Abstract
The cyclin-dependent kinase inhibitor p21(WAF1/CIP1) inhibits proliferation both in vitro and in vivo, and overexpression of p21 in normal and tumor cell lines results in cell cycle arrest. In contrast, ectopic expression of Myc alleviates G1 cell cycle arrest. Recent studies showed that Myc can repress p21 transcription, thereby overriding a p21-mediated cell cycle checkpoint. We found that activation of a Myc-estrogen receptor fusion protein by 4-hydroxytamoxifen in mouse cells resulted in suppression of endogenous p21 transcription. This effect was observed in the absence of de novo protein synthesis and was independent of histone deacetylase activity. In transient transfection studies, Myc effectively repressed p21 promoter constructs containing only 119 bp of sequence upstream of the transcription start site. This region contains multiple Sp1-binding sites and a potential initiator element, but no canonical Myc DNA-binding sites. Deletion of the potential initiator element does not affect repression of the p21 promoter by c-Myc. Coimmunoprecipitation and glutathione S-transferase pull-down experiments demonstrate that c-Myc may form complexes with Sp1/Sp3. We found that the central region of c-Myc interacts with the zinc finger domain of Sp1. Because Sp1 is required for p21 transcription, it is possible that Myc may down-regulate p21 transcription, at least in part, by sequestering Sp1. Repression of the p21 promoter may contribute to the ability of c-Myc to promote cell proliferation.
Keywords: transcriptional repression
The Myc family of protooncogenes includes three evolutionarily conserved genes: c-, N-, and L-Myc. Overexpression of Myc diminishes growth-factor requirements, hinders cell cycle arrest by a variety of growth-inhibitory signals, and can block differentiation (1). Myc belongs to a family of the basic helix–loop–helix leucine zipper (bHLH-Zip) transcription factors. It must dimerize with the bHLH-Zip protein Max, to bind the E-box sequence CACGTG and activate transcription of a number of genes. Targets of Myc include α-prothymosin (2), ornithine decarboxylase (3), Cdc25A (4), p53 (5), and others (reviewed in ref. 6). Max also forms alternative heterodimers with the bHLH-Zip proteins Mad1, Mxi-1, Mad3, Mad4, and Mnt (reviewed in ref. 6). These alternative dimers can compete for binding with Myc–Max heterodimers and repress transcription, hence antagonizing the transcriptional and transforming activities of Myc. In addition to its function as a transactivator, Myc has also been shown to repress transcription of several genes, such as C/EBPα (7), gadd45 (8), gas1 (9), and p15Ink4b (10).
Cyclin kinase inhibitors (CKIs) bind and inhibit cyclin-dependent kinases (CDKs). p21 is a CKI that has broad specificity for cyclin/CDK complexes (11, 12), and the p21 gene is a transcriptional target of the tumor suppressor p53 (13). p21 may induce growth arrest by different mechanisms, including inhibition of CDKs, or the activity of proliferating cell nuclear antigen. Expression of p21 is controlled at the transcriptional level by both p53-dependent and -independent mechanisms. After DNA damage, p21 transcription is activated by p53 (14). However, a variety of agents that promote growth arrest and differentiation also activate p21 transcription by p53-independent mechanisms by means of different transcription factors, such as Sp1, Sp3, AP2, STATs, C/EBPα, C/EBPβ, and the bHLH proteins BETA2 and Myo D (reviewed in ref. 15).
In wild-type mouse embryonic fibroblasts, Myc is able to activate p21 transcription in a p53-dependent manner by inducing expression of p19ARF, leading to stabilization of the tumor suppressor p53 (16). A positive role for c-Myc in regulation of p21 expression has also been suggested by studies in c-Myc-null mouse cells, in which p21 expression is decreased (17), and in c-Myc-overexpressing fibroblasts, where c-Myc induced p53- and p21-dependent G2 arrest (18). In contrast, other studies indicate that Myc negatively regulates p21 expression. After exposure of human breast and prostate cancer cell lines to phorbol 12-myristate 13-acetate (PMA), overexpression of c-Myc inhibited p21 expression and overcame PMA-induced growth arrest (19). By using oligonucleotide microarray analysis of 6,416 genes and expressed sequence tags in primary human fibroblasts, it was shown that p21 transcription is repressed by c-Myc (20). Transforming growth factor-β-induced cell cycle arrest was inhibited by c-Myc by means of repression of induction of p21 transcription (21).
We report that c-Myc can repress p21 transcription in p53-null mouse cells and in a human adenocarcinoma cell line. This repression could be mediated through interactions between c-Myc and Sp1/Sp3. The repression of a growth arrest gene such as p21 may contribute to the oncogenic properties of c-Myc.
Materials and Methods
Plasmid Constructs.
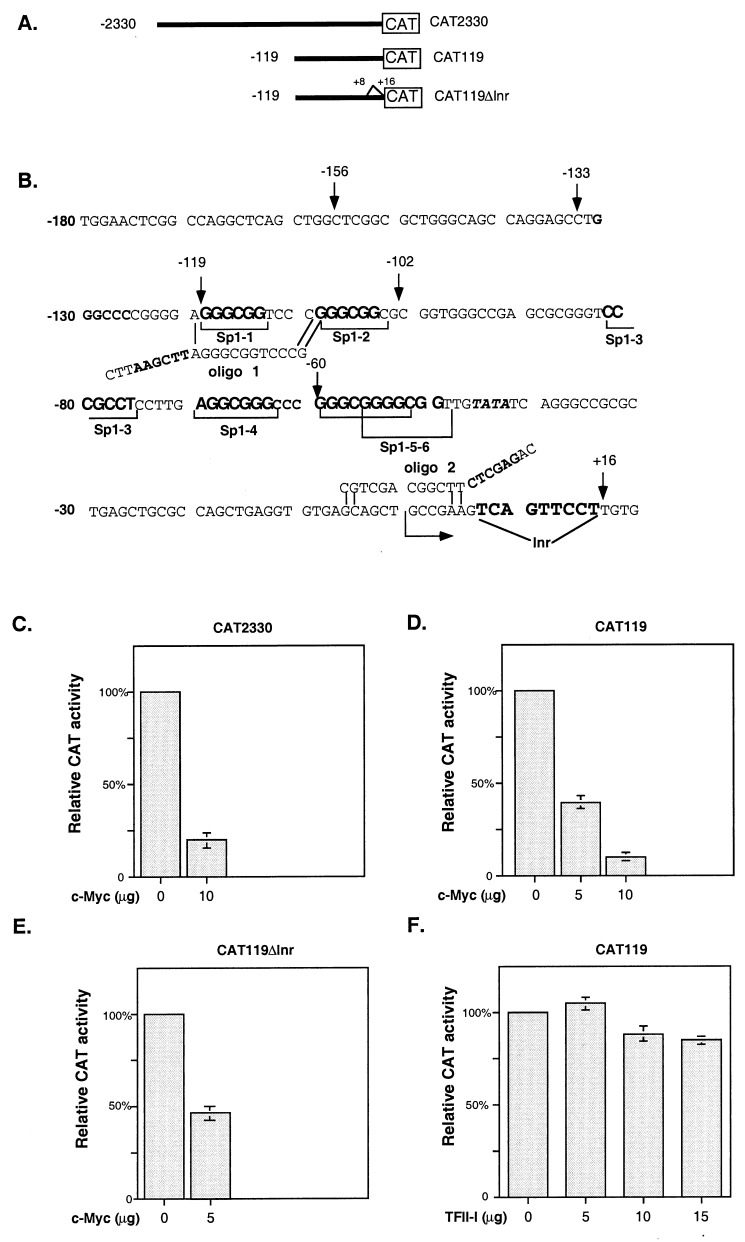
The human p21WAF1/CIP1 promoter deletion–chloramphenicol acetyltransferase (CAT) reporter construct CAT2330 in pJFCAT1 (22) (see Fig. 2A) was a gift from Wafik El-Deiry (University of Pennsylvania School of Medicine, Philadelphia). The plasmid CAT119 (see Fig. 2B) was a gift from Joseph Biggs (University of Colorado Health Sciences Center, Denver). These plasmids contain different fragments of the p21 promoter, with CAT2330 extending from −2330 to +16 bp and CAT119 from −119 to +16 bp. We digested CAT119 with XhoI–HindIII and purified the vector pJFCAT1 and insert. We used primers 1 (5′-CTTAAGCTTAGGGCGGTCCCG-3′) and 2 (5′-CAGAGCTCTTCGGCAGCTGC-3′) with HindIII and XhoI sites in bold (see Fig. 2B) and the insert for amplification of the p21 promoter fragment with deletion of an initiator element. The product of PCR amplification was digested with HindIII and XhoI enzymes and cloned back into HindIII and XhoI sites of pJFCAT1 (construct CAT119ΔInr; see Fig. 2B). Glutathione S-transferase (GST)-Sp1(1–612) and GST-Sp1(612–778) expression constructs (23) were a gift from Jane Clifford (Medical College of Pennsylvania Hahnemann School of Medicine, Philadelphia).
Figure 2.
Myc represses the p21 promoter in Caco-2 cells. (A) The human p21 promoter CAT reporter deletion constructs are diagrammed. (B) The proximal p21 promoter contains six Sp1 binding sites, a TATA box, and a potential initiator element (Inr). Oligo 1, 5′-CTTAAGCTTAGGGCGGTCCCG-3′, and oligo 2, 5′-CAGAGCTCTTCGGCAGCTGC-3′, with HindIII and XhoI sites in bold, were used for amplification of the p21 promoter fragment with deletion of a potential initiator element (construct CAT119ΔInr). (C–E) CAT assays were performed with extracts from Caco-2 cells transfected with empty vector or with different amounts of the expression constructs encoding c-Myc or TFII-I and the p21 promoter–reporter constructs CAT2330, CAT119, and CAT119ΔInr. The amount of extract that was loaded for each CAT assay was normalized for expression of β-galactosidase. All transfections were repeated at least three times in duplicate. Graphs represent the results of three independent experiments, with bars indicating SD. (C and D) Myc represses the CAT2330 and CAT119 reporter constructs in Caco-2 cells. (E) Deletion of the potential initiator site from the p21 promoter does not affect its repression by c-Myc. (F) Overexpression of TFII-I in Caco-2 cells does not lead to induction of the p21 promoter.
T7-tagged c-Myc and expression constructs encoding the Myc deletions Δ7–91, Δ106–143, and Δ347–439, and the Myc fragment 250–352 were created by PCR using BamHI (5′) and EcoRI (3′) restriction site-engineered PCR products. CMV5 c-Myc, CMV5 c-Myc Δ7–91, and CMV5 c-Myc Δ106–143 plasmids were described elsewhere (24) and were used to create respective T7-tagged expression constructs. T7 tag (5′) sequences (25) were engineered into the primers to facilitate cloning. Sequences of primers are as follows: c-Myc upstream primer, CGGGATCCATGGCTAGCATGACCGGCGGACAGCAGATGGGCATGCCCCTCAACGTTAGCTTC; c-Myc downstream primer, CGGAATTCTTACGCACAAGAGTTCCGTAG; Δ347–439 downstream primer, CGGAATTCTCTATGTTCGCCTCTTGACATTCTCCTC; 250–352 upstream primer, CGGGATCCATGGCTAGCATGACCGGCGGACAGCAGATGGGCAGCGACTCTGAGGAGGAA; 250–352 downstream primer, CGGAATTCTTACTCCTCGGTGTCCGAGGA. PCR fragments were then cloned into pcDNA3 (Invitrogen) cut with BamHI and EcoRI. The constructs were sequenced.
Cell Culture, Transfections, and CAT Assays.
The BALB/c mouse immortalized fibroblast cell line (10.1)-MycER, which contains an inducible fusion protein of Myc and the estrogen receptor (ER), does not express p53 (26). These cells were maintained in DMEM without phenol red (GIBCO/BRL) with 10% FBS with certified low estrogen content (Atlanta Biologicals, Norcross, GA). 293 human embryonal kidney cells were grown in DMEM containing 10% FBS under a 5% CO2/95% air atmosphere. The Caco-2 human colon adenocarcinoma cell line was obtained from the American Type Culture Collection and grown in DMEM/F12 with 20% FCS. Transfections and CAT assays were performed as described (27).
RNA Preparation and Reverse Transcription (RT)–PCR.
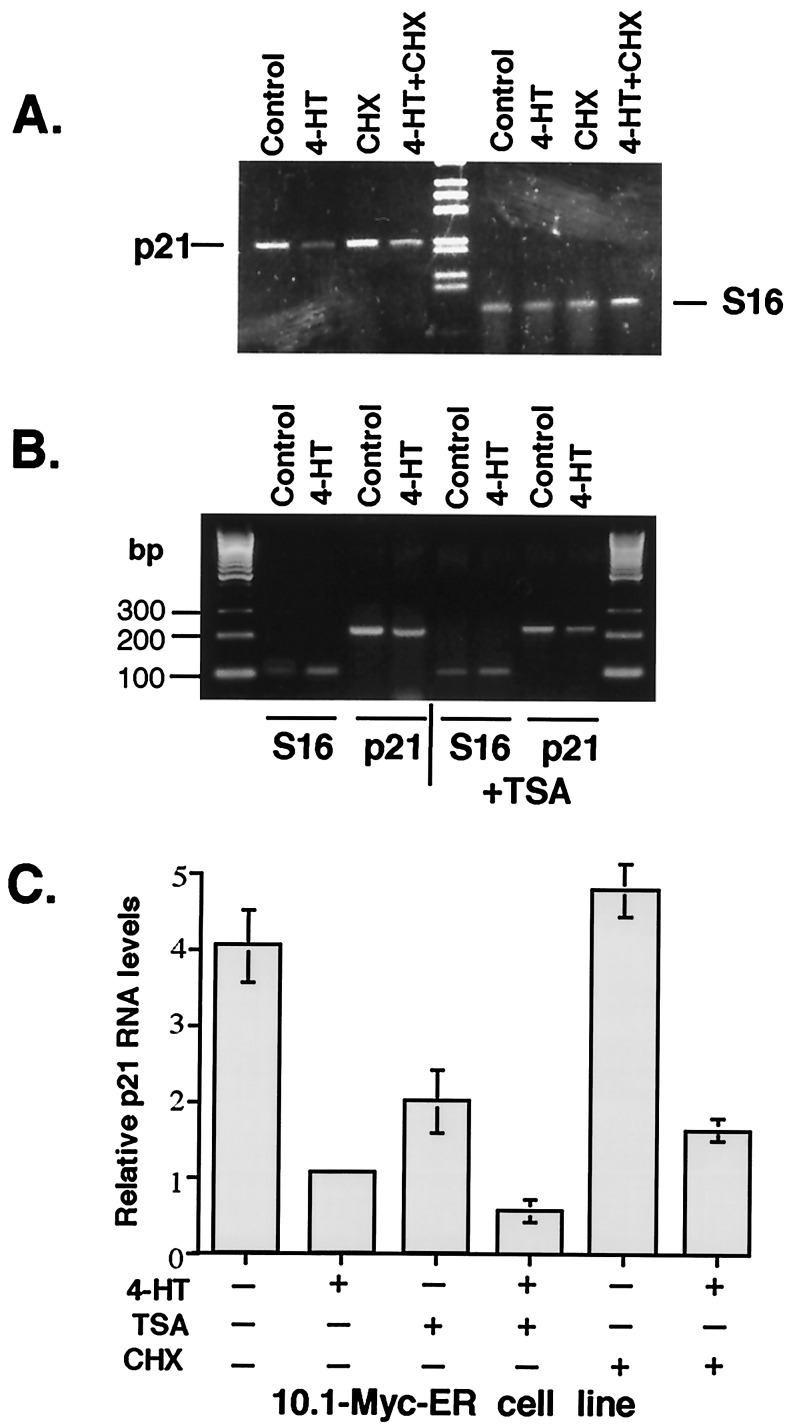
Total RNA was prepared from (10.1)-MycER cells before and after addition of 4-hydroxytamoxifen (4-HT) by using Trizol reagent (GIBCO/BRL). RT-PCR was performed by using Ready To Go RT-PCR Beads (Amersham Pharmacia). Mouse S16 ribosomal protein expression was used as an internal control. Amplification of the p21 cDNA included 27 cycles of PCR with 45 sec of denaturation at 94°C, 45 sec of primer annealing at 62°C, and 1 min of extension/synthesis at 72°C with primers to mp21, AGCCTGAAGACTGTGATGGG and AAAGTTCCACCGTTCTCGG, and with product size 228 bp. Amplification of the S16 ribosomal protein cDNA included 23 cycles of PCR with 45 sec of denaturation at 94°C, 45 sec of primer annealing at 70°C, and 1 min of extension/synthesis at 72°C with primers AGGAGCGATTTGCTGGTGTGGA and GCTACCAGGCCTTTGAGATGGA, making a 102-bp product (28). The same conditions were used after simultaneous addition of cycloheximide or trichostatin A (TSA) for 12 h. Amplification of p21 cDNA after addition of TSA was performed for 25 cycles. PCR products were separated on a 2% agarose gel and stained with ethidium bromide.
GST Pull-Down Assay, Coimmunoprecipitation, and Immunoblotting.
For immunoprecipitation with T7 antibodies, 293 cells were transfected with T7-Myc constructs and harvested 48 h after transfection. Cell lysates were incubated with agarose-linked T7 antibody (Novagen) for 2 h at 4°C. Beads were collected by centrifugation and washed, and bound proteins were subjected to immunoblotting with Sp1 antibody (PEP2; Santa Cruz Biotechnology).
For immunoprecipitation with Sp1 or Sp3 antibodies, 500 μg of cell lysate was incubated with 5 μl of Sp1 antibody (PEP2) or with 5 μl of Sp3 antibody (D-20; Santa Cruz Biotechnology). Immunoblotting was performed with monoclonal anti-c-Myc antibody (9E10) and anti-mouse antibodies conjugated to horseradish peroxidase (Zymed).
For GST pull-down assays, 293 cells were transfected with T7-Myc constructs and harvested 48 h after transfection. Cell lysates were precleared by incubation with GST-saturated glutathione beads for 1 h. Lysates were incubated with GST-Sp1, GST-Sp1(1–612), or GST-Sp1(612–778) for 1 h at 4°C followed by incubation with glutathione beads for 1 h. Bound proteins were eluted with sample buffer and subjected to SDS/PAGE followed by immunoblotting with anti-T7 antibody.
Results
Myc Negatively Regulates p21 Expression in the Absence of de Novo Protein Synthesis in Vivo.
We examined whether c-Myc can regulate p21 transcription in a p53-null immortalized mouse fibroblast cell line, (10.1)-MycER, that contains an inducible human Myc-ER fusion protein (26). The MycER fusion protein is constitutively expressed in this cell line, but it remains inactive until stimulated by addition of 4-HT, which derepresses the hormone-binding domain of the fusion protein. We compared levels of p21 mRNA in control (10.1)-MycER cells and in cells where Myc was induced by addition of 4-HT. By using RT-PCR, we found that endogenous p21 transcription is repressed 2- to 3-fold by activation of MycER. Simultaneous treatment of these cells with 4-HT and the protein synthesis inhibitor cycloheximide also led to repression of the p21 transcription (Fig. 1A). These data suggest that p21 expression may be down-regulated upon activation of preexisting MycER in the absence of new protein synthesis.
Figure 1.
Myc represses endogenous p21 transcription in p53-null cells in the absence of new protein synthesis. Immortalized (10.1)-MycER mouse fibroblast cells were grown to 30% confluence, and 4-HT was added for 5 h. RT-PCR was performed to examine p21 (27 cycles) and mouse S16 ribosomal protein mRNA expression (internal control). The experiment was performed under the same conditions with simultaneous addition of cycloheximide (CHX) or TSA for 12 h. Amplification of mouse p21 cDNA after addition of cycloheximide was performed for 27 cycles and after addition of TSA for 25 cycles. Typical ethidium bromide-stained 2% agarose gels with bands corresponding to p21, S16, and the 100-bp or 1-kb DNA ladders (GIBCO/BRL) are shown. (A) Myc negatively regulates p21 expression in the absence of de novo protein synthesis in vivo. (B) Myc does not recruit histone deacetylase for repression of the p21 promoter. (C) Graphs showing the results of 4–6 independent experiments described in A and B with bars indicating SD.
One possible mechanism of transcriptional repression includes recruitment of histone deacetylase to a promoter. To test whether Myc may recruit histone deacetylase for repression of the p21 promoter, we used a specific inhibitor of histone deacetylase, TSA, that activates p21 transcription (29). We found that addition of TSA to these cells did not alter repression of p21 transcription by c-Myc (Fig. 1B). This finding suggests that Myc does not recruit histone deacetylase for repression of the p21 promoter.
Myc Represses the p21 Promoter by Its Proximal Region in Human Caco-2 Cells.
p21 expression is induced during differentiation of Caco-2 colon adenocarcinoma cell line (28). In these cells, basal p21 transcription is controlled primarily by the transcription factors Sp1 and Sp3 through the region of the p21 promoter between −119 and +16 bp that contains six Sp1 binding sites (Fig. 2B) (27). To determine whether c-Myc repression of p21 transcription occurs at the promoter level, we performed transient transfection experiments in Caco-2 cells with different p21-promoter reporters (Fig. 2A) in the presence of a c-Myc expression construct. We found that c-Myc is able to repress the minimal p21 promoter–reporter construct CAT119 containing sequences from −119 to +16 bp as efficiently as a 2.3-kb p21 promoter construct CAT2330 (Fig. 2 C and D).
It has been shown that Myc is capable of binding and repressing the albumin, C/EBPα (30), and the adenovirus major late promoters (30, 31) by means of initiator (Inr) elements. The Inr element is usually defined as a weak consensus YYCAYYYYY, where Y is a pyrimidine base (32). The initiator binding protein TFII-I stimulates basal transcription from the Inr element (33). Myc can bind TFII-I and form a complex associated with the Inr sequence that prevents transcriptional activation from Inr (34). After analysis of the p21 promoter, we found a previously undescribed potential initiator site TCAGTTCCT located between +7 and +16 bp (Fig. 2B), and we decided to test whether repression of the p21 promoter by Myc is mediated through interaction between Myc and the initiator binding protein TFII-I. By using PCR, we deleted the putative initiator sequence in the p21 promoter (see Materials and Methods; Fig. 2 A and B) and determined that Myc could repress CAT119ΔInr, a p21 promoter-reporter without the initiator sequence (Fig. 2D). Cotransfection of increasing amounts of TFII-I together with the CAT119 reporter did not lead to induction of the p21 promoter in Caco-2 cells (Fig. 2E). These data suggest that repression of the p21 promoter by Myc does not involve an Inr sequence and is not based on interaction between Myc and TFII-I.
Myc Represses the p21 Promoter by Interactions with Transcription Factors Sp1/Sp3.
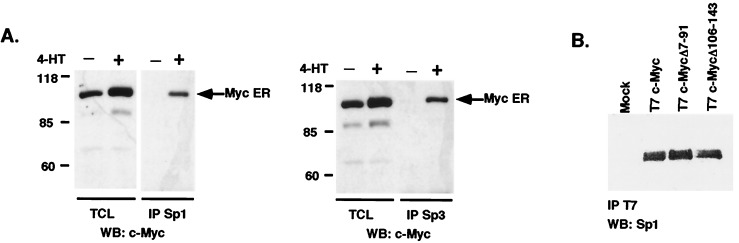
c-Myc repressed the p21 CAT119 construct in Caco-2 cells (Fig. 2D) and was able to repress Sp1/Sp3-induced expression of CAT119 in SL2 Drosophila cells that lack Sp1 and related factors (data not shown). To determine whether c-Myc repression of the p21 promoter is based on interactions between c-Myc and the transcription factors Sp1/Sp3, we induced c-Myc activity in the (10.1)-MycER cell line by addition of 4-HT, and performed immunoprecipitations using antibodies against Sp1 (PEP2) and Sp3 (D-20) followed by immunoblotting with antibodies against human c-Myc. We observed association between c-Myc and Sp1, and c-Myc and Sp3 only after activation of c-Myc by addition of 4-HT (Fig. 3 A and B). We also transfected 293 cells with plasmids expressing T7 epitope-tagged c-Myc or empty vector. Cell extracts were prepared and subjected to immunoprecipitation with T7 antibody that was covalently linked to beads, followed by immunoblotting with Sp1 antibody. Using this method, we also observed association between c-Myc and Sp1 (Fig. 3B).
Figure 3.
c-Myc is associated with Sp1 and Sp3. (A) Immunoprecipitation (IP) with Sp1 or Sp3 antibodies was performed with cellular extracts from 10.1-MycER cells before and after addition of 4-HT, followed by immunoblotting (WB) with monoclonal anti-c-Myc antibody (9E10). (B) 293 cells were transiently transfected with mammalian expression constructs encoding T7-tagged c-Myc and Myc Δ7-91 and Myc Δ106–143 and harvested after 48 h. Cell lysates were incubated with agarose-linked T7 antibody for 2 h at 4°C. Bound proteins were subjected to immunoblotting with Sp1 antibody PEP2 (Santa Cruz Biotechnology). TCL, total cell lysate.
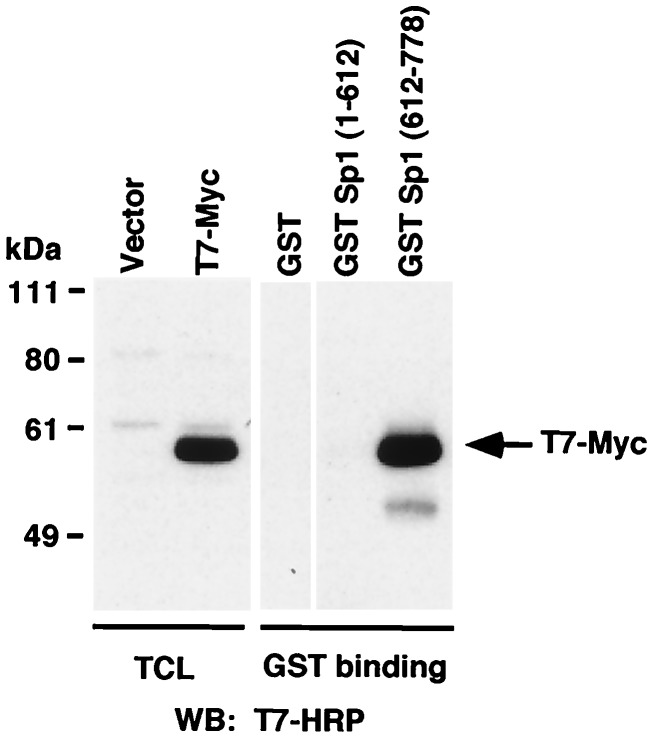
To identify the region of Sp1 that is responsible for interaction with c-Myc, we used GST-fusion proteins containing the N-terminal transactivation domain, GST-Sp1(1–612) and the C-terminal DNA-binding zinc finger motif GST-Sp1(612–778). These GST-fusion proteins were used in pull-down experiments with extracts from 293 cells transfected with plasmids expressing T7 epitope-tagged c-Myc. We found that c-Myc can interact with the C-terminal zinc finger-containing region of Sp1, but not with the N-terminal part of the protein (Fig. 4).
Figure 4.
c-Myc is associated with C-terminal zinc finger domain of Sp1. 293 cells were transfected with the T7-Myc construct and harvested 48 h after transfection. Total cell lysates were incubated with GST, GST-Sp1(1–612), or GST-Sp1(612–778) for 1 h at 4°C, followed by incubation with glutathione beads for 1 h. Total cell lysate or bound proteins were eluted with sample buffer and subjected to SDS/PAGE followed by immunoblotting with anti-T7 antibody.
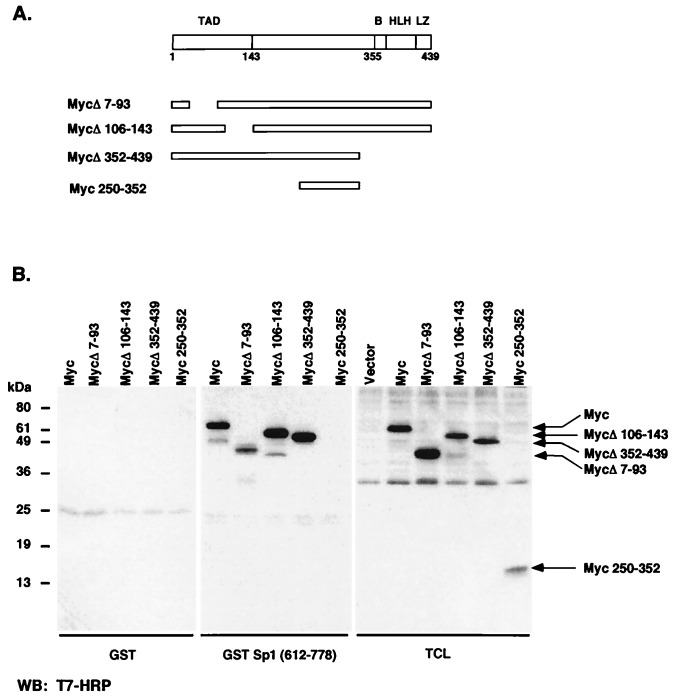
To identify the region of c-Myc that is responsible for interaction with Sp1, we used GST-Sp1(612–778) in GST pull-down experiments with extracts from 293 cells transfected with the plasmids expressing different T7 epitope-tagged c-Myc constructs. Deletions of the N- or C-terminal parts of c-Myc did not affect Myc interaction with Sp1 (Fig. 5B), suggesting that the central region of c-Myc (amino acids 143–352) is involved in interaction with Sp1. After transfection of 293 cells, we also immunoprecipitated T7 epitope-tagged c-Myc, Myc Δ7–91, and MycΔ106–143 from total cell lysates and subjected immunoprecipitated protein to immunoblotting with Sp1 antibody. Sp1 coimmunoprecipitated with both Myc Δ7–91 and MycΔ106–143, suggesting that the N-terminal region of c-Myc is not important for interactions between c-Myc and Sp1 (Fig. 3B).
Figure 5.
The central region of c-Myc protein between amino acids 143 and 352 is involved in interaction with Sp1. (A) The structural domains of c-Myc and the c-Myc mutants used in this work are diagrammed. Domains include the transcriptional activation domain (TAD), a basic region (B), a helix–loop–helix motif (HLH), and leucine zipper domain (LZ). (B) 293 cells were transfected with expression constructs for T7-tagged c-Myc, the Myc deletion mutants MycΔ7–91, MycΔ106–143, and MycΔ352–439, and the Myc fragment 250–352 and harvested 48 h after transfection. Pull-down assays were performed by incubating lysates with GST or GST-Sp1-(612–778) for 1 h at 4°C followed by incubation with glutathione beads for 1 h. Bound proteins were subjected to SDS/PAGE followed by immunoblotting with anti-T7 antibody.
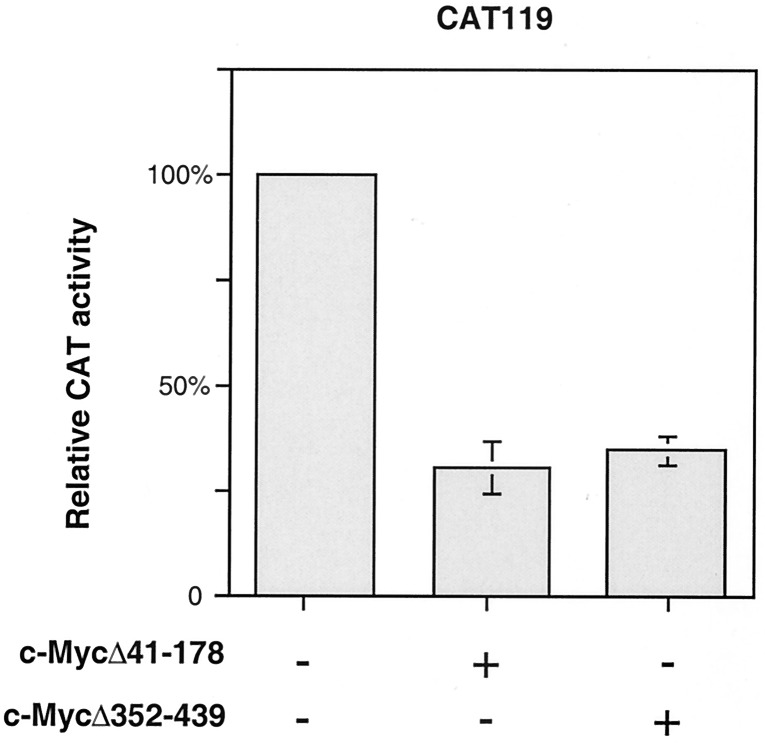
To further examine the roles of N- and C-terminal regions of c-Myc in p21 repression, the constructs with N- or C-terminal deletions that did not affect binding of c-Myc to Sp1 were cotransfected into Caco-2 cells together with p21 promoter–reporter construct CAT119. We found that these expression constructs efficiently repress p21 promoter activity in Caco-2 cells (Fig. 6), indicating that N- and C-terminal regions of c-Myc are dispensable, not only for interactions with Sp1, but also for repression of the p21 promoter.
Figure 6.
N- or C-terminal deletions of c-Myc do not affect its ability to repress the p21 promoter. CAT assays were performed with extracts from Caco-2 cells transfected with the empty vector or with expression plasmids encoding the c-Myc deletion mutants c-MycΔ41–178 or c-MycΔ352–439 and the p21 promoter–reporter construct CAT119. The amount of extract that was loaded for each CAT assay was normalized for expression of the β-galactosidase control. All transfections were repeated at least three times in duplicate. Graphs represent the results of three independent experiments, with bars indicating SD.
Discussion
We examined the mechanism by which c-Myc represses p21 transcription. By using transient transfections, we found that Myc represses the p21 promoter through the short GC-rich region just upstream of the transcription start site, although Myc does not appear to bind to the promoter. We found that c-Myc does not need to heterodimerize with Max for repression of the p21 promoter (data not shown). This proximal region of the p21 promoter lacks canonical Myc-binding sites, but contains multiple Sp1-binding sites and a potential Inr site. We tested whether repression of p21 by c-Myc involved the Inr site or recruitment of histone deacetylase. Our data indicate that c-Myc repression of p21 transcription is not based on interaction between the Inr-binding protein TFII-I and c-Myc, and it is independent of histone deacetylase activity. Claassen and Hann (21) obtained similar results with NIH 3T3 cells stably transfected with a p21 promoter-LUC reporter construct. They showed that c-Myc represses the p21 promoter through a region near transcription start site, and this repression is independent of histone deacetylase activity.
Previously, we found that Sp1 and Sp3 regulate basal transcription of the p21 gene in Caco-2 cells through sequences located between −119 and +16 bp of the p21 promoter (27), and, here, we show that c-Myc represses the CAT119 construct containing these sequences. c-Myc deletion mutants that retain the ability to associate with Sp1 were able to repress the p21 promoter, suggesting that interactions between Sp1/Sp3 and c-Myc may be responsible for repression. We found that c-Myc can associate with Sp1/Sp3 and that C-terminal DNA-binding zinc finger domain of Sp1 interacts with central region of c-Myc. Further studies are needed to determine whether the interactions between c-Myc and Sp1/Sp3 are direct.
These data provide evidence of possible interactions between Sp1/Sp3 and c-Myc, and suggest that c-Myc may bind to the DNA-binding domain of Sp1 and titrate Sp1 proteins from the p21 promoter. It has been found that promoters with multiple Sp1-binding sites may be very sensitive to mutations in a single Sp1-binding site (35, 36), which may dramatically reduce their activity. In these cases, minor changes in Sp1 levels can severely affect activity of such promoters. Similarly, the p21 promoter with six Sp1-binding sites may be sensitive to subtle alterations in Sp1/Sp3 levels because of sequestration of Sp1/Sp3 by c-Myc. Overexpression of c-Myc may lead to a decrease of Sp1/Sp3 binding to the p21 promoter and to repression of p21 promoter activity. It will be interesting to determine whether other promoters that contain multiple Sp1 sites are repressed by c-Myc by a similar mechanism. Because the c-Myc promoter contains multiple Sp1-binding sites (36) and it can repress its own transcription (37), it is plausible that Myc repression is also based on interactions between Sp1 and c-Myc.
It has been reported that c-Myc may induce (16–18) or repress (19–21) p21 transcription. Data from these reports suggested that induction of p21 by c-Myc overexpression in normal human and mouse fibroblasts is p53-dependent (16, 18). However, in primary human fibroblasts, the p21 gene was identified as one of the targets for repression by c-Myc by oligonucleotide microarray analysis (20). It has been shown that c-Myc bypasses TPA- (19) or transforming growth factor-β-induced (21) growth arrest by transcriptional repression of p21. Claassen and Hann (21) and this paper showed that a region immediately upstream of the site of transcriptional initiation is sufficient for repression of the p21 promoter by c-Myc. Our data suggest that repression of the p21 promoter by c-Myc is based on interactions between c-Myc and transcription factors Sp1/Sp3.
c-Myc may regulate p21 transcription by two different mechanisms. It may indirectly activate p53 expression through p19/ARF and induce p53-dependent induction of p21 by means of p53 binding sites at −2301 and −1394 bp. Alternatively, Myc may repress p21 transcription by interaction with Sp1/Sp3 through the −119-bp to +16-bp region containing multiple Sp1-binding sites. These opposite effects of c-Myc on p21 transcription may reflect a dual function of p21. On one hand, p21 may be required for cell cycle progression, whereas, on the other hand, it can induce cell cycle arrest depending on its threshold levels (38). In cells with mutant p53, c-Myc will repress p21, and, in these cells, overexpression of c-Myc will induce cell proliferation. In contrast, the overall effect of repression and activation will be additive in cells with wild-type p53.
Overexpression of c-Myc in quiescent cells induces cell cycle entry, and deregulated expression of c-Myc induces tumorigenesis. The mechanisms by which c-Myc regulates cell cycle progression are not fully understood, even though it has been found that c-Myc may activate and repress transcription of certain cell cycle-regulatory genes. Recent evidence suggests that gene repression may play a vital role in Myc-induced cell cycle progression and cellular transformation. Repression of cell cycle inhibitory genes provides a way to promote cell growth and c-Myc represses several growth arrest genes, such as gadd34, gadd45, gadd153 (8, 39), gas1 (9), and p15Ink4b (10). Different lines of evidence suggest that c-Myc-mediated repression, rather than transactivation, correlates with cellular transformation. For example, a naturally occurring truncated form of Myc, MycS, lacks a functional transactivation domain, but retains the ability to repress promoters of the growth arrest genes and transform Rat1a fibroblasts (40). An ability of c-Myc to repress the checkpoint gene p21 may contribute to its ability to promote proliferation and oncogenesis.
Acknowledgments
We thank Charlotte Hurth for excellent technical assistance, Wafik El-Deiry and Joseph Biggs for the p21 promoter-deletion constructs, and Jane Clifford for GST-Sp1(1–612) and GST-Sp1(612–778). This work was supported by Award 98-28 from the American Cancer Society, Illinois Division, and a Charles E. Culpeper Biomedical Pilot Initiative Grant (to A.L.G.), National Institutes of Health Grant RO1 DK56283 (to A.L.T.), National Institutes of Health Grant RO1 CA71874 (to N.H.), and by the National Cancer Institute Oncology Research Faculty Development Program (to E.G.).
Abbreviations
- bHLH
basic helix–loop–helix
- GST
glutathione S-transferase
- CAT
chloramphenicol acetyltransferase
- ER
estrogen receptor
- RT-PCR
reverse transcription–PCR
- 4-HT
4-hydroxytamoxifen
- TSA
trichostatin A
- Inr
initiator
References
- 1.Grandori C, Cowley S M, James L P, Eisenman R N. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 2.Eilers M, Schirm S, Bishop J M. EMBO J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner A J, Meyers C, Laimins L A, Hay N. Cell Growth Differ. 1993;4:879–883. [PubMed] [Google Scholar]
- 4.Galaktionov K, Chen X, Beach D. Nature (London) 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 5.Reisman D, Elkind N B, Roy B, Beamon J, Rotter V. Cell Growth Differ. 1993;4:57–65. [PubMed] [Google Scholar]
- 6.Dang C V, Resar L M, Emison E, Kim S, Li Q, Prescott J E, Wonsey D, Zeller K. Exp Cell Res. 1999;253:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- 7.Mink S, Mutschler B, Weiskirchen R, Bister K, Klempnauer K H. Proc Natl Acad Sci USA. 1996;93:6635–6640. doi: 10.1073/pnas.93.13.6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marhin W W, Chen S, Facchini L M, Fornace A, Jr, Penn L Z. Oncogene. 1997;14:2825–2834. doi: 10.1038/sj.onc.1201138. [DOI] [PubMed] [Google Scholar]
- 9.Lee T C, Li L, Philipson L, Ziff E B. Proc Natl Acad Sci USA. 1997;94:12886–12891. doi: 10.1073/pnas.94.24.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warner B J, Blain S W, Seoane J, Massague J. Mol Cell Biol. 1999;19:5913–5922. doi: 10.1128/mcb.19.9.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harper J W, Adami G, Wei N, Keyomarsi K, Elledge S. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 12.Xiong Y, Hannon G, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 13.El-Deiry W, Tokino T, Velculescu V, Levy D, Parsons R, Trent J, Lin D, Mercer W, Kinzler K, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 14.El-Deiry W S, Harper W J, O'Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, et al. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 15.Gartel A L, Tyner A L. Exp Cell Res. 1999;246:280–289. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- 16.Zindy F, Eischen C M, Randle D H, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mateyak M K, Obaya A J, Sedivy J M. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsher D W, Zetterberg A, Zhu J, Tlsty T, Bishop J M. Proc Natl Acad Sci USA. 2000;97:10544–10548. doi: 10.1073/pnas.190327097. . (First Published August 29, 2000; 10.1073/pnas.190327097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell K O, El-Deiry W S. Cell Growth Differ. 1999;10:223–230. [PubMed] [Google Scholar]
- 20.Coller H A, Grandori C, Tamayo P, Colbert T, Lander E S, Eisenman R N, Golub T R. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claassen G F, Hann S R. Proc Natl Acad Sci USA. 2000;97:9498–9503. doi: 10.1073/pnas.150006697. . (First Published August 1, 2000; 10.1073/pnas.150006697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fridovich-Keil J L, Gudas J M, Bryan I B, Pardee A B. BioTechniques. 1991;11:572–579. [PubMed] [Google Scholar]
- 23.Black A R, Jensen D, Lin S Y, Azizkhan J C. J Biol Chem. 1999;274:1207–1215. doi: 10.1074/jbc.274.3.1207. [DOI] [PubMed] [Google Scholar]
- 24.Amin C, Wagner A J, Hay N. Mol Cell Biol. 1993;13:383–390. doi: 10.1128/mcb.13.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studier F W. J Mol Biol. 1981;153:493–502. doi: 10.1016/0022-2836(81)90404-6. [DOI] [PubMed] [Google Scholar]
- 26.Wagner A J, Kokontis J M, Hay N. Genes Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- 27.Gartel A L, Goufman E, Najmabadi F, Tyner A L. Oncogene. 2000;19:5182–5188. doi: 10.1038/sj.onc.1203900. [DOI] [PubMed] [Google Scholar]
- 28.Gartel A L, Serfas M S, Gartel M, Goufman E, Wu G S, El-Deiry W S, Tyner A L. Exp Cell Res. 1996;227:171–181. doi: 10.1006/excr.1996.0264. [DOI] [PubMed] [Google Scholar]
- 29.Sowa Y, Orita T, Minamikawa S, Nakano K, Mizuno T, Nomura H, Sakai T. Biochem Biophys Res Commun. 1997;241:142–150. doi: 10.1006/bbrc.1997.7786. [DOI] [PubMed] [Google Scholar]
- 30.Li L H, Nerlov C, Prendergast G, MacGregor D, Ziff E B. EMBO J. 1994;13:4070–4079. doi: 10.1002/j.1460-2075.1994.tb06724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee L A, Dolde C, Barrett J, Wu C S, Dang C V. J Clin Invest. 1996;97:1687–1695. doi: 10.1172/JCI118595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smale S T, Baltimore D. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 33.Roy A L, Meisterernst M, Pognonec P, Roeder R G. Nature (London) 1991;354:245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- 34.Roy A L, Carruthers C, Gutjahr T, Roeder R G. Nature (London) 1993;365:359–361. doi: 10.1038/365359a0. [DOI] [PubMed] [Google Scholar]
- 35.Gidoni D, Kadonaga J T, Barrera-Saldana H, Takahashi K, Chambon P, Tjian R. Science. 1985;230:511–517. doi: 10.1126/science.2996137. [DOI] [PubMed] [Google Scholar]
- 36.DesJardins E, Hay N. Mol Cell Biol. 1993;13:5710–5724. doi: 10.1128/mcb.13.9.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penn L J, Brooks M W, Laufer E M, Land H. EMBO J. 1990;9:1113–1121. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng M, Olivier P, Diehl J A, Fero M, Roussel M F, Roberts J M, Sherr C J. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amundson S A, Zhan Q, Penn L Z, Fornace A., Jr Oncogene. 1998;17:2149–2154. doi: 10.1038/sj.onc.1202136. [DOI] [PubMed] [Google Scholar]
- 40.Xiao Q, Claassen G, Shi J, Adachi S, Sedivy J, Hann S R. Genes Dev. 1998;12:3803–3808. doi: 10.1101/gad.12.24.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]