Abstract
The 18-amino-acid cationic, tryptophan-rich ApoEdpL-W peptide derived from human ApoE apolipoprotein was shown to have antifungal activity against pathogenic yeasts of the Candida genus (except C. glabrata). ApoEdpL-W was active against planktonic cells and early-stage biofilms but less active against mature biofilms, possibly because of its affinity for extracellular matrix beta-glucans. Moreover, ApoEdpL-W absorbed to medically relevant materials partially prevented the formation of biofilms on these materials. The exposure of C. albicans cells to sublethal doses of ApoEdpL-W triggered a transcriptional response reminiscent of that associated with the inactivation of the MYO5 gene required for endocytosis as well as the upregulation of amino acid transporter genes. A fluorescent derivative of ApoEdpL-W accumulated at the cytoplasmic membrane and subsequently was translocated to the vacuole. Strikingly, the inactivation of MYO5 or addition of latrunculin, an inhibitor of endocytosis, prevented the vacuolar accumulation of fluorescein-labeled ApoEdpL-W and reduced the antifungal activity of ApoEdpL-W. This, together with the insensitivity of ApoEdpL-W to alterations in membrane fluidity and high salt, suggested that the ApoEdpL-W mode of action was dependent upon vacuolar targeting and differed significantly from that of other antifungal peptides, such as Histatin-5 and Magainin 2.
INTRODUCTION
The increasing number of immune-suppressed patients or patients in intensive care for extended time periods leads to an inflating incidence of opportunistic Candida infections (55). Moreover, the tolerance of Candida biofilms to usual antifungals such as azoles makes infections associated with invasive medical procedures and the use of medical devices (such as catheters, prostheses, and implantable ports) a recurring problem in health care settings (11). Today there is an urgent need for new antifungal therapy, and antimicrobial peptides have emerged as a possible alternative (32). This class of molecules has been identified from a large panel of organisms, ranging from invertebrates to amphibians to humans (32). They are considered to be less subject to the possible emergence of resistance (32) and can have a broad spectrum of anti-infective activity. The latter property is essential when addressing Candida infections, as infections by Candida species other than Candida albicans species are becoming increasingly prevalent, and the ability of a drug to target the most pathogenic Candida species is a prerequisite for development (53, 55).
Several antimicrobial peptides have been extensively studied for their efficacy toward various pathogenic species, including fungi. In particular, histatins, a family of low-molecular-weight histidine-rich cationic peptides present in saliva (52), have been the focus of numerous studies (66). Other antifungal peptides, such as Magainin 2, a natural antibiotic from frog skin (67), or derivatives of the human salivary mucin MUC7 (23) also are well documented. Recent progress has been made in understanding the mode of action of Histatin-5 (Hst-5), a member of the histatin family of antifungal peptides that shows the highest antifungal activity toward C. albicans (66). Briefly, Hst-5 has been shown to bind C. albicans cell wall components, including beta-glucans and the heat shock protein Ssa2, prior to being internalized at specific sites on the plasma membrane (30, 31, 38, 60). Hst-5 internalization is thought to trigger ionic imbalance through ionic efflux, resulting in cell death (30), although it also has been proposed that Hst-5 entry ruptures the plasma membrane (42). As a consequence of this mode of action, altering Ssa2 or reducing cell wall beta-glucans through treatment with echinocandin antifungals, increasing the rigidity of membranes through ATP depletion with sodium azide or carbonyl cyanide m-chlorophenylhydrazone (CCCP), or increasing osmolarity with external NaCl protects against Hst-5 antifungal activity (30, 60). Noticeably, Hst-5 appears to accumulate in the cytoplasm and in the vacuole, the latter through both endocytosis and autophagy (30, 42). However, altering the vacuolar accumulation of Hst-5 does not interfere with its antifungal activity, which is consistent with an activity of Hst-5 being mainly associated with cytosolic translocation (30). In contrast, the mode of action of other cationic antifungal peptides (CAFPs) is less well understood and appears to involve cytoplasmic membrane permeabilization by pore-forming mechanisms and/or the targeting of intracellular components (reviewed in reference 9). It is becoming increasingly clear that CAFPs, and antimicrobial peptides in general, have different modes of action that are or are not correlated with the sharing of some important features, such as their high content of cationic amino acids and amphipathy.
Human-derived antimicrobial peptides have attracted interest as, intuitively, they should have reduced toxicity, thus representing promising leads for drug development. Kelly et al. (35) recently described a series of peptides with low hemolytic activity and a broad spectrum of antimicrobial and antiviral activities. These peptides were derived from the receptor-binding region of human apolipoprotein E (ApoE), which has been shown to possess anti-infective properties (13). One of these peptides, ApoEdpL-W, an 18-mer highly cationic and tryptophan-rich peptide (WRKWRKRWWWRKWRKRWW), proved to be particularly effective against C. albicans, the most prevalent clinical Candida species. To obtain an exhaustive view of the antifungal spectrum of this new class of antimicrobial peptides, we first investigated the antifungal properties of ApoEdpL-W against planktonic and sessile forms of C. albicans as well as its efficacy against clinical strains of the most prevalent Candida species. To get insight into the mode of action of ApoEdpL-W, we used microarray analysis to compare the transcript profiles of planktonic C. albicans cells left unexposed or exposed to sub-MIC concentrations of ApoEdpL-W. Additionally, we used a fluorescein-labeled ApoEdpL-W derivative as well as C. albicans mutant strains or chemical inhibitors in an attempt to characterize the mode of action of ApoEdpL-W at the cellular level. The transcriptional and physiological responses of ApoEdpL-W-exposed C. albicans cells suggest that endocytosis and the vacuolar accumulation of the peptide are necessary for optimized antifungal activity. Overall, responses to ApoEdpL-W differ from those to other CAFPs, such as Hst-5 and Magainin 2, suggesting that ApoEdpL-W has a distinct mode of action.
MATERIALS AND METHODS
Strains and media.
All C. albicans laboratory strains used in this study are listed in Table 1. Ninety-one clinical Candida strains from a subset of the CandiRea collection (8) (see Table S1 in the supplemental material) were used for MIC determination. Strains were routinely grown at 30°C on YPD medium (1% yeast extract, 2% peptone, 2% glucose) or SD minimal medium (0.67% yeast nitrogen base without amino acids [Difco] plus 0.4 or 2% glucose) supplemented with uridine (40 mg/liter), arginine (20 mg/liter), or histidine (20 mg/liter), when needed, and 2% agar for solid media. RPMI 1640 (Gibco) medium complemented with 2% glucose and buffered with 0.165 M morpholinepropanesulfonic acid (MOPS), pH 7, as recommended for the EUCAST method (17) was used for MIC determination and checkerboard assays. SD with 2% glucose at pH 7 (SD medium with 0.165 M MOPS buffered at pH 7 with NaOH) was used for MIC determination and fluorescence microscopy experiments. Overexpression experiments were performed in S-casa medium (0.67% yeast nitrogen base without amino acids [Difco], 2% Casamino Acids). RPMI 1640 (Gibco) medium buffered with 50 mM HEPES was used for biofilm experiments in 96-well plates.
Table 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SC5314 | C. albicans wild type | 20 |
| BWP17U | ura3Δ::λimm434/URA3 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG | 44 |
| BWP17AHU | ura3Δ::λimm434/URA3 arg4Δ::hisG/ARG4 his1Δ::hisG/HIS1 | 18 |
| CEC361 | ura3Δ::λimm434/URA3 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG YAK1/yak1Δ::HIS1 RPS10/rps10::CIp10-URA3 | 22 |
| CEC161 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/HIS1 arg4::hisG/ARG4 | 16 |
| CEC1088 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/HIS1 arg4::hisG/ARG4 RPS1/RPS1::CIp10::(URA3 PCK1p-RTA2-TAPtag) | This study |
| CEC1545 | ura3Δ::λimm434/URA3 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG rta2Δ::HIS1/rta2Δ::ARG4 | This study |
| CAI4 | ura3Δ::λimm434/ura3Δ::λimm434 | 19 |
| CEC915 | ura3Δ::λimm434/ura3Δ::λimm434 RPS1/rps1::CIp10::URA3 | 5 |
| COU73 | ura3Δ::λimm434/ura3Δ::λimm434 myo5::hisG/myo5::hisG::(URA3 MYO5) | 50 |
| COU46 | ura3Δ::λimm434/ura3Δ::λimm434 myo5::hisG/myo5::hisG | 50 |
| ssa2Δ | ura3Δ::λimm434/ura3Δ::λimm434 SSA1/ssa1::FRT Δssa2/ssa2::FRT | 38 |
| ssa2Δ/SSA2 | ura3Δ::λimm434/ura3Δ::λimm434 SSA1/ssa1::FRT Δssa2/ssa2::FRT/RPS10::SSA2 | 38 |
| CEC290 | ptr21Δ::HIS1/ptr21Δ::ARG4 ura3Δ::λimm434/URA3 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG | 7 |
| CEC305 | can1Δ::HIS1/can1Δ::ARG4 ura3Δ::λimm434/URA3 arg4Δ::hisG/arg4Δ::hisG his1Δ::hisG/his1Δ::hisG | 7 |
C. albicans strain construction.
The C. albicans RTA2 deletion strain is derived from strain BWP17U (Table 1). The lithium acetate procedure was used to transform C. albicans as described previously (65). The replacement of the entire open reading frames (ORFs) of both alleles of RTA2 was performed through sequential transformation using PCR-generated ARG4 and HIS1 disruption cassettes flanked by 100 bp of target homology region as described by Gola et al. (21). Primers RTA2S1 and RTA2S2 (Table 2) were used for generating disruption cassettes using the pFA-HIS and pFA-ARG plasmids as templates (21). The occurrence of the appropriate gene replacements in transformants was verified by PCR using primers V5-RTA2 and V3-RTA2 (Table 2).
Table 2.
Oligonucleotides used in this study
| Name | Sequence (5′–3′) |
|---|---|
| RTA2S1 | TGACTTTTTTCTAGCCAACCTAGAGTATTAGATTTGTTTCTCCATCATCAACAACTAAGAAACTTTTCTTGGTGTACCAGTTCATTCCCACCCTTCAACTGAAGCTTCGTACGCTGCAGGTC |
| RTA2S2 | GACAAATATCGAAATTGGTCCTAATATTAACTCTATCAGAATAAAAACTGTTTATTGCATTTACTATCTATCTAGAACAATACTTAATCTAATGCATAACTCTGATATCATCGATGAATTCGAG |
| V5-RTA2 | TCAGGATCACCACAAACACAG |
| V3-RTA2 | TGGTTGCTTTTGCTGGATCT |
| RTA2overF | GGGACAAGTTTGTACAAAAAAGCAGGCTTGATGAGTGAAATCTTGAATTATTTATCTTCG |
| RTA2overR | GGGACCACTTTGTACAAGAAAGCTGGGTTAACTTACTATTTAAACAATGTATGTCATT |
| CEF3_F | GATCACAATTGGGTCCAAGG |
| CEF3_R | AGCAGCGGCAATCTTGTTAC |
| FRE7_F | TCGTTACGCCGATACTTGTG |
| FRE7_R | AATCTCCGTTTGCACCCTTC |
| MET15_F | GGAGATTGCCTCAAACTTGG |
| MET15_R | TGGTAACACCAGAAGCCAAC |
| LYS4_F | CGAAAACCAAACCAGGTGAC |
| LYS4_R | TGTGGCTGCTTGTTCTCTTG |
| GNP1_F | CGACTGATGCATTGGGTTAC |
| GNP1_R | AGCATCTGGTTTGGAAGCAC |
| GAP1_F | ATGGCACCTAAATGGACAGG |
| GAP1_R | TGAAAGCAACCACCTGTTTC |
| CAN1_F | GCCTTTACTGCTGCATTTGG |
| CAN1_R | TGAGATTAACCCTGCGGTTG |
| RTA2_F | GGGGTGGATATTTGTTCACG |
| RTA2_R | ACCCAAACTGGATGGAATGG |
| PTR2_F | GGGTTCATGTTTGGATCTGG |
| PTR2_R | AACATGGACCGGCTTTGTAG |
| orf19.1799_F | TATCTGTGGGGTCATCATCG |
| orf19.1799_R | AGCACGATCAACGTATCCAC |
The C. albicans RTA2 overexpression strain was constructed according to the procedure of Cabral et al. (10). The RTA2 ORF was amplified using oligonucleotides RTA2overF and RTA2overR (Table 2). The PCR product was cloned in plasmid pDONR207 and transferred by Gateway-mediated recombination into plasmid CIp10::PCK1p-GTW-TAPtag (10), a derivative of the C. albicans CIp10 integrative plasmid (46), thus placing the RTA2 ORF under the Casamino Acid-inducible promoter of the C. albicans PCK1 gene (37) and in frame with a sequence encoding a TAP tag (58). The resulting RTA2 overexpression plasmid was linearized with StuI to promote targeting at the C. albicans RPS1 locus and transformed into C. albicans strain CEC161 (16), yielding strain CEC1088 (Table 1).
Peptides and antifungals.
ApoEdpL-W (WRKWRKRWWWRKWRKRWW), fluorescein-labeled ApoEdpL-W (ApoEdpL-W-Fluo), Histatin-5 (DSHAKRHHGYKRKFHEKHHSHRGY), and Magainin 2 (GIGKFLHSAKKFGKAFVGEIMNS) used in this study were manufactured by Alta Bioscience (Birmingham, United Kingdom). According to the supplier's data, all peptides had a purity ranging from 95 to 99% and molecular masses validated by mass spectrometry. Peptides were solubilized in 5% dimethylsulfoxide (DMSO) at a final concentration of 1 mM.
Evaluation of ApoEdpL-W antifungal activity on planktonic cells.
The MICs at which 90% of the isolates tested were inhibited (MIC90s) were determined in microtiter plates according to the EUCAST method (17) with an inoculum of 1 × 105 cells/ml, and plates were read 24 h after exposure. MICs were evaluated in several laboratory media. For MICs determined in PPB (10 mM potassium phosphate buffer, pH 7), 1 × 105 cells/ml were washed in 10 mM PPB, exposed for 1 h to peptides, diluted 200 times in phosphate-buffered saline (PBS), and plated on YPD plates for CFU counting (28). When applied, P values for MIC differences were calculated using Wilcoxon's test using the R program.
To test the fungicidal capacity of ApoEdpL-W, an overnight culture of C. albicans strain SC5314 was diluted to an optical density (OD) of 0.05 and grown to an OD of 0.4 in SD 2% glucose medium, and the culture was treated with increasing concentrations of the ApoEdpL-W peptide or an equal volume of solvent. Cells were sampled at various times, diluted 1 × 105 times, and plated on YPD plates for CFU counting.
The potential of NaN3 to protect C. albicans against the antifungal activity of ApoEdpL-W was tested as follows. An overnight culture of C. albicans strain SC5314 was diluted to an OD of 0.05, grown to an OD of 0.4 in SD 2% glucose medium, washed in 10 mM PPB, and exposed to 10 mM NaN3 for 1 h before being exposed to ApoEdpL-W for an additional hour. For testing the salt sensitivity of the ApoEdpL-W antifungal activity, cells of C. albicans strain SC5314 in 10 mM PPB as described above were exposed to various concentrations of the ApoEdpL-W peptide and to 0, 10, and 100 mM NaCl for 1 h. These tests were performed by adding the ApoEdpL-W peptide and then salt, or vice versa. To evaluate whether beta-glucans had a protective effect, various amounts of the ApoEdpL-W peptide were incubated with 5 mg · ml−1 laminarin (Sigma) for 1 h in 10 mM PPB. C. albicans SC5314 cells obtained as described above and resuspended in 10 mM PPB were added to the peptide-glucan mix for 1 h. For each test, cells were diluted 1 × 105-fold and plated on YPD plates for CFU counting. Each test was performed in duplicate using independent cultures of C. albicans. Growth inhibition kinetics by latrunculin was performed by coincubating cells with 10 to 50 μM latrunculin A (Sigma) and ApoEdpL-W at various concentrations. Growth was performed in 96-well plates at 30°C with agitation, and ODs at 600 nm were recorded every 20 min using a microtiter plate reader (Tecan) for 18 h.
Checkerboard assays were performed by serial dilutions of the two antimicrobial agents in a microtiter plate using ApoEdpL-W in conjunction with fluconazole (Sigma), amphotericin B (Sigma), and caspofungin (Merck). The fractional inhibitory concentration (FIC) index and synergy or antagonist effects were calculated as described in reference 43.
Evaluation of ApoEdpL-W antifungal activity on biofilm cells.
C. albicans biofilms were developed in 96-well plates as described previously (59). ApoEdpL-W was added at different times after C. albicans cells had been adhered to the surface, and the metabolic activity of treated and untreated biofilms was quantified using the FDA method 24 h after peptide addition (59). Biofilms on squares of materials were obtained as follows. One-centimeter squares of polyurethane (PU) and polydimethylsiloxane (PDMS), two materials used for medical devices, were soaked overnight in a 200 μM solution of ApoEdpL-W for coating. Coated materials were washed and submerged in a culture of C. albicans strain SC5314 diluted to an OD of 1 for 1 h to promote C. albicans adherence. Materials were washed to remove nonadherent cells, placed in 12-well culture plates with fresh SD medium, and incubated at 37°C with gentle agitation for 72 h. Biofilm formation was evaluated by direct observation after 24 and 72 h. Materials then were washed to remove nonattached cells, and biofilm quantification was performed using the FDA method (59). Experiments were performed in duplicate separately.
Microarray experiments.
Total RNA was isolated using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. The concentration, purity, and integrity of the isolated RNA were evaluated using a Nanodrop spectrophotometer and an Agilent 2100 Bioanalyzer. Ten μg of total RNA was used for cDNA synthesis using the Superscript indirect cDNA labeling kit (Invitrogen) according to the manufacturer's instructions. cDNA samples were labeled with Cy3 or Cy5 dye (Amersham Biosciences). Purified fluorescent cDNAs were hybridized to C. albicans oligonucleotide microarrays (Eurogentec) according to the manufacturer's instructions. Arrays were scanned with an Axon 4000A scanner (Molecular Devices), and data were acquired and analyzed using Genepix Pro 5.0 (Molecular Devices). For each comparison, two biological replicates were used, and each biological replicate was subjected to technical replicates with dye swaps. Data normalization (Lowess) and statistical analysis (Student t test) were performed using Genespring GX 7.3 (Agilent Technologies). Genes regulated by at least a 1.5-fold with a P value of <0.05 were considered significant. Microarray data have been deposited at ArrayExpress under accession number E-MEXP-3084. Normalized data are available in Table S2 in the supplemental material. Gene ontology analyses were performed using tools available at the Candida Genome Database (2). Pearson correlation coefficients between ApoEdpL-W and myo5Δ microarrays experiments were calculated using R software based on ratios of all genes for which a ratio was available for each experiment.
qRT-PCR experiments.
Total RNA was isolated using an RNeasy minikit (Qiagen) according to the manufacturer's instructions and treated with Turbo DNase kit (Ambion, Applied Biosystems) to ensure the absence of genomic DNA contamination. The concentration, purity, and integrity of the isolated RNA were evaluated using a Nanodrop spectrophotometer and an Agilent 2100 Bioanalyzer. cDNA was synthesized from 2 μg of purified RNA using the SuperScript II reverse transcriptase kit (Invitrogen). Quantitative reverse transcription-PCR (qRT-PCR) was carried using a Mastercycler EP Realplex real-time PCR system (Eppendorf) with SYBR green fluorescence probes (Applied Biosystems). Cycling was for 2 min at 50°C and then 10 min at 95°C, followed by 40 cycles (95°C for 15 s, 60°C for 1 min, and 95°C for 15 s), and finally 15 s at 95°C, 20 s at 60°C, and 15 s at 95°C. For each condition, two to three biological replicates were used. Relative expression levels were calculated using the ΔΔCT method, using CEF3 transcripts levels for normalization. All oligonucleotides used for qRT-PCR are listed in Table 2.
Fluorescence microscopy.
Fluorescence microscopy was performed using a Nikon Eclipse E 600 microscope with 4′,6′-diamidino-2-phenylindole (DAPI), Texas Red, and a fluorescein isothiocyanate (FITC) filter or a Leica DRMXA with DsRed and an FITC filter. Images were taken using 40× or 100× objectives. C. albicans strain SC5314 was exposed to ApoEdpL-W-Fluo in SD 2% glucose (pH 7) or RPMI medium at different concentrations for various durations. For nucleus staining, cells were observed in Vectashield mounting medium containing DAPI at 1.5 μg/ml (Vector Laboratories). Vacuole staining was performed using the FM4-64 dye (Molecular Probes) with a pulse-chase method. One μl of 1 μg/ml of FM4-64 was added to 50 μl of cells at an OD of 5 for 30 min at 30°C with agitation for the pulse. One ml of medium then was added, and cells were centrifuged, resuspended in 5 ml of medium, and incubated at 30°C for 90 min under agitation for the chase. After 90 min of chase, ApoEdpL-W-Fluo was added and cells were observed by fluorescence microscopy after an extra 30 min of incubation. For evaluating cell permeability, cells were exposed to ApoEdpL-W-Fluo in the presence of propidium iodide (PI) at 10 μg/ml (Sigma) and visualized after 1 h at 30°C. The percentage of cells with different peptide accumulation profiles and colabeled with PI were calculated on at least 100 cells. The inhibition of endocytosis was performed by preincubating cells with 10 to 50 μM latrunculin A (Sigma) for 1 h prior to exposure to ApoEdpL-W-Fluo or by coincubation.
Microarray accession number.
Microarray data have been deposited at ArrayExpress under accession number E-MEXP-3084.
RESULTS
Antifungal activity of ApoEdpL-W on planktonic Candida.
Previous reports have shown that ApoEdpL-W has a strong antiviral and antibacterial activity (35). Here, we investigated the antifungal activity of ApoEdpL-W toward the C. albicans standard laboratory strain SC5314 by determining the MIC using the broth dilution method in RPMI medium and SD 2% glucose medium, with or without buffering to pH 7. Results summarized in Table 3 showed MIC90s ranging from 3.75 to 7.5 μM in these media, suggesting that medium and pH had little influence on the antifungal activity of ApoEdpL-W. These MICs were compared to those of other CAFPs, namely, Hst-5 and Magainin 2. Because of their salt sensitivity, Hst-5 and Magainin 2 are inefficient in usual laboratory media, such as SD 2% glucose or RPMI (27, 63, 66). Therefore, MICs were determined through the incubation of C. albicans SC5314 cells in 10 mM PPB with increasing concentrations of peptides followed by CFU counting. Results presented in Table 3 showed that, under these conditions, the MIC90 of ApoEdpL-W was similar to those in growth media and was four to eight times lower than MIC90s obtained for Hst-5 and Magainin 2. For these peptides, MIC90s of 10 to 20 μM were obtained, similarly to what has been reported by others (24, 29, 56). In addition, ApoEdpL-W was fungicidal to C. albicans cells. Indeed, killing curves showed that, at the MIC90, killing was very rapid, with more than 95% of the C. albicans cells being killed after 5 min of incubation and no viable cells being detected after 30 min of exposure (Fig. 1). In contrast, when the concentration was reduced 2-fold, ApoEdpL-W remained fungistatic (Fig. 1).
Table 3.
MIC90 of various antifungal peptides on C. albicans strain SC5314
| Peptide | MIC90b (μM) |
|||
|---|---|---|---|---|
| ApoEdpL-W | ApoEdpL-W-Fluo | Histatin 5 | Magainin 2 | |
| SD 2% glucose | 3.75 | ND | ND | ND |
| SD 2% glucose (pH 7) | 5 | 7.5 | ND | ND |
| RPMI 2% glucose (pH 7) | 7.5 | 5 | ND | ND |
| 10 mM PPBa | 2.5 | ND | 10–20 | 10 |
MICs were determined by CFU counting.
ND, not done.
Fig. 1.
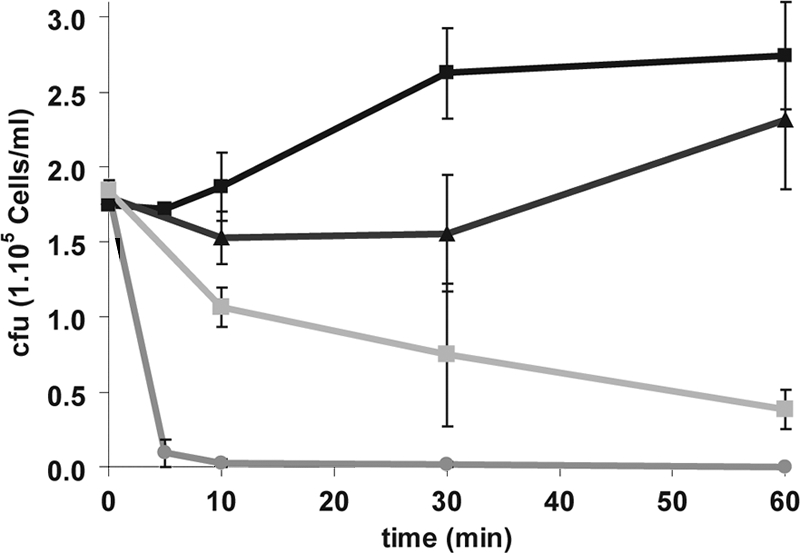
ApoEdpL-W peptide is fungicidal to C. albicans at concentrations above the MIC90. C. albicans strain SC5314 cells were exposed to solvent (black squares), 2.5 μM ApoEdpL-W (black triangles), 5 μM ApoEdpL-W (gray circles), or 20 μM ApoEdpL-W-Fluo (gray squares) for the indicated times, and the number of cells remaining viable in each sample was determined through plating on YPD medium. Data are average values obtained from two independent experiments, with bars indicating the range of values.
ApoEdpL-W also was tested for its efficacy toward a panel of 91 clinical strains of various pathogenic Candida species (C. albicans, Candida parapsilosis, Candida tropicalis, and Candida glabrata) with a wide range of resistance/sensitivity to antifungals in clinical use (see Table S1 in the supplemental material) (8). Results presented in Table 4 showed that ApoEdpL-W was effective against a majority of the tested strains. However, ApoEdpL-W was much less effective against C. glabrata, with most of the MICs against these isolates being more than 20 μM (Table 4), whereas C. tropicalis isolates appeared to be exquisitely sensitive to ApoEdpL-W. Overall, a chi-square test confirmed a significant difference in MIC distributions between species (P < 2.2 × 10−16). However, no correlation with the MICs of the antifungals in clinical use already determined for these strains (8) was observed. Taken together, these data showed that ApoEdpL-W was fungicidal against C. albicans and other pathogenic yeasts, with the exception of C. glabrata, and was more potent than other antifungal peptides, such as Hst-5 and Magainin 2.
Table 4.
Distribution of ApoEdpL-W MIC90s on a panel of clinical Candida strains
| Species | Strain no. | No. of strains in each ApoEdpL-W MIC90 distribution groupa |
||||
|---|---|---|---|---|---|---|
| >20 μM | 20 μM | 10 μM | 5 μM | 2.5 μM | ||
| C. albicans | 42 | 2 | 9 | 30 | 1 | 0 |
| C. glabrata | 28 | 24 | 3 | 1 | 0 | 0 |
| C. tropicalis | 11 | 0 | 0 | 0 | 8 | 3 |
| C. parapsilosis | 10 | 2 | 6 | 1 | 1 | 0 |
MICs were determined twice per strain.
Antifungal activity of ApoEdpL-W on Candida albicans biofilms.
Biofilm formation by Candida species represents a major threat in the clinical setting. Thus, the antifungal activity of ApoEdpL-W was tested on C. albicans preformed biofilms. Biofilms developed for 1, 5, or 18 h in microtiter plates in the absence of antifungal peptide were exposed to increasing levels of ApoEdpL-W for an additional 24 h. When applied on adhesion-stage biofilms for 1 h, ApoEdpL-W was able to completely inhibit biofilm growth at concentrations equal to or above the MIC90 determined for planktonic cells (data not shown). On early-stage biofilms after 5 h, ApoEdpL-W was able to partially inhibit biofilm formation (Fig. 2A). The biofilm metabolic activity measured after 24 h of incubation with ApoEdpL-W was reduced to 60% of the control at the planktonic MIC90 and down to 40% by increasing the concentration to four times the planktonic MIC90. The exposure of mature biofilms after 18 h to ApoEdpL-W resulted in a slight reduction of biofilm metabolic activity, corresponding to 80% of the control when four times the planktonic MIC90 was used (data not shown).
Fig. 2.
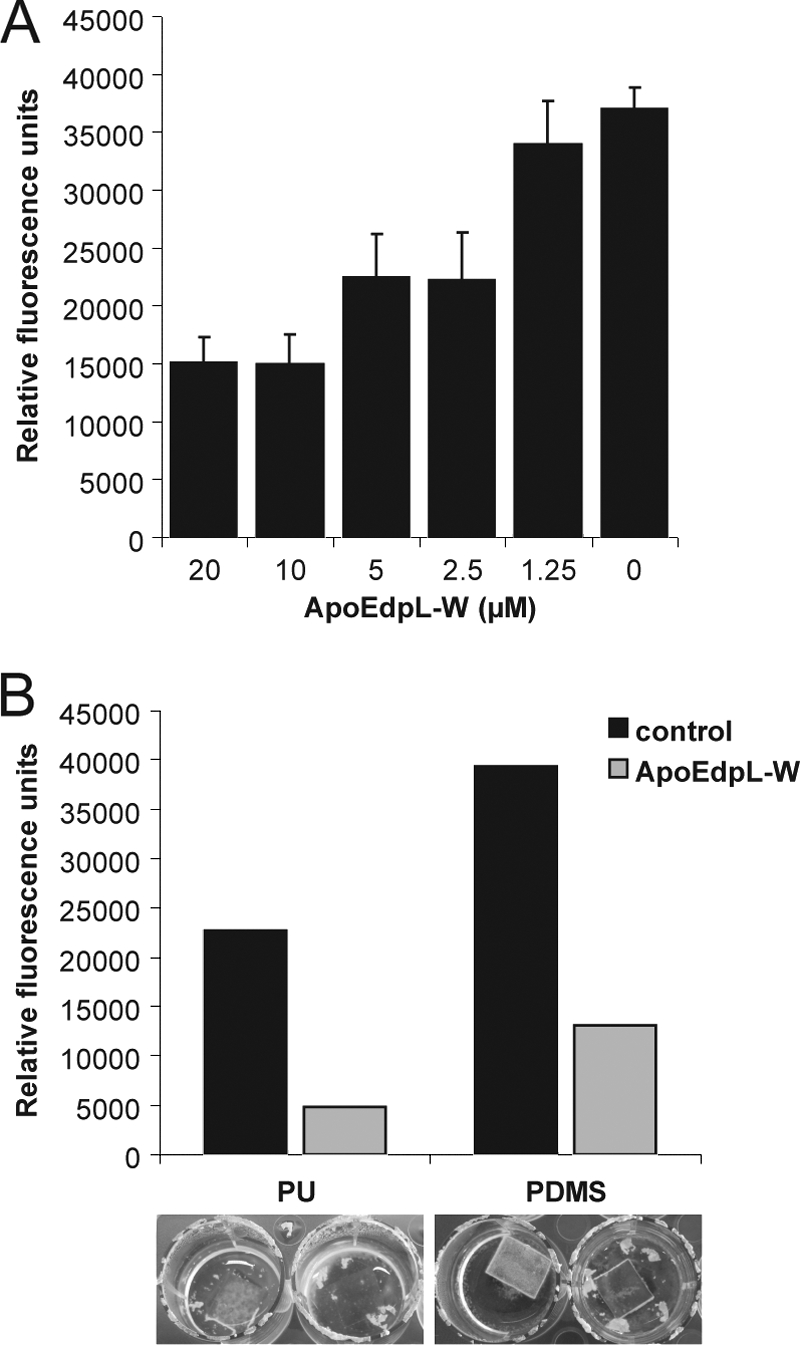
ApoEdpL-W peptide prevents biofilm formation. (A) Biofilms of C. albicans strain SC5314 were developed in 96-well microtiter plates (1 h of adherence followed by 4 h of growth), exposed to increasing concentrations of ApoEdpL-W, and incubated for 24 h. The metabolic activities of biofilms were quantified using the FDA method and are shown. Data presented are from three to five replicates. Means and standard deviations are displayed. (B) Biofilms of C. albicans strain SC5314 were developed on ApoEdpL-W-coated or noncoated polyurethane and polydimethylsilloxane squares for 72 h and quantified using the FDA method. Biofilms were washed prior to quantification. Photographs show examples of the biofilms prior to washing.
ApoE-derived antimicrobial peptides could be a promising antimicrobial coating agent for medical devices such as catheters. Thus, the antifungal activity of ApoEdpL-W on biofilms formed on two polymer materials used in medical devices, namely, polyurethane (PU) and polydimethylsiloxane (PDMS), also was tested. These materials were coated with ApoEdpL-W as described in Materials and Methods and used as substrates for C. albicans biofilm formation. After 24 h of incubation, macroscopic observation revealed that C. albicans strain SC5314 was able to form a biofilm on the coated materials, but the biofilms had a strong tendency to detach (data not shown). After 72 h of incubation, an increased tendency of the biofilms to detach from the coated materials was observed (Fig. 2B, lower panel). This was apparent in the 3- to 5-fold reduction of the metabolic activity of biofilms formed on the ApoEdpL-W-coated materials relative to uncoated materials (Fig. 2B). Therefore, the coating of PU and PDMS with ApoEdpL-W appeared to promote the formation of loose C. albicans biofilms rather than fully prevent biofilm formation. Similar experiments were performed with an FITC-labeled derivative of ApoEdpL-W (ApoEdpL-W-Fluo) without cells to evaluate the possible release of the fluorescent peptide from the materials. Fluorescent quantification revealed a stable fluorescence of the materials after up to 48 h, even if the release of the peptide in the medium was observed (data not shown). This suggested that ApoEdpL-W remained attached mainly on the materials during the course of incubation but did not rule out a possible role of released peptides in the observed biofilm detachment.
Usual antagonists of antifungal peptide activity do not affect ApoEdpL-W efficiency.
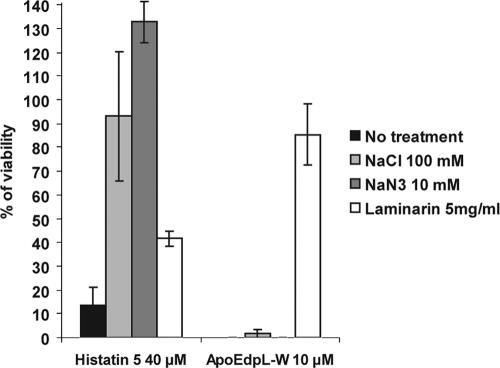
The antifungal activity of Hst-5 is susceptible to a variety of treatments that interfere with its binding to the cell wall, cytoplasmic translocation, or eventual killing of C. albicans cells through ionic imbalance (14, 30, 36, 60, 66). These treatments also have been shown to interfere with the activity of other CAFPs. Therefore, we tested whether they influenced ApoEdpL-W antifungal activity.
The binding of Hst-5 to both cell wall beta-glucans and the cell wall-bound Ssa2 heat shock protein has been shown to contribute to its activity, as C. albicans SSA2 knockout mutants and caspofungin-treated C. albicans cells show decreased sensitivity to this CAFP (30, 60). ApoEdpL-W MICs against a C. albicans SSA2 knockout mutant (ssa2Δ) (Table 1) and a complemented strain (ssa2Δ/SSA2) (Table 1) were determined. They were similar to those obtained for wild-type C. albicans strains (data not shown), suggesting that Ssa2 did not contribute significantly to C. albicans susceptibility to ApoEdpL-W. In contrast, the preincubation of ApoEdpL-W with laminarin beta-(1,3)-glucan prior to incubation with C. albicans cells fully inhibited the antifungal activity of ApoEdpL-W (Fig. 3). Laminarin was more potent at inhibiting ApoEdpL-W activity than Hst-5 activity (Fig. 3), and the inhibition was dose dependent, with increased levels of laminarin required to inhibit increased levels of ApoEdpL-W (data not shown). Importantly, ApoEdpL-W showed some synergy with caspofungin, an inhibitor of cell wall beta-glucan synthesis (FIC of 0.465). This was in contrast to what has been observed for Hst-5, whose activity is impaired by caspofungin treatment (30), suggesting that beta-glucans contribute to trap ApoEdpL-W in the cell wall rather than provide a path for ApoEdpL-W entry into C. albicans cells.
Fig. 3.
ApoEdpL-W antifungal activity is insensitive to treatments that impair histatin-5 antifungal activity. C. albicans strain SC5314 was incubated with 10 μM ApoEdpL-W or 5 μM Histatin-5 (Hst-5) in the presence of 100 mM NaCL, 10 mM NaN3, or 5 mg/ml laminarin for 1 h, and cells were plated on YPD, except for cells treated with laminarin, where ApoEdpL-W was preincubated with laminarin for 1 h and the mixture was subsequently added to cells and incubated for 1 h. The ratios obtained for treated versus untreated cells are shown. Data are averages obtained from two independent experiments, with bars indicating the range of values. These data are representative of the protective effect observed in other experiments performed individually.
Results presented above suggested that ApoEdpL-W was less susceptible to physiochemical variations than other CAFPs, as it was active in a variety of media (Table 3). Therefore, the impact of known antagonists of CAFP activity on ApoEdpL-W activity was tested. Results presented in Fig. 3 showed that ApoEdpL-W antifungal activity was not prevented by 100 mM NaCl, while this was the case for Hst-5 activity (14, 66), suggesting that ionic imbalance was not a determining factor for ApoEdpL-W antifungal action on C. albicans cells. Similarly, NaN3, which has been proposed to protect against Hst-5 by triggering ATP depletion and altering membrane fluidity (36), did not have a protective effect against ApoEdpL-W (Fig. 3). Taken together, these data suggested that the mode of action of ApoEdpL-W differed from that of Hst-5 and possibly that of other CAFPs.
Intracellular localization of ApoEdpL-W.
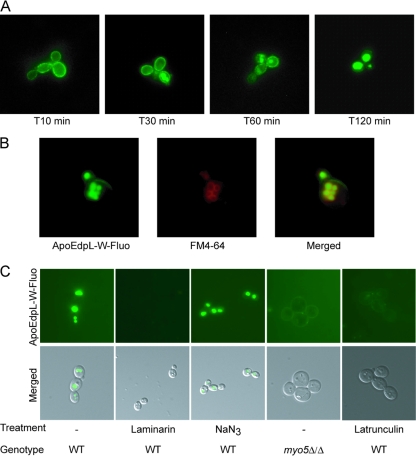
To investigate the mode of action of ApoEdpL-W, we assessed its localization in C. albicans cells using ApoEdpL-W-Fluo, which showed MIC90s similar to those of native ApoEdpL-W against C. albicans SC5314 (Table 3). C. albicans SC5314 cells were exposed to subinhibitory concentrations of ApoEdpL-W-Fluo (5 μM) for different durations and were observed by fluorescence microscopy. The fluorescent peptide was shown originally to bind to the cell surface, accumulate within intracellular organelles after 30 min for 33% of the cells, reaching 66 to 87% after 60 min, and be exclusively localized to these organelles after 2 h (Fig. 4A). Increasing concentrations of ApoEdpL-W-Fluo resulted in the faster accumulation of the peptide within these organelles but did not modify the overall intracellular distribution of the peptide. Only when the concentration was above 20 μM could we observe that 10 to 25% of the cells showed cytoplasmic labeling after 60 min of incubation (data not shown). Noticeably, a majority of the cells with cytoplasmic ApoEdpL-W-Fluo labeling showed intense propidium iodide staining, while cells with vacuolar ApoEdpL-W-Fluo staining did not show PI labeling (data not shown). Intracellular ApoEdpL-W-Fluo did not colocalize with the nucleus stained with DAPI (data not shown). In contrast, colocalization with FM4-64 was observed (Fig. 4B), indicating that the structures where ApoEdpL-W-Fluo accumulated were vacuoles. Therefore, ApoEdpL-W-Fluo appeared to localize to the vacuole following binding to the cell surface irrespective of the concentration used. As shown in Fig. 4C, laminarin was able to inhibit the vacuole accumulation of ApoEdpL-W-Fluo, whereas NaN3 was not, in agreement with their respective abilities to protect against ApoEdpL-W. Overall, these data indicated that ApoEdpL-W was targeted to the vacuoles independently of cytoplasmic membrane permeabilization, suggesting that the antifungal activity of ApoEdpL-W was not primarily by disrupting membrane homeostasis.
Fig. 4.
ApoEdpL-W is targeted to vacuoles through endocytosis. (A) Kinetic localization of ApoEdpL-W-Fluo in C. albicans SC5314 cells exposed to 5 μM ApoEdpL-W-Fluo. Images are representative of labeling distribution at each time point. (B) C. albicans SC5314 cells exposed to ApoEdpL-W-Fluo (40 μM) and stained with FM4-64. (C) C. albicans MYO5/MYO5 or myo5Δ/myo5Δ cells exposed to ApoEdpL-W-Fluo (5 or 20 μM) for 30 to 60 min. In the laminarin panel, ApoEdpL-W-Fluo was preincubated with 5 mg/ml laminarin for 1 h prior to being applied to cells. In the NaN3 panel, cells were incubated with 10 mM NaN3 for 1 h prior to being exposed to ApoEdpL-W-Fluo. In the latrunculin A panel, cells were incubated with 50 μM latrunculin A for 1 h prior to being exposed to ApoEdpL-W-Fluo.
Transcriptional analysis of C. albicans exposed to sub-MIC concentrations of ApoEdpL-W.
To get further insights into the antifungal mode of action of ApoEdpL-W, we performed a gene expression analysis of C. albicans SC5314 planktonic cells exposed to ApoEdpL-W. Exponentially growing C. albicans SC5314 cells in SD 2% glucose medium at 30°C were exposed to 2.5 μM ApoEdpL-W, and samples were collected after 10 and 30 min for transcript profiling. The concentration of ApoEdpL-W that was used was sublethal, i.e., it led to a reduction in the C. albicans growth rate but did not result in any massive cell killing (Fig. 1), thus minimizing changes in gene expression due to cell death and enabling us to focus our gene profiling study on cellular responses elicited by ApoEdpL-W.
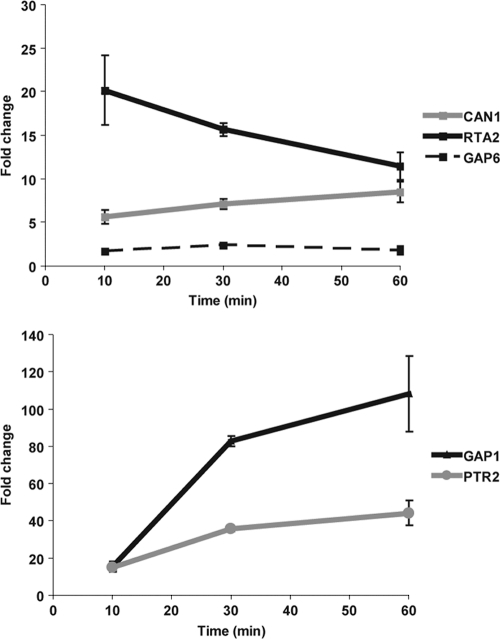
A comparison of the transcriptomes of ApoEdpL-W-treated an untreated cells identified 63 genes upregulated and 47 genes downregulated after 10 min of exposure to ApoEdpL-W (see Table S2 in the supplemental material). After 30 min of exposure, 48 genes were upregulated and 26 were downregulated in response to ApoEdpL-W (see Table S2). Fifteen genes were upregulated and five were downregulated at both time points and are listed in Table 5. Gene ontology analysis of the differentially regulated genes indicated a statistically significant overrepresentation of genes encoding amino acid transporters. These genes represented one-third of the genes upregulated at both time points (GAP6, CAN1, GNP1, ORF19.4940, and GAP1) and were among the genes that showed the highest upregulation in response to ApoEdpL-W (Table 5). PTR2, a putative oligopeptide transporter, also was strongly induced at both time points, as well as RTA2, which encodes a putative long-chain base (LCBs) floppase translocating LCBs from the inner to the outer leaflet of the plasma membrane, and it may have a role in membrane integrity (33). Noticeably, 27 of the 63 genes upregulated after 10 min of exposure to ApoEdpL-W, 26 of the 48 genes upregulated after 30 min of exposure, and 9 of the 15 genes upregulated at both time points were predicted to encode a protein with at least one membrane-spanning domain. The upregulation of CAN1, GAP1, GAP6, PTR2, and RTA2 was confirmed by qRT-PCR after 10, 30, and 60 min of exposure to 2.5 μM ApoEdpL-W (Fig. 5). These data suggested that sublethal concentrations of ApoEdpL-W exerted a major effect on the expression of genes encoding membrane proteins. Interestingly, none of the published studies that have analyzed the transcriptional response of C. albicans to cationic peptides have observed an upregulation of genes encoding membrane proteins, again suggesting that ApoEdpL-W has a specific mode of action.
Table 5.
Genes identified as regulated at both 10 and 30 min in response to ApoEdpL-Wd
| Gene name and regulation status | Name | Regulation at: |
myo5Δ/Δ expression ratioc | Description | |
|---|---|---|---|---|---|
| 10 mina | 30 minb | ||||
| Upregulated | |||||
| orf19.675 | 11.970 | 2.745 | 23.279 | Similar to cell wall proteins | |
| orf19.2959.1 | 6.862 | 3.214 | 10.169 | ORF predicted by annotation working group | |
| orf19.6659 | GAP6 | 6.470 | 28.310 | 0.909 | Putative general amino acid permease |
| orf19.4082 | DDR48 | 5.568 | 1.897 | 31.251 | Immunogenic stress-associated protein |
| orf19.6937 | PTR2 | 5.199 | 12.830 | 0.729 | Putative oligopeptide transporter |
| orf19.24 | RTA2 | 4.543 | 2.230 | 17.373 | Putative floppase |
| orf19.97 | CAN1 | 3.209 | 4.484 | 0.980 | Amino acid permease, transports basic amino acids |
| orf19.6877 | 3.107 | 2.181 | 4.479 | Predicted ORF in assemblies 19, 20, and 21 | |
| orf19.1193 | GNP1 | 2.762 | 9.543 | 1.242 | Protein described as similar to asparagine and glutamine permease |
| orf19.2125 | 2.534 | 2.146 | 13.254 | Predicted ORF in assemblies 19, 20, and 21 | |
| orf19.4940 | 2.188 | 5.554 | 0.964 | Putative histidine permease | |
| orf19.4304 | GAP1 | 2.084 | 5.618 | 0.796 | General amino acid permease |
| orf19.3548.1 | WH11 | 2.020 | 1.886 | 17.890 | Cytoplasmic protein expressed specifically in white-phase yeast-form cells |
| orf19.986 | GLY1 | 1.686 | 2.365 | 1.129 | L-Threonine aldolase |
| orf19.1353 | 1.659 | 3.013 | 0.806 | Predicted ORF in assemblies 19, 20, and 21 | |
| Downregulated | |||||
| orf19.6844 | ICL1 | 0.663 | 0.442 | 1.766 | Isocitrate lyase |
| orf19.541 | 0.536 | 0.591 | 0.814 | Predicted ORF in assemblies 19, 20, and 21 | |
| orf19.2693 | 0.503 | 0.622 | 1.572 | Increased transcription is observed upon benomyl treatment | |
| orf19.4063 | GPT1 | 0.365 | 0.458 | 1.134 | GABA/polyamine transporter |
| orf19.1608 | 0.300 | 0.304 | 1.033 | Predicted ORF in assemblies 19, 20, and 21 | |
Fold expression change for cells exposed to subinhibitory concentrations of ApoEdpL-W after 10 min versus unexposed cells.
Fold expression change for cells exposed to subinhibitory concentrations of ApoEdpL-W after 30 min versus unexposed cells.
Fold expression change of the myo5Δ/Δ mutant to the wild type.
Only genes regulated by a factor of 1.5 with a P value of <0.05 have been considered.
Fig. 5.
C. albicans amino acid transporter genes are upregulated upon exposure to ApoEdpL-W. Shown are qRT-PCR results for genes identified as upregulated by ApoEdpL-W by microarray experiments, exposed to 2.5 μM ApoEdpL-W, and sampled at 10, 30, and 60 min of exposure. Fold changes were determined by the ΔΔCT method, using CEF3 transcript levels for normalization.
The upregulation of genes encoding amino acid transporters upon exposure to ApoEdpL-W could reflect peptide degradation and assimilation by C. albicans, as the concentration that was used in transcript profiling experiments was sublethal. Indeed, it has already been shown that C. albicans is able to degrade CAFPs such as Hst-5 through the Sap9-secreted aspartyl protease (41). However, protease- or peptidase-encoding genes as well as genes related to nitrogen catabolite repression and amino acid biosynthesis, with the exception of methionine biosynthesis genes, did not show any differential gene expression in response to ApoEdpL-W. The transcription of some amino acid transporter genes increased when arginine but not tryptophan was added to the medium to a concentration equivalent to what is present in ApoEdpL-W (data not shown). However, the inability of the C. albicans arginine-auxotrophic strain CEC361 to grow in the absence of arginine and the presence of sublethal levels of ApoEdpL-W indicated that the degradation of the peptide was unlikely to account for the upregulation of amino acid transporters (data not shown).
The CAN1, PTR2, and RTA2 genes do not contribute to tolerance or resistance to ApoEdpL-W.
As mentioned above, several of the amino acid transporter-encoding genes, namely, CAN1, PTR2, GAP1, and GAP6, were upregulated in response to ApoEdpL-W or arginine. In contrast, RTA2 appeared induced upon ApoEdpL-W exposure and, to a lower extent, Hst-5, but not in response to arginine (data not shown). Therefore, we tested whether RTA2 could contribute to modulate the susceptibility of C. albicans to ApoEdpL-W. Strains with a deletion of the two RTA2 alleles or overexpressing RTA2 were constructed and tested for their susceptibility to ApoEdpL-W (strains CEC1545 and CEC1088, respectively). ApoEdpL-W MICs for these strains were similar to those obtained for wild-type C. albicans strains, suggesting that RTA2 did not contribute to C. albicans susceptibility to ApoEdpL-W. In addition, strains deleted for both alleles of CAN1 or PTR2 (CEC290 and CEC305, respectively) (7) did not show any increased resistance or sensitivity to ApoEdpL-W, indicating that neither CAN1 nor PTR2 contributed to C. albicans susceptibility to ApoEdpL-W on their own (data not shown).
Endocytosis of ApoEdpL-W contributes to its antifungal activity.
Localization data presented above suggested that ApoEdpL-W was endocytosed and targeted to the vacuole. Interestingly, half of the most upregulated genes upon the exposure of C. albicans cells to ApoEdpL-W showed a strong upregulation upon the inactivation of the MYO5 gene (Table 5) that encodes a class I myosin necessary for efficient endocytosis in C. albicans (49). Moreover, the overall microarray data at 10 min showed a positive correlation with myo5 mutant transcriptome experiments (Pearson correlation coefficient, 0.33; P < 2.2 × 10−16) (51), which was particularly obvious for the most upregulated genes. However, genes encoding amino acid or oligopeptide transporters were not regulated in the myo5 mutant. Therefore, we tested the susceptibility to ApoEdpL-W of a C. albicans strain defective for the MYO5 gene (COU46) (Table 1). The MIC90 of ApoEdpL-W against this strain was 2-fold increased (P = 0.046) relative to that against strain COU73, which harbors a functional MYO5 gene (Table 1). Moreover, ApoEdpL-W-Fluo persisted at the cell surface and did not accumulate in vacuoles of the COU46 strain (Fig. 4C). This was in contrast to what was observed with COU73 and SC5314 strains that accumulated ApoEdpL-W-Fluo in the vacuole following binding to the cell surface. However, a limited number of cells of the COU46 strain showed the accumulation of ApoEdpL-W-Fluo in the vacuole (data not shown). However, the fluorescence intensity associated with the vacuole in these cells was lower than that in the MYO5-complemented COU73 cells or in the wild-type SC5314 cells. These results suggested that Myo5-dependent processes are necessary for vacuolar targeting and the efficient antifungal activity of ApoEdpL-W.
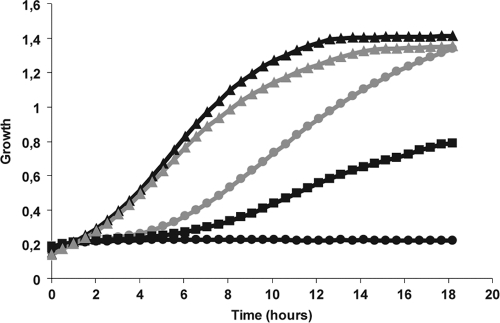
Latrunculin A is an inhibitor of actin polymerization and is known to inhibit endocytosis in yeasts. As shown in Fig. 6, latrunculin A protected C. albicans SC5314 cells against the antifungal activity of ApoEdpL-W. Even though the doubling time of C. albicans incubated in the presence of 5 μM ApoEdpL-W and 50 μM latrunculin A was higher than that of cells grown in the absence of ApoEdpL-W, the final cell densities of the two cultures were identical. Moreover, a dose-dependent protection of ApoEdpL-W by latrunculin A was observed (Fig. 6). Consistently with this observation, when cells were incubated with 10 or 50 μM latrunculin A for 1 h and subsequently exposed to ApoEdpL-W, the ApoEdpL-W MIC90 increased by 2-fold. Similarly to what was observed upon the genetic alteration of endocytosis, ApoEdpL-W-Fluo did not accumulate in vacuoles of latrunculin A-treated cells, and only a faint staining of the cell surface was observed (Fig. 4C). Thus, blocking endocytosis through latrunculin A treatment prevented the accumulation of ApoEdpL-W-Fluo in vacuoles and provided partial protection against the antifungal effect of ApoEdpL-W.
Fig. 6.
Latrunculin A prevents the antifungal activity of ApoEdpL-W. Shown are the growth kinetics in 96-well microtiter plates of C. albicans strain SC5314 exposed to 5 μM ApoEdpL-W plus 0 μM (black circles), 10 μM (black squares), or 50 μM (gray circles) latrunculin A. Results for cells exposed to 50 μM latrunculin A only (gray triangles) and for untreated cells (black triangles) are shown as well. For clarity, only one-third of the values are represented on the graph. Preincubation for 1 h with latrunculin A before exposure to ApoEdpL-W gave similar profiles.
DISCUSSION
Although antimicrobial peptides have long been thought to exert their antimicrobial activity through insertion into biological membranes and their subsequent permeabilization, increasing amounts of evidence suggest that they act through other mechanisms targeting intracellular components (9). In this regard, several antifungal peptides have been shown to associate primarily with mitochondria (26), vacuoles (30, 42), or the nucleus (45), suggesting that the alteration of membrane homeostasis could be secondary to the action of these peptides on intracellular targets. However, the precise modes of action of the different antifungal peptides remain to be elucidated. In this study, we have investigated a novel antifungal peptide derived from human apoliprotein E. Strikingly, the sensitivity or resistance of ApoEdpL-W to a variety of treatments suggests that its mode of action is significantly different from that of other cationic antifungal peptides (CAFPs) that have been investigated in detail, such as Hst-5, Magainin 2, or MUC7-20. Indeed, we have shown that ApoEdpL-W activity does not require binding to the cell wall heat shock protein Ssa2 and to beta-glucans, and it is insensitive to azide, high salt, and changes in pH. Furthermore, we have shown that ApoEdpL-W accumulates primarily in vacuoles, and that vacuolar targeting contributes to ApoEdpL-W activity. This is in marked contrast to observations made with Hst-5, which requires binding to glucans or Ssa2 (15, 30, 38, 60) and shows azide, salt, and pH sensitivity (3, 36, 62). Finally, Hst5 targeting to the vacuole is observed at low concentrations only and is not required for activity (30, 42). These requirements for Hst-5 activity are all thought to reflect its mode of action at the plasma membrane. Therefore, the differences in behavior that we have observed between Hst-5 and ApoEdpL-W suggest in particular that ApoEdpL-W uses other routes than Hst-5 to reach the plasma membrane, to penetrate cells, and to exert its antifungal activity. This also appears to be the case for other antifungal peptides. While Magainin 2 and Muc7-20 also show salt sensitivity, other antifungal peptides, such as arenicin, proved to be salt insensitive (54). Differences also are observed regarding azide sensitivity, with arenicin being sensitive (54) and Muc7-20 being insensitive, even though it is translocated intracellularly (6). Finally, human β-defensins (hBD) show fungicidal activity in an energy-dependent manner and are salt sensitive but do not cause membrane disruption (64). Moreover, an additional energy-independent antifungal mechanism has been revealed at higher concentrations of hBD-3 (64). Taken together, these data underscore the heterogeneity of behaviors of antifungal peptides and show that ApoEdpL-W is not an exception. This might reflect differences in the amino acid content, the positive charge, and the hydropathy of the different CAFPs at physiological pH, even though rules still need to emerge. Noticeably, ApoEdpL-W is composed mainly of tryptophan, arginine, and lysine residues and is likely to have a higher net positive charge than Hst-5 or Magainin 2.
Another major difference between Hst-5 and ApoEdpL-W lies in their apparent targeting to different cellular compartments, which is exemplified through the use of fluorescently labeled derivatives. While Hst-5 was shown to accumulate in the vacuole at low, nonlethal concentrations, it is targeted to the cytoplasm at higher concentrations (42). This contrasts with ApoEdpL-W, which we have shown to associate with the cell surface and subsequently be targeted to the vacuole at all concentrations tested, even though we cannot formally exclude that the observed fluorescence is not due in part to free fluorescent label released from degraded ApoEdpL-W. Moreover, our data using a C. albicans strain defective for the MYO5 gene and consequently endocytosis or latrunculin A, an inhibitor of actin-mediated endocytosis, have shown that endocytosis is necessary for ApoEdpL-W targeting to the vacuole and full activity, while it is not needed for Hst-5 activity. A role of active endocytosis in the internalization of the Tat(47-58) peptide of HIV by mammalian cells has been proposed (57). Tat(47-58) is another arginine-rich, lysine-rich, highly cationic peptide that does not form pores in artificial liposomes and whose antifungal activity against Trichosporon beigelii is insensitive to azide and salt, similarly to what we have observed for ApoEdpL-W in C. albicans (34). Binding to the cell surface, possibly through interaction with phosphomannan, followed by vacuolar targeting, also has been observed for another antifungal peptide, namely, dermaseptin S3(1-16) (25). However, the results of a fitness profiling study using a collection of Saccharomyces cerevisiae deletion mutants showed that impairing vacuolar function increased the toxicity of dermaseptin S3(1-16), suggesting that vacuolar targeting is more likely to provide S. cerevisiae cells a defense mechanism toward this peptide. Similar results have been obtained through fitness profiling in S. cerevisiae of Magainin 2 (45) and MUC2-7 (39), suggesting that peptide degradation in the vacuole represents a frequent defense mechanism against antifungal peptides, even though it is not used to protect cells against ApoEdpL-W, as indicated by the decreased rather than increased sensitivity of C. albicans cells to this peptide when vacuolar targeting is impaired.
The transcript profiling of ApoEdpL-W-exposed C. albicans cells revealed additional specificities to the mode of action of this antifungal peptide. Indeed, the exposure of C. albicans to ApoEdpL-W triggered the upregulation of a range of genes encoding plasma membrane amino acid transporters (GAP1, MEP2, PTR2, and CAN1) as well as transcriptional responses similar to those resulting from the inactivation of C. albicans MYO5 and, consequently, endocytosis. Among those genes that are upregulated in response to ApoEdpL-W, DDR48 and RTA2 showed elevated expression in C. albicans strains resistant to amphotericin B and fluconazole (4), while orthologues of GAP1 and MEP2 are induced in response to different classes of antifungal drugs (ketoconazole, 5-fluorocytosine, amphotericin B, and caspofungin) in S. cerevisiae (1). The induction of these genes in response to ApoEdpL-W might reflect a general and nonspecific stress response. However, the upregulation of these genes in response to other antifungal peptides has never been reported, possibly indicating an original mode of action for ApoEdpL-W. However, the inactivation of CAN1, PTR2, and RTA2 or the overexpression of RTA2 did not alter the sensitivity of C. albicans to ApoEdpL-W, suggesting that the change in expression of only one of these genes does not explain ApoEdpL-W toxicity. Transcript profiling of C. albicans cells exposed to other antifungal peptides has revealed a variety of responses. For instance, the exposure of C. albicans cells to MUC7-20 resulted in the activation of the calcineurin pathway and the 20S and 26S proteasome and in the downregulation of genes involved in iron metabolism (40), with the latter also being observed in ApoEdpL-W-treated cells. On the other hand, Morton et al. (45) have shown that the exposure of C. albicans cells to Magainin 2 triggered the upregulation of genes related to DNA replication and repair. Finally, the exposure of C. albicans cells to Hst-5 triggered the expression of genes involved in adaptation to osmotic stress and the general stress response (63). Again, these data underscore the heterogeneous behavior of antifungal peptides, even though differences might in part reflect the use of concentrations below or above the MIC in transcript profiling experiments.
Our study does not provide a definitive mode of action for ApoEdpL-W but suggests that one component of this mode of action is at the level of the vacuole. Indeed, we have shown that ApoEdpL-W is preferentially targeted to the vacuole through endocytosis, and that the exposure of C. albicans cells to ApoEdpL-W triggers a transcriptional response that is reminiscent of that of an endocytosis-defective mutant. The additional upregulation of genes encoding amino acid transporters that are subject to endocytosis-dependent recycling might reflect a general stress response or the consequence of an alteration of endocytosis triggered by the interaction between ApoEdpL-W and plasma membrane components. However, blocking the endocytosis of ApoEdpL-W is not sufficient to fully prevent its antifungal activity, suggesting that ApoEdpL-W penetrates cells through means other than endocytosis. We speculate, based on the observation that most propidium iodide-positive cells show cytoplasmic rather than vacuolar ApoEdpL-W, that the accumulation of ApoEpL-W in the vacuole facilitates its subsequent release in the cytoplasm, where it exerts its antifungal activity. This does not exclude that the antifungal activity of ApoEdpL-W is exerted in part at the vacuolar level.
Biofilms play an important role in the persistence of fungal infections, and antifungal molecules that act on both biofilm cells and planktonic cells would prove highly useful. Very few studies have investigated the potential of antifungal peptides toward biofilms. Here, we have shown that ApoEdpL-W could prevent biofilm formation when applied at early stages of biofilm development but was less efficient when applied to mature biofilms. This relative inefficiency might reflect the affinity of ApoEdpL-W for beta-glucans. Indeed, it has been shown that soluble beta-(1,3)-glucans are major constituents of the extracellular matrix of C. albicans biofilms that trap several antifungals, thus contributing to the intrinsic tolerance of biofilms toward these antifungals (47, 48, 61). Therefore, it is likely that the same applies to ApoEdpL-W, explaining the tolerance of mature C. albicans biofilms to this CAFP. Another application of antifungal peptides could be in the coating of medical devices. Here, we have shown that coating with ApoEdpL-W of polymers used in the design of medical devices, namely, PDMS and PU, promotes the formation of C. albicans biofilms that have a strong tendency to detach from the polymer, thus reducing the metabolic activity of the biofilm that remains attached to the material by 60 to 70%. A similar reduction in biofilm formation has been observed when PDMS grafted with Hst-5 was used, while a higher reduction was observed when a derivative of Hst-5 or low-molecular-weight poly-d-lysine was used (12). Whether a detachment of the biofilm from the grafted surface was observed in that study is not known. These data suggest that the coating of materials with cationic antifungal peptides will prove useful.
In summary, we have characterized a novel cationic antifungal peptide that appears to have an original mode of action relative to other CAFPs, such as Hst-5. The further understanding of the mode of action of ApoEdpL-W, in particular the role of endocytosis and the mechanisms underlying cellular toxicity, will be necessary to develop further this peptide for usual antifungal applications as well as the coating of materials used in the production of medical devices.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mike Birch, Lionel Ferrières, Jean-Marc Ghigo, and members of the NPARI consortium for fruitful discussions during the course of this project. ssa2Δ and ssa2Δ/SSA2 strains were kindly provided by M. Edgerton. COU46 and COU73 strains were kindly provided M. Whiteway.
This work was supported by a grant from the European Commission (LSHE-CT-2006-037692). T.R. was the recipient of a postdoctoral fellowship in the framework of the NPARI consortium (LSHE-CT-2006-037692).
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 1 August 2011.
REFERENCES
- 1. Agarwal A. K., et al. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278:34998–35015 [DOI] [PubMed] [Google Scholar]
- 2. Arnaud M. B., Costanzo M. C., Shah P., Skrzypek M. S., Sherlock G. 2009. Gene ontology and the annotation of pathogen genomes: the case of Candida albicans. Trends Microbiol. 17:295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baev D., et al. 2004. The TRK1 potassium transporter is the critical effector for killing of Candida albicans by the cationic protein, Histatin 5. J. Biol. Chem. 279:55060–55072 [DOI] [PubMed] [Google Scholar]
- 4. Barker K. S., et al. 2004. Genome-wide expression profiling reveals genes associated with amphotericin B and fluconazole resistance in experimentally induced antifungal resistant isolates of Candida albicans. J. Antimicrob. Chemother. 54:376–385 [DOI] [PubMed] [Google Scholar]
- 5. Bates S., et al. 2005. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 280:23408–23415 [DOI] [PubMed] [Google Scholar]
- 6. Bobek L. A., Situ H. 2003. MUC7 20-Mer: investigation of antimicrobial activity, secondary structure, and possible mechanism of antifungal action. Antimicrob. Agents Chemother. 47:643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bonhomme J., et al. 2011. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol. Microbiol. 80:995–1013 [DOI] [PubMed] [Google Scholar]
- 8. Bougnoux M. E., Kac G., Aegerter P., d'Enfert C., Fagon J. Y. 2008. Candidemia and candiduria in critically ill patients admitted to intensive care units in France: incidence, molecular diversity, management and outcome. Intensive Care Med. 34:292–299 [DOI] [PubMed] [Google Scholar]
- 9. Brogden K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
- 10. Cabral V., et al. Modular gene over-expression strategies for Candida albicans. Methods Mol. Biol., in press [DOI] [PubMed] [Google Scholar]
- 11. d'Enfert C. 2006. Biofilms and their role in the resistance of pathogenic Candida to antifungal agents. Curr. Drug Targets 7:465–470 [DOI] [PubMed] [Google Scholar]
- 12. De Prijck K., et al. 2010. Candida albicans biofilm formation on peptide functionalized polydimethylsiloxane. Biofouling 26:269–275 [DOI] [PubMed] [Google Scholar]
- 13. Dobson C. B., Sales S. D., Hoggard P., Wozniak M. A., Crutcher K. A. 2006. The receptor-binding region of human apolipoprotein E has direct anti-infective activity. J. Infect. Dis. 193:442–450 [DOI] [PubMed] [Google Scholar]
- 14. Dong J., Vylkova S., Li X. S., Edgerton M. 2003. Calcium blocks fungicidal activity of human salivary histatin 5 through disruption of binding with Candida albicans. J. Dental Res. 82:748–752 [DOI] [PubMed] [Google Scholar]
- 15. Edgerton M., et al. 1998. Candidacidal activity of salivary histatins. Identification of a histatin 5-binding protein on Candida albicans. J. Biol. Chem. 273:20438–20447 [DOI] [PubMed] [Google Scholar]
- 16. Enjalbert B., et al. 2009. A multifunctional, synthetic Gaussia princeps luciferase reporter for live imaging of Candida albicans infections. Infect. Immun. 77:4847–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. EUCAST 2008. EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 14:398–405 [DOI] [PubMed] [Google Scholar]
- 18. Firon A., et al. 2007. The SUN41 and SUN42 genes are essential for cell separation in Candida albicans. Mol. Microbiol. 66:1256–1275 [DOI] [PubMed] [Google Scholar]
- 19. Fonzi W. A., Irwin M. Y. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gillum A. M., Tsay E. Y., Kirsch D. R. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 21. Gola S., Martin R., Walther A., Dunkler A., Wendland J. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20:1339–1347 [DOI] [PubMed] [Google Scholar]
- 22. Goyard S., et al. 2008. The Yak1 kinase is involved in the initiation and maintenance of hyphal growth in Candida albicans. Mol. Biol. Cell 19:2251–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gururaja T. L., et al. 1999. Candidacidal activity prompted by N-terminus histatin-like domain of human salivary mucin (MUC7). Biochim. Biophys. Acta Prot. Struct. Mol. Enzymol. 1431:107–119 [DOI] [PubMed] [Google Scholar]
- 24. Gyurko C., Lendenmann U., Troxler R. F., Oppenheim F. G. 2000. Candida albicans mutants deficient in respiration are resistant to the small cationic salivary antimicrobial peptide Histatin 5. Antimicrob. Agents Chemother. 44:348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harris M., Mora-Montes H. M., Gow N. A. R., Coote P. J. 2009. Loss of mannosylphosphate from Candida albicans cell wall proteins results in enhanced resistance to the inhibitory effect of a cationic antimicrobial peptide via reduced peptide binding to the cell surface. Microbiology 155:1058–1070 [DOI] [PubMed] [Google Scholar]
- 26. Helmerhorst E. J., et al. 1999. The cellular target of Histatin 5 on Candida albicans is the energized mitochondrion. J. Biol. Chem. 274:7286–7291 [DOI] [PubMed] [Google Scholar]
- 27. Helmerhorst E. J., Reijnders I. M., van't Hof W., Veerman E. C., Nieuw Amerongen A. V. 1999. A critical comparison of the hemolytic and fungicidal activities of cationic antimicrobial peptides. FEBS Lett. 449:105–110 [DOI] [PubMed] [Google Scholar]
- 28. Helmerhorst E. J., van't Hof W., Veerman E. C., Simoons-Smit I., Nieuw Amerongen A. V. 1997. Synthetic histatin analogues with broad-spectrum antimicrobial activity. Biochem. J. 326:39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hong S. Y., et al. 1998. Identification and characterization of novel antimicrobial decapeptides generated by combinatorial chemistry. Antimicrob. Agents Chemother. 42:2534–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jang W. S., Bajwa J. S., Sun J. N., Edgerton M. 2010. Salivary histatin 5 internalization by translocation, but not endocytosis, is required for fungicidal activity in Candida albicans. Mol. Microbiol. 77:354–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jang W. S., Li X. S., Sun J. N., Edgerton M. 2008. The P-113 fragment of Histatin 5 requires a specific peptide sequence for intracellular translocation in Candida albicans, which is independent of cell wall binding. Antimicrob. Agents Chemother. 52:497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jenssen H., Hamill P., Hancock R. E. 2006. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jia X. M., et al. 2008. RTA2, a novel gene involved in azole resistance in Candida albicans. Biochem. Biophys. Res. Commun. 373:631–636 [DOI] [PubMed] [Google Scholar]
- 34. Jung H. J., Park Y., Hahm K.-S., Lee D. G. 2006. Biological activity of Tat (47–58) peptide on human pathogenic fungi. Biochem. Biophys. Res. Commun. 345:222–228 [DOI] [PubMed] [Google Scholar]
- 35. Kelly B. A., et al. 2007. Apolipoprotein E-derived antimicrobial peptide analogues with altered membrane affinity and increased potency and breadth of activity. FEBS J. 274:4511–4525 [DOI] [PubMed] [Google Scholar]
- 36. Koshlukova S. E., Lloyd T. L., Araujo M. W. B., Edgerton M. 1999. Salivary Histatin 5 induces non-lytic release of ATP from Candida albicans leading to cell death. J. Biol. Chem. 274:18872–18879 [DOI] [PubMed] [Google Scholar]
- 37. Leuker C. E., Sonneborn A., Delbruck S., Ernst J. F. 1997. Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene 192:235–240 [DOI] [PubMed] [Google Scholar]
- 38. Li X. S., Sun J. N., Okamoto-Shibayama K., Edgerton M. 2006. Candida albicans cell wall Ssa proteins bind and facilitate import of salivary Histatin 5 required for toxicity. J. Biol. Chem. 281:22453–22463 [DOI] [PubMed] [Google Scholar]
- 39. Lis M., Fuss J. R., Bobek L. A. 2009. Exploring the mode of action of antimicrobial peptide MUC7 12-mer by fitness profiling of Saccharomyces cerevisiae genomewide mutant collection. Antimicrob. Agents Chemother. 53:3762–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lis M., Liu T. T., Barker K. S., Rogers P. D., Bobek L. A. 2010. Antimicrobial peptide MUC7 12-mer activates the calcium/calcineurin pathway in Candida albicans. FEMS Yeast Res. 10:579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meiller T. F., et al. 2009. A novel immune evasion strategy of Candida albicans: proteolytic cleavage of a salivary antimicrobial peptide. PLoS One 4:e5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mochon A. B., Liu H. 2008. The antimicrobial peptide Histatin-5 causes a spatially restricted disruption on the Candida albicans surface, allowing rapid entry of the peptide into the cytoplasm. PLoS Pathog. 4:e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moody J. 2004. Synergism testing: broth microdilution checkerboard and broth macrodilution methods, p. 5.12.12–15.12.23 In Isenberg H. D. (ed.), Clinical microbiology procedures handbook, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 44. Moreno-Ruiz E., et al. 2009. The GPI-modified proteins Pga59 and Pga62 of Candida albicans are required for cell wall integrity. Microbiology 155:2004–2020 [DOI] [PubMed] [Google Scholar]
- 45. Morton C. O., et al. 2007. Global phenotype screening and transcript analysis outlines the inhibitory mode (s) of action of two amphibian-derived, alpha-helical, cationic peptides on Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 51:3948–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murad A. M., Lee P. R., Broadbent I. D., Barelle C. J., Brown A. J. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325–327 [DOI] [PubMed] [Google Scholar]
- 47. Nett J. E., Crawford K., Marchillo K., Andes D. R. 2010. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob. Agents Chemother. 54:3505–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nett J. E., Sanchez H., Cain M. T., Andes D. R. 2010. Genetic basis of Candida biofilm resistance due to drug-sequestering matrix glucan. J. Infect. Dis. 202:171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oberholzer U., Iouk T. L., Thomas D. Y., Whiteway M. 2004. Functional characterization of myosin I tail regions in Candida albicans. Eukaryot. Cell 3:1272–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Oberholzer U., Marcil A., Leberer E., Thomas D. Y., Whiteway M. 2002. Myosin I is required for hypha formation in Candida albicans. Eukaryot. Cell 1:213–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oberholzer U., Nantel A., Berman J., Whiteway M. 2006. Transcript profiles of Candida albicans cortical actin patch mutants reflect their cellular defects: contribution of the Hog1p and Mkc1p signaling pathways. Eukaryot. Cell 5:1252–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oppenheim F. G., et al. 1988. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J. Biol. Chem. 263:7472–7477 [PubMed] [Google Scholar]
- 53. Ostrosky-Zeichner L., Casadevall A., Galgiani J. N., Odds F. C., Rex J. H. 2010. An insight into the antifungal pipeline: selected new molecules and beyond. Nat. Rev. Drug Discov. 9:719–727 [DOI] [PubMed] [Google Scholar]
- 54. Park C., Lee D. G. 2009. Fungicidal effect of antimicrobial peptide arenicin-1. Biochim. Biophys. Acta Biomembranes 1788:1790–1796 [DOI] [PubMed] [Google Scholar]
- 55. Pfaller M. A., Diekema D. J. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36:1–53 [DOI] [PubMed] [Google Scholar]
- 56. Raj P. A., Edgerton M., Levine M. J. 1990. Salivary histatin 5: dependence of sequence, chain length, and helical conformation for candidacidal activity. J. Biol. Chem. 265:3898–3905 [PubMed] [Google Scholar]
- 57. Richard J. P., et al. 2003. Cell-penetrating peptides. J. Biol. Chem. 278:585–590 [DOI] [PubMed] [Google Scholar]
- 58. Rigaut G., et al. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032 [DOI] [PubMed] [Google Scholar]
- 59. Rossignol T., et al. 2009. Correlation between biofilm formation and the hypoxic response in Candida parapsilosis. Eukaryot. Cell 8:550–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sun J. N., et al. 2008. Uptake of the antifungal cationic peptide Histatin 5 by Candida albicans Ssa2p requires binding to non-conventional sites within the ATPase domain. Mol. Microbiol. 70:1246–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vediyappan G., Rossignol T., d'Enfert C. 2010. Interaction of Candida albicans biofilms with antifungals: transcriptional response and binding of antifungals to beta-glucans. Antimicrob. Agents Chemother. 54:2096–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Veerman E. C. I., et al. 2007. Energy depletion protects Candida albicans against antimicrobial peptides by rigidifying its cell membrane. J. Biol. Chem. 282:18831–18841 [DOI] [PubMed] [Google Scholar]
- 63. Vylkova S., Jang W. S., Li W., Nayyar N., Edgerton M. 2007. Histatin 5 initiates osmotic stress response in Candida albicans via activation of the Hog1 mitogen-activated protein kinase pathway. Eukaryot. Cell 6:1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vylkova S., Nayyar N., Li W., Edgerton M. 2007. Human beta-defensins kill Candida albicans in an energy-dependent and salt-sensitive manner without causing membrane disruption. Antimicrob. Agents Chemother. 51:154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Walther A., Wendland J. 2003. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 42:339–343 [DOI] [PubMed] [Google Scholar]
- 66. Xu T., Levitz S. M., Diamond R. D., Oppenheim F. G. 1991. Anticandidal activity of major human salivary histatins. Infect. Immun. 59:2549–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zasloff M. 1987. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. U. S. A. 84:5449–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.