live cells, but the fusion protein appeared to be cleaved primarily by proteolysis during apoptosis. Caspases-12, -9, and -3 were specifically activated in the apoptotic cells (Fig. 2B) (15). The expression of active ATF6 in live cells appeared to be insufficient to induce caspase activation or apoptosis. However, expression of immunoglobulin heavy chain binding protein (BiP), a representative of the proteins induced by ATF6, substantially increased in both live and dead cells compared with control cells (Fig. 2B). This result suggested that the small amount of GFP-ATF6(1–360) in live cells was sufficient to up-regulate BiP. Our previous study showed that active ATF6 was detectable in dying cells by Western blot, but active ATF6 was below the detection level in live cells treated with ER stressors or differentiation medium (4). Taken together, these findings suggested that relatively low levels of active ATF6 may mediate the UPR in self-defense, but higher levels of ATF6 may be required to mediate apoptosis.
FIGURE 2.
Active ATF6 induced caspase activation. A, C2C12 cells were electroporated with GFP vector or GFP-ATF6(1–360). After 24 h, dead and live cells were separated and subjected to Western blot analysis. Arrow indicates GFP-ATF6(1–360). α-Tubulin was the loading control. B, the same blot shown in A was sequentially probed with anti-caspase-12, anti-caspase-9, anti-caspase-3, and anti-BiP antibodies. Black arrows show caspase precursors; gray arrows show cleaved caspase products. IB, immunoblot.
Apoptosis Induction Required Transcription Activation by ATF6
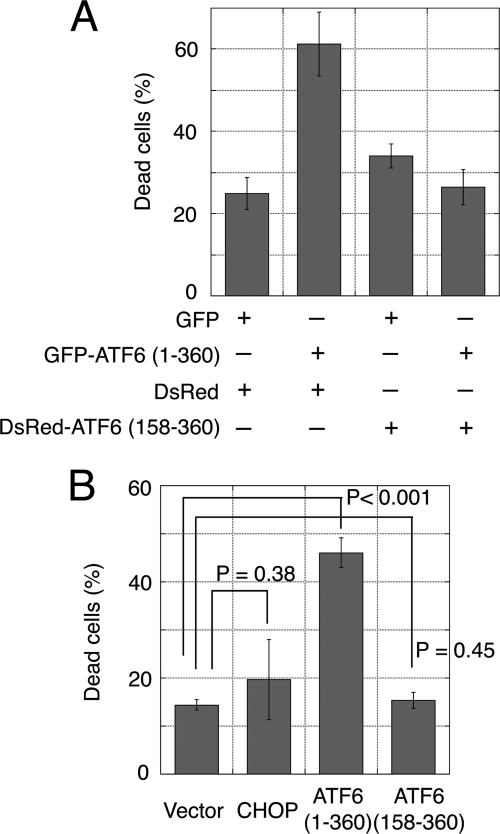
Apoptosis was induced by the expression of active GFP-ATF6(1–360) but not by the dominant negative GFP-ATF6(158–360) (Fig. 1A). In fact, apoptosis induced by active ATF6 was efficiently suppressed by overexpression of the dominant negative form tagged with DsRed (Fig. 3A). This might be explained by the efficient DNA binding of the ATF6 mutant, which lacked the transcription activation domain. This binding would competitively inhibit the binding of active ATF6 (19, 20). Nearly all the GFP-positive cells were also DsRed-positive; this indicated that both the active form and the dominant negative form of ATF6 were successfully co-expressed. Thus, the results were not due to competition between the CMV promoters within transfected cells (data not shown). These results suggested that apoptosis induction required transcription activated by ATF6.
FIGURE 3.
Dominant negative form of ATF6 suppressed apoptosis induced by active ATF6. A, C2C12 cells grown in six-well plates were transiently co-transfected with GFP or GFP-ATF6(1–360) (0.3 μg) and either the pDsRed vector (control) or DsRed-ATF6(158–360) (dominant negative) (1.8 μg). Apoptosis was assessed by cell morphology. B, CHOP did not induce apoptosis in C2C12 cells. C2C12 cells were transiently transfected with GFP or GFP-tagged CHOP. The apoptotic efficiencies of GFP-ATF6(1–360) and the dominant negative GFP-ATF6(158–360) are included in the bar graph.
We reasoned that CHOP might mediate the proapoptotic function of ATF6 because the CHOP gene promoter contains a binding site for ATF6 (also sites for ATF4 and XBP1 (22)), and CHOP induction was detected in C2C12 cells treated with ER stressors (4, 16). However, overexpression of GFP-CHOP in C2C12 cells did not induce significant apoptosis (Fig. 3B). Furthermore, CHOP was not detected on Western blots of C2C12 cells transfected with GFP-ATF6(1–360) (data not shown). Therefore, CHOP expression is likely to be regulated by factor(s) other than ATF6 in C2C12 cells. These results suggested that the pro-apoptotic function of ATF6 was not dependent on CHOP. Therefore, ATF6-induced apoptosis must be dependent on another downstream factor(s) that is up-regulated by active ATF6.
Mediator of ATF6-induced Apoptosis
To identify genes that were up-regulated by ATF6, we performed a whole-genome microarray analysis of RNA samples prepared from C2C12 cells that had been electroporated with GFP-ATF6(1–360). Control cells were electroporated with the GFP vector alone. Fluorescence microscopy showed that the gene transfer efficiency was >90% for the GFP vector, but only ∼20% for GFP-ATF6(1–360) at 24 h post-electroporation. Approximately 50% of the cells transfected with GFP-ATF6(1–360) underwent apoptosis, which was consistent with the transfection assay (Fig. 1A). Live and dead cells were combined for the microarray analysis.
To identify proapoptotic factors that function downstream of ATF6, we first selected genes that showed at least 1.5-fold higher expression in cells transfected with GFP-ATF6(1–360) compared with controls. After excluding genes with relatively low expression levels (<1% of the GAPDH expression level), we identified 44 up-regulated genes (supplemental Table 1). One was CHOP, and 12 others were reported previously to be UPR-related; thus, they participated in protein folding and/or vesicle transport. Of note, the CHOP expression level was only one-twentieth of the GAPDH and BiP expression levels. In addition, 24 of 44 up-regulated genes were related to phenomena apparently irrelevant to apoptosis. The remaining seven genes had not been evaluated previously for their involvement in apoptosis; thus, we analyzed them further.
To examine whether these seven genes could induce apoptosis, the cDNAs were GFP-tagged and transfected into C2C12 cells. Among the seven candidates, only WBP1 induced apoptosis. WBP1 was originally identified in vitro as a protein that bound to the WW domain of Yes kinase-associated protein (23), but its function had not been determined. The WW domain comprises 38 to 40 semiconserved amino acids, a motif shared by proteins of diverse functions, including structural, regulatory, and signaling proteins (24).
Apoptosis-inducing Activity of WBP1
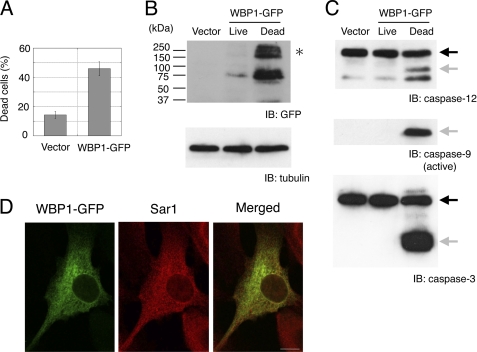
Sequence analysis strongly suggested that WBP1 was a type I transmembrane protein with an N terminus that contained a putative signal sequence (see “Experimental Procedures”). Thus, we fused WBP1 to a C-terminal GFP tag. Similar to GFP-ATF6(1–360), transfection of C2C12 cells with the GFP-tagged WBP1 cDNA caused apoptosis in ∼45% of transfected cells (Fig. 4A). Western blot analysis showed that apoptosis was dependent on the level of WBP1 (Fig. 4B). We also found that, in apoptotic cells, caspases-12, -9, and -3 were processed for activation (Fig. 4C). Confocal microscopy of live cells showed that WBP1-GFP was localized primarily on the ER membrane. We detected co-localization of WBP1-GFP and Sar1 by immunofluorescence (Fig. 4D).
FIGURE 4.
WBP1 induced apoptosis in C2C12 cells. A, transfection of WBP1-GFP caused apoptosis in C2C12 cells. B, Western blot shows WBP1 expression in apoptotic cells after electroporation with WBP1-GFP. High molecular mass complexes of WBP-GFP (asterisk) that were resistant to denaturation were reproducibly detected. C, activation of caspases in apoptotic cells. The same blot in B was sequentially probed with anti-caspase-12, anti-active caspase-9, and anti-caspase-3. Black arrows show caspase precursors; gray arrows show cleaved caspase products. D, co-localization of WBP1-GFP with Sar1. C2C12 cells were transfected with WBP1-GFP, fixed, and immunostained with anti-Sar1 antibody. Scale bar, 10 μm. IB, immunoblot.
Up-regulation of WBP1 during Apoptosis Induced by ER Stressors
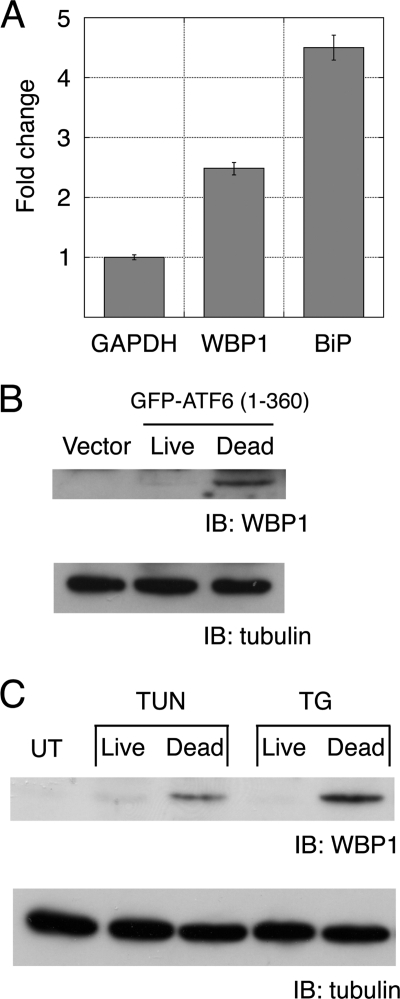
The up-regulation of WBP1 by active ATF6 was confirmed by quantitative PCR. Under conditions where active ATF6 was expressed in ∼20% of cells, WBP1 expression increased ∼2.5-fold, and BiP expression increased ∼4.5-fold compared with controls (Fig. 5A). Endogenous WBP1 protein was detected in cells that overexpressed active ATF6 and underwent apoptosis (Fig. 5B); again, this indicated that WBP1 was up-regulated in apoptotic cells. WBP1 was also detected in apoptotic cells treated with the ER stressors, tunicamycin (inhibitor of N-glycosylation) and thapsigargin (inhibitor of ER-specific calcium ATPase). In contrast, WBP1 was nearly undetectable in C2C12 that grew or survived after treatment with ER stressors (Fig. 5C). These results suggested that WBP1 might serve as a marker of ER stress-induced apoptosis.
FIGURE 5.
WBP1 was induced in apoptotic cells. A, real-time quantitative PCR analysis. Fold changes in WBP1 and BiP expression were determined by comparing the GAPDH-normalized ratios of mRNAs from active ATF6-induced samples versus vector-transfected samples. Bars represent an average of three independent experiments. B, active ATF6 induced WBP1 expression in apoptotic cells. C2C12 cells were electroporated with GFP-ATF6(1–360). After 24 h, dead and live cells were separated and subjected to Western blot analysis. C, WBP1 was induced in apoptotic cells treated with ER stressors tunicamycin (TUN) and thapsigargin (TG). UT, untreated; IB, immunoblot.
ATF6-WBP1 Apoptotic Pathway
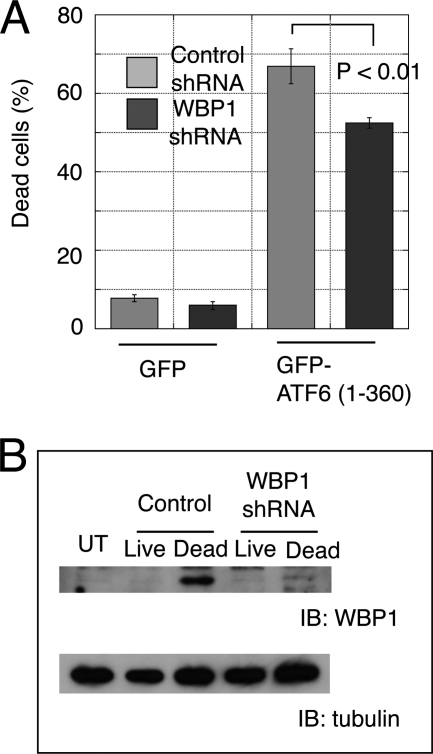
The results described above suggested that WBP1 mediated the apoptosis induced by active ATF6. To confirm that WBP1 functioned downstream of ATF6 in apoptosis, we constructed an shRNA plasmid that targeted WBP1. When WBP1-GFP was expressed in MCF-7 cells, it did not induce apoptosis (data not shown); this was consistent with the result that MCF-7 cells resisted apoptosis induction by active ATF6 (Fig. 1D). When the WBP1 shRNA plasmid was transiently co-transfected with WBP1-GFP in MCF-7 cells, WBP1 expression was suppressed (supplemental Fig. 2A); this confirmed the efficacy of WBP1 shRNA. Next, C2C12 cells were co-transfected with ATF6 and shRNA. The shRNA silencing of WBP1 caused partial but significant (p < 0.01) suppression of ATF6-induced apoptosis (Fig. 6A). This finding supported the notion that WBP1 acted downstream of ATF6 in the induction of apoptosis. Although the specific shRNA efficiently suppressed up-regulation of WBP1 (Fig. 6B), WBP1 expression was detected in dead cells; this suggested that cells may undergo apoptosis only when the WBP1 protein reaches a threshold level. Alternatively, a factor other than WBP1 may also mediate ATF6-induced apoptosis.
FIGURE 6.
WBP1 knockdown suppressed apoptosis induced by active ATF6. A, C2C12 cells were electroporated with GFP-ATF6(1–360) and WBP1 shRNA, version A, at a ratio of 1:3 (w/w). After 24 h, apoptosis was assessed by cell morphology. B, WBP1 levels were analyzed by Western blot (IB). UT, untreated.
Possible Involvement of Mcl-1 in Apoptosis Induced by Active ATF6 or WBP1
We previously showed that a proapoptotic member of the Bcl-2 family, Bim, was involved in ER stress-induced apoptosis in C2C12 cells. Furthermore, Bcl-xL overexpression appeared to antagonize Bim activity and thus efficiently suppressed ER stress-induced apoptosis (16). Here, we examined the expression of anti-apoptotic members of the Bcl-2 family to evaluate their involvement in ATF6- and WBP1-induced apoptosis.
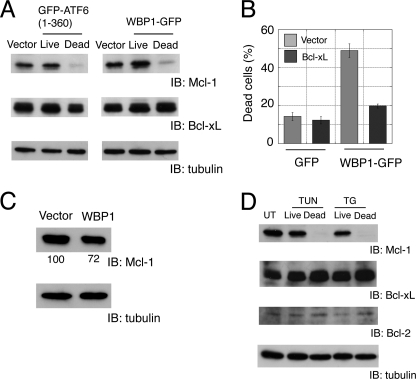
Overexpression of GFP-ATF6(1–360) and WBP1-GFP resulted in a reduction of Mcl-1 but not Bcl-xL levels in apoptotic cells (Fig. 7A). This finding suggested that activation of the ATF6-WBP1 pathway in response to ER stress could suppress Mcl-1 expression and result in apoptosis. Note that Bcl-2, the founder member of the Bcl-2 family, was expressed in myoblast cells at much lower levels than Bcl-xL (25). Quantitative PCR analysis showed that active ATF6 alone did not down-regulate Mcl-1 expression at the transcription level (data not shown). This suggested that Mcl-1 expression might be suppressed by post-translational regulation (see “Discussion“).
FIGURE 7.
Analysis of Bcl-2 family proteins in apoptotic cells. A, specific reduction of Mcl-1 expression in apoptotic cells transfected with active ATF6 or WBP1. B, co-expression of Bcl-xL suppressed apoptosis induced by WBP1. C2C12 cells were grown in six-well plates and transiently co-transfected with GFP or WBP1-GFP (0.3 μg) and pcDNA3.1 vector or Bcl-xL (1.8 μg). C, down-regulation of Mcl-1 in cells that overexpressed Bcl-xL. C2C12 cells were electroporated with WBP1-GFP and Bcl-xL at a 1:1 ratio (w/w). Mcl-1 was analyzed by Western blot. The band intensity was normalized with α-tubulin as a standard. Representative data from three independent experiments are shown. D, specific reduction of Mcl-1 in apoptotic cells treated with ER stressors tunicamycin (TUN) and thapsigargin (TG). UT, untreated; IB, immunoblot.
To examine whether the reduction in Mcl-1 expression was a consequence of apoptosis, we inhibited apoptosis in C2C12 cells by co-transfecting Bcl-xL and WBP1-GFP. Bcl-xL efficiently suppressed apoptosis induced by WBP1-GFP (Fig. 7B). Mcl-1 was reduced by 25% (75 ± 3.5%, n = 3) in live C2C12 cells co-electroporated with WBP1-GFP and Bcl-xL (Fig. 7C). Considering that WBP1-GFP was detected by fluorescence microscopy in ∼50% of transfected cells, this Mcl-1 reduction was not negligible. This suggested that Mcl-1 expression was reduced independent of cell destruction during apoptosis.
Mcl-1 expression was also specifically reduced in ER stressor-induced apoptosis. When C2C12 cells were exposed to ER stressors, 40–50% of cells underwent apoptosis (15). Mcl-1 expression was nearly undetectable in apoptotic cells, but Bcl-2 and Bcl-xL expression remained unaltered (Fig. 7D). Taken together, these results suggested that Mcl-1 may be an important regulator of ER stress-induced apoptosis in myoblasts. Interestingly, Bcl-xL predominantly localizes to the mitochondria (26), but Mcl-1 is present on the ER and mitochondria (27). We reasoned that a specific reduction in anti-apoptotic Mcl-1 might shift the balance between anti-apoptotic and proapoptotic factors present on the ER.
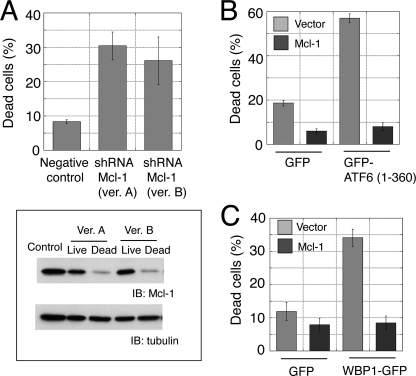
To investigate whether Mcl-1 was a cell fate determinant during ER stress in C2C12 cells, we expressed shRNAs specific for Mcl-1 (supplemental Fig. 2B). Mcl-1 knockdown caused a significant increase in apoptosis, but the effect was not comparable with that observed with ATF6 or WBP1 overexpression (Fig. 8A). Nevertheless, the amount of Mcl-1 in apoptotic cells was much lower than that in live cells (Fig. 8A). These results suggested that the Mcl-1 level was a critical determinant of apoptosis in C2C12 cells. Conversely, overexpression of Mcl-1 suppressed apoptosis induced by active ATF6 (Fig. 8B) or WBP1 (Fig. 8C). This confirmed the antagonistic effect of Mcl-1 on the ATF6/WBP1 pathway.
FIGURE 8.
Mcl-1 was a cell fate determinant in C2C12 cells. A, Mcl-1 knockdown caused apoptosis. C2C12 cells were transfected with Mcl-1 shRNA; dead cells were counted after 24 h. Mcl-1 was analyzed by Western blot (lower panel). B and C, coexpression of Mcl-1 caused suppression of the apoptosis induced by active ATF6 (B) or WBP1 (C). C2C12 cells were grown in six-well plates and transiently co-transfected with GFP, GFP-ATF6(1–360), or WBP1-GFP (0.3 μg) and pcDNA3.1 vector or Mcl-1 (1.8 μg).
DISCUSSION
The present study showed that active ATF6 can induce apoptosis at least partly by up-regulating WBP1. Activation of ATF6-WBP1 appeared to cause specific decreases in Mcl-1 that rendered cells vulnerable to apoptosis. Up-regulation of WBP1 and specific reduction of Mcl-1 levels were observed in both ER stressor-induced apoptosis and with the forced expression of active ATF6. These results suggested that ATF6 mediated ER stress-induced apoptosis.
Furthermore, our results suggested that ATF6 may play an important role in the transition from self-defense to self-destruction of cells during ER stress. At relatively low levels, ATF6 appeared to activate the UPR for self-defense; but at higher levels, ATF6 mediated apoptosis (Fig. 2). Future studies should focus on the mechanism underlying these apparently opposite activities of ATF6.
We found it difficult to overexpress ATF6 in growing C2C12 cells, which suggested that ATF6 may have an ephemeral nature. Initially, we transfected C2C12 cells with ATF6 or FLAG-tagged ATF6 placed under the control of a strong cytomegalovirus promoter. However, we failed to detect high levels of ATF6 by immunoblot analysis and immunostaining (data not shown). Next, we fused the N terminus of ATF6 to GFP and placed it under the control of the cytomegalovirus promoter. This strategy allowed successful detection with a fluorescent microscope. Improved expression by GFP tagging the N terminus has been observed for other proteins (28). Previous studies showed that ATF6 has a relatively short half-life (∼40 min) in HeLa cells. This may be due to degradation signals in the N-terminal domain of ATF6 (18). Strict control of the active ATF6 content in the cell may be part of the mechanism that controls switching the cell fate from self-defense to self-destruction.
A recent study suggested the importance of ATF6 expression levels in the induction of cell death under pathological conditions (29). They found that an ER membrane protein, WFS1 (Wolfram syndrome 1 gene product), stabilized an E3 ubiquitin ligase, HRD1, on the ER. This enhanced the ubiquitination and degradation of ATF6. Conversely, inactive WFS1 may lead to stabilization of ATF6, which in turn might cause excess cell death (29). WFS1 mutations have been linked to Wolfram syndrome, a genetic form of diabetes, optic atrophy, neurodegeneration, and psychiatric illness (30, 31). The present study suggested potential therapeutic targets for treating these pathological conditions.
We previously showed that activation of PERK or IRE1 was detected in surviving cells after treatment with ER stressors when the active ATF6 was absent (4). This observation indicated that activation of PERK or IRE1 did not cause WBP1 induction in live cells (Fig. 5C) and supported the notion that WBP1 is specifically induced by ATF6.
To further confirm that WBP1 was specifically induced by ATF6, we attempted to knockdown ATF6 in ER stress-induced apoptosis. However, we failed to completely knock down ATF6, because the levels of active ATF6 (and WBP1) in dead cells was comparable with that in dead cells transfected with a negative control plasmid (data not shown). However, it is possible that apoptosis only occurred in cells with inefficient ATF6 knockdown. This observation would support our previous suggestion that ER stress-induced apoptosis in C2C12 cells required activation of ATF6 (4).
Interestingly, 10 kb of the genomic noncoding region upstream of the WBP1 coding sequence lacked the well defined ATF6 binding motif (ER stress response elements) (19, 20). This region also lacked the ER stress response element binding motif for the transcription factor, NF-Y, which assists ATF6 binding to ER stress response elements (32, 33). Instead, the 10-kb region contained the whole coding sequence of the adjacent gene Ino80B. Although the promoter of WBP1 has not been determined, it is unlikely that active ATF6 directly binds to the promoter of WBP1. Perhaps ATF6 indirectly up-regulates WBP1 by inducing the expression of another transcription factor(s).
Activation of the ATF6-WBP1 pathway appeared to reduce Mcl-1 expression. This may shift the balance between anti-apoptotic and proapoptotic proteins (e.g. members of the Bcl-2 family, including Bim (16, 34)) on the ER membrane. DNA damage or growth factor withdrawal also caused a considerable reduction in Mcl-1 that was associated with the apoptotic response (35–37). However, the reduction in Mcl-1 may differentially impact the sensitivity to active ATF6 in different cell types, depending on the expression pattern of Bcl-2 family proteins (38). For example, the presence of Bcl-2 on the ER and mitochondria was both tissue and cell type-specific (39).
The apoptosis-inducing activity of ATF6 has not been widely recognized. This is at least partly due to the fact that ATF6 does not induce apoptosis in cell lines commonly used in biological and medical research (e.g. COS-1). Our results cannot exclude the possibility that factors other than ATF6 play major roles in ER stress-induced apoptosis in different cell types. For instance, dephosphorylation of eIF2α, a target of PERK, has been suggested to play a critical role in ER stress-induced apoptosis signaling in PC12 cells (40). Also, an anti-apoptotic protein (e.g. Bcl-2), not Mcl-1, is possibly a major player on the ER that regulates ER stress-induced apoptosis in cells other than myoblasts. In that case, the ATF6 to Mcl-1 pathway may not necessarily be important for cell fate decisions. Accordingly, it would be interesting to compare the expression of Bcl-2 family members among different cell types and determine their involvement in each branch of the UPR (ATF6, PERK, or IRE1) during apoptosis.
Active ATF6 did not down-regulate Mcl-1 at the transcriptional level, whereas the amount of Mcl-1 protein decreased (Fig. 7A), suggesting that Mcl-1 might be suppressed by post-translational regulation; e.g. ubiquitin-mediated degradation, as occurs in DNA damage-induced apoptosis (41). Indeed, the NEDD4 family of E3 ubiquitin ligases forms a branch of the family of WW domain containing proteins, which are potential targets of WBP1 (42).
Further studies are necessary to define the specific pathway that links active ATF6 to reduced Mcl-1 expression via the induction of WBP1. Identifying specific WBP1 binding proteins might elucidate the molecular mechanism involved. WW domains bind to proline-rich peptide motifs in proteins that act in a wide variety of signaling pathways (43). WW domains have been identified in over 200 non-redundant proteins (44). Our results suggested that a specific WBP1 binding protein is likely to interact with WBP1 on the ER membrane. Currently, we are investigating candidate proteins in the microsomal fraction of C2C12 cells. Identification of the WBP1 binding protein might also clarify the function of WBP1 in the regulation of apoptosis.
Unraveling the mechanism of ER stress-induced apoptosis is important for understanding physiological apoptosis and cell death under pathological conditions, including diabetes mellitus, neurodegenerative disorders, and prion diseases (1). In this study, we explored the mechanism of apoptosis and identified a potential link between active ATF6, a component of the UPR, and Mcl-1, an apoptotic regulator. This mechanism determines whether cells initiate apoptosis in response to ER stress induced by physiological and pathological conditions.
Supplementary Material
Acknowledgments
We thank Ron Prywes for providing human ATF6 cDNA, and Junying Yuan for providing the anti-caspase-12 monoclonal antibody. We also thank Rie Nakazawa and Yasue Ichikawa for DNA sequencing and Keisuke Fukumoto (the Support Unit for Biomaterial Analysis, RIKEN BSI Research Resources Center) for assistance with GeneChip and real-time PCR analyses.
This work was supported in part by grants from the RIKEN Bioarchitect Research Project and by the Japan Society for the Promotion of Science (to N. M).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
- ER
- endoplasmic reticulum
- ATF6
- activating transcription factor-6
- BiP
- binding immunoglobulin protein
- CHOP
- CCAAT-enhancer-binding protein homologous protein
- PERK
- double-stranded RNA-dependent protein kinase (PKR)-like ER kinase
- UPR
- unfolded protein response.
REFERENCES
- 1. Kim I., Xu W., Reed J. C. (2008) Nat. Rev. Drug Discov. 7, 1013–1030 [DOI] [PubMed] [Google Scholar]
- 2. Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 3. Schröder M., Kaufman R. J. (2005) Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 4. Nakanishi K., Sudo T., Morishima N. (2005) J. Cell Biol. 169, 555–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rutkowski D. T., Kaufman R. J. (2004) Trends Cell Biol. 14, 20–28 [DOI] [PubMed] [Google Scholar]
- 6. Haze K., Yoshida H., Yanagi H., Yura T., Mori K. (1999) Mol. Biol. Cell 10, 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rutkowski D. T., Hegde R. S. (2010) J. Cell Biol. 189, 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu C., Bailly-Maitre B., Reed J. C. (2005) J. Clin. Invest. 115, 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. (1998) Genes Dev. 12, 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCullough K. D., Martindale J. L., Klotz L. O., Aw T. Y., Holbrook N. J. (2001) Mol. Cell Biol. 21, 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsujii H., Eguchi Y., Chenchik A., Mizutani T., Yamada K., Tsujimoto Y. (2010) J. Biochem. 148, 157–170 [DOI] [PubMed] [Google Scholar]
- 12. Fidzianska A., Goebel H. H. (1991) Acta Neuropathol. 81, 572–577 [DOI] [PubMed] [Google Scholar]
- 13. Glücksmann A. (1951) Biol. Rev. Camb. Philos. Soc. 26, 59–86 [DOI] [PubMed] [Google Scholar]
- 14. The FANTOM Consortium, Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kodzius R., Shimokawa K., Bajic V. B., Brenner S. E., Batalov S., Forrest A. R., Zavolan M., Davis M. J., Wilming L. G., Aidinis V., Allen J. E., Ambesi-Impiombato A., Apweiler R., Aturaliya R. N., Bailey T. L., Bansal M., Baxter L., Beisel K. W., Bersano T., Bono H., Chalk A. M., Chiu K. P., Choudhary V., Christoffels A., Clutterbuck D. R., Crowe M. L., Dalla E., Dalrymple B. P., de Bono B., Della Gatta G., di Bernardo D., Down T., Engstrom P., Fagiolini M., Faulkner G., Fletcher C. F., Fukushima T., Furuno M., Futaki S., Gariboldi M., Georgii-Hemming P., Gingeras T. R., Gojobori T., Green R. E., Gustincich S., Harbers M., Hayashi Y., Hensch T. K., Hirokawa N., Hill D., Huminiecki L., Iacono M., Ikeo K., Iwama A., Ishikawa T., Jakt M., Kanapin A., Katoh M., Kawasawa Y., Kelso J., Kitamura H., Kitano H., Kollias G., Krishnan S. P., Kruger A., Kummerfeld S. K., Kurochkin I. V., Lareau L. F., Lazarevic D., Lipovich L., Liu J., Liuni S., McWilliam S., Madan Babu M., Madera M., Marchionni L., Matsuda H., Matsuzawa S., Miki H., Mignone F., Miyake S., Morris K., Mottagui-Tabar S., Mulder N., Nakano N., Nakauchi H., Ng P., Nilsson R., Nishiguchi S., Nishikawa S., Nori F., Ohara O., Okazaki Y., Orlando V., Pang K. C., Pavan W. J., Pavesi G., Pesole G., Petrovsky N., Piazza S., Reed J., Reid J. F., Ring B. Z., Ringwald M., Rost B., Ruan Y., Salzberg S. L., Sandelin A., Schneider C., Schönbach C., Sekiguchi K., Semple C. A., Seno S., Sessa L., Sheng Y., Shibata Y., Shimada H., Shimada K., Silva D., Sinclair B., Sperling S., Stupka E., Sugiura K., Sultana R., Takenaka Y., Taki K., Tammoja K., Tan S. L., Tang S., Taylor M. S., Tegner J., Teichmann S. A., Ueda H. R., van Nimwegen E., Verardo R., Wei C. L., Yagi K., Yamanishi H., Zabarovsky E., Zhu S., Zimmer A., Hide W., Bult C., Grimmond S. M., Teasdale R. D., Liu E. T., Brusic V., Quackenbush J., Wahlestedt C., Mattick J. S., Hume D. A., Kai C., Sasaki D., Tomaru Y., Fukuda S., Kanamori-Katayama M., Suzuki M., Aoki J., Arakawa T., Iida J., Imamura K., Itoh M., Kato T., Kawaji H., Kawagashira N., Kawashima T., Kojima M., Kondo S., Konno H., Nakano K., Ninomiya N., Nishio T., Okada M., Plessy C., Shibata K., Shiraki T., Suzuki S., Tagami M., Waki K., Watahiki A., Okamura-Oho Y., Suzuki H., Kawai J., Hayashizaki Y. (2005) Science 309, 1559–1563 16141072 [Google Scholar]
- 15. Morishima N., Nakanishi K., Takenouchi H., Shibata T., Yasuhiko Y. (2002) J. Biol. Chem. 277, 34287–34294 [DOI] [PubMed] [Google Scholar]
- 16. Morishima N., Nakanishi K., Tsuchiya K., Shibata T., Seiwa E. (2004) J. Biol. Chem. 279, 50375–50381 [DOI] [PubMed] [Google Scholar]
- 17. Nakagawa T., Zhu H., Morishima N., Li E., Xu J., Yankner B. A., Yuan J. (2000) Nature 403, 98–103 [DOI] [PubMed] [Google Scholar]
- 18. Thuerauf D. J., Morrison L. E., Hoover H., Glembotski C. C. (2002) J. Biol. Chem. 277, 20734–20739 [DOI] [PubMed] [Google Scholar]
- 19. Wang Y., Shen J., Arenzana N., Tirasophon W., Kaufman R. J., Prywes R. (2000) J. Biol. Chem. 275, 27013–27020 [DOI] [PubMed] [Google Scholar]
- 20. Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M., Mori K. (2000) Mol. Cell Biol. 20, 6755–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu C., Johansen F. E., Prywes R. (1997) Mol. Cell Biol. 17, 4957–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oyadomari S., Mori M. (2004) Cell Death Differ. 11, 381–389 [DOI] [PubMed] [Google Scholar]
- 23. Chen H. I., Einbond A., Kwak S. J., Linn H., Koepf E., Peterson S., Kelly J. W., Sudol M. (1997) J. Biol. Chem. 272, 17070–17077 [DOI] [PubMed] [Google Scholar]
- 24. Ilsley J. L., Sudol M., Winder S. J. (2002) Cell Signal. 14, 183–189 [DOI] [PubMed] [Google Scholar]
- 25. Dominov J. A., Houlihan-Kawamoto C. A., Swap C. J., Miller J. B. (2001) Dev. Dyn. 220, 18–26 [DOI] [PubMed] [Google Scholar]
- 26. Kaufmann T., Schlipf S., Sanz J., Neubert K., Stein R., Borner C. (2003) J. Cell Biol. 160, 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang T., Kozopas K. M., Craig R. W. (1995) J. Cell Biol. 128, 1173–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaba S. A., Nene V., Musoke A. J., Vlak J. M., van Oers M. M. (2002) Parasitology 125, 497–505 [DOI] [PubMed] [Google Scholar]
- 29. Fonseca S. G., Ishigaki S., Oslowski C. M., Lu S., Lipson K. L., Ghosh R., Hayashi E., Ishihara H., Oka Y., Permutt M. A., Urano F. (2010) J. Clin. Invest. 120, 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Strom T. M., Hörtnagel K., Hofmann S., Gekeler F., Scharfe C., Rabl W., Gerbitz K. D., Meitinger T. (1998) Hum. Mol. Genet. 7, 2021–2028 [DOI] [PubMed] [Google Scholar]
- 31. Inoue H., Tanizawa Y., Wasson J., Behn P., Kalidas K., Bernal-Mizrachi E., Mueckler M., Marshall H., Donis-Keller H., Crock P., Rogers D., Mikuni M., Kumashiro H., Higashi K., Sobue G., Oka Y., Permutt M. A. (1998) Nat. Genet. 20, 143–148 [DOI] [PubMed] [Google Scholar]
- 32. Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M., Mori K. (2001) Mol. Cell Biol. 21, 1239–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamamoto K., Yoshida H., Kokame K., Kaufman R. J., Mori K. (2004) J. Biochem. 136, 343–350 [DOI] [PubMed] [Google Scholar]
- 34. Puthalakath H., O'Reilly L. A., Gunn P., Lee L., Kelly P. N., Huntington N. D., Hughes P. D., Michalak E. M., McKimm-Breschkin J., Motoyama N., Gotoh T., Akira S., Bouillet P., Strasser A. (2007) Cell 129, 1337–1349 [DOI] [PubMed] [Google Scholar]
- 35. Cuconati A., Mukherjee C., Perez D., White E. (2003) Genes Dev. 17, 2922–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nijhawan D., Fang M., Traer E., Zhong Q., Gao W., Du F., Wang X. (2003) Genes Dev. 17, 1475–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maurer U., Charvet C., Wagman A. S., Dejardin E., Green D. R. (2006) Mol. Cell 21, 749–760 [DOI] [PubMed] [Google Scholar]
- 38. Chipuk J. E., Moldoveanu T., Llambi F., Parsons M. J., Green D. R. (2010) Mol. Cell 37, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Negrini M., Silini E., Kozak C., Tsujimoto Y., Croce C. M. (1987) Cell 49, 455–463 [DOI] [PubMed] [Google Scholar]
- 40. Boyce M., Bryant K. F., Jousse C., Long K., Harding H. P., Scheuner D., Kaufman R. J., Ma D., Coen D. M., Ron D., Yuan J. (2005) Science 307, 935–939 [DOI] [PubMed] [Google Scholar]
- 41. Zhong Q., Gao W., Du F., Wang X. (2005) Cell 121, 1085–1095 [DOI] [PubMed] [Google Scholar]
- 42. Ingham R. J., Gish G., Pawson T. (2004) Oncogene 23, 1972–1984 [DOI] [PubMed] [Google Scholar]
- 43. Sudol M., Hunter T. (2000) Cell 103, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 44. Kasanov J., Pirozzi G., Uveges A. J., Kay B. K. (2001) Chem. Biol. 8, 231–241 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.