Abstract
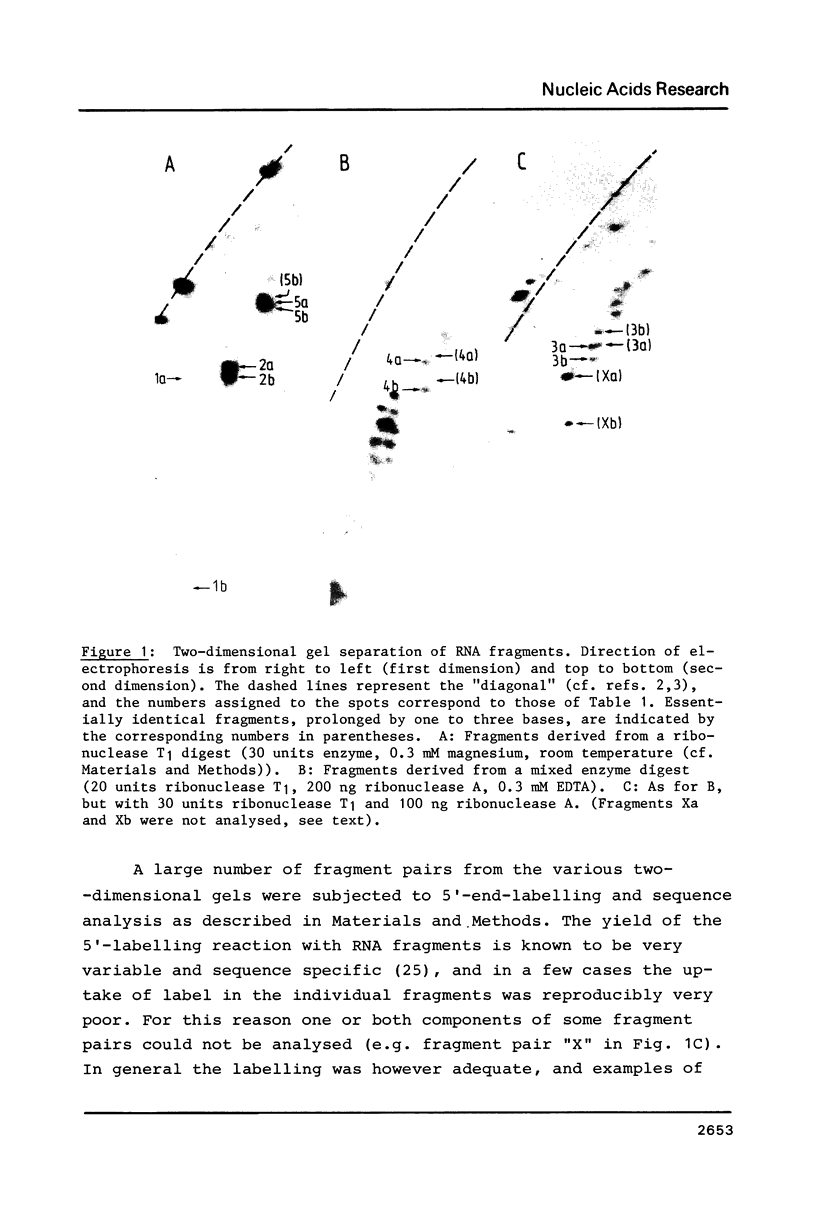
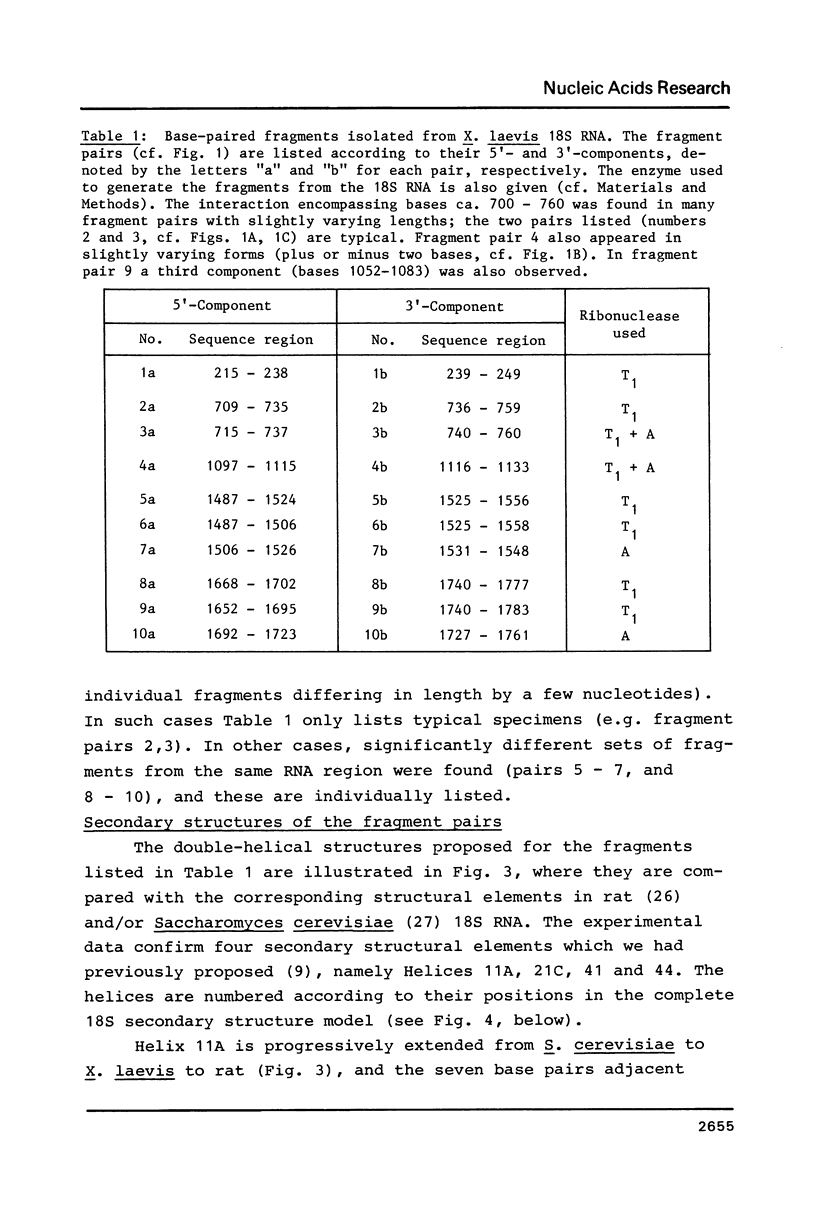
18S ribosomal RNA from X. laevis was subjected to partial digestion with ribonucleases A or T1 under a variety of conditions, and base-paired fragments were isolated. Sequence analysis of the fragments enabled five base-paired secondary structural elements of the 18S RNA to be established. Four of these elements (covering bases 221-256, 713-757, 1494-1555 and 1669-1779) confirm our previous secondary structure predictions, whereas the fifth (comprising bases 1103-1125) represents a phylogenetically conserved "switch" structure, which can also form in prokaryotic 16S RNA. The results are incorporated into a refined model of the 18S RNA secondary structure, which also includes the locations of the many methyl groups in X. laevis 18S RNA. In general the methyl groups occur in non-helical regions, at hairpin loop ends, or at helix boundaries and imperfections. One large cluster of 2'-O-methyl groups occurs in a region of complicated secondary structure in the 5'-one third of the molecule.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brand R. C., Klootwijk J., Planta R. J., Maden B. E. Biosynthesis of a hypermodified nucleotide in Saccharomyces carlsbergensis 17S and HeLa-cell 18S ribosomal ribonucleic acid. Biochem J. 1978 Jan 1;169(1):71–77. doi: 10.1042/bj1690071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe R., Maly P., Zwieb C. The structure of ribosomal RNA and its organization relative to ribosomal protein. Prog Nucleic Acid Res Mol Biol. 1983;28:1–48. doi: 10.1016/s0079-6603(08)60081-1. [DOI] [PubMed] [Google Scholar]
- Carbon P., Ehresmann C., Ehresmann B., Ebel J. P. The complete nucleotide sequence of the ribosomal 16-S RNA from Excherichia coli. Experimental details and cistron heterogeneities. Eur J Biochem. 1979 Oct 15;100(2):399–410. doi: 10.1111/j.1432-1033.1979.tb04183.x. [DOI] [PubMed] [Google Scholar]
- Chan Y. L., Gutell R., Noller H. F., Wool I. G. The nucleotide sequence of a rat 18 S ribosomal ribonucleic acid gene and a proposal for the secondary structure of 18 S ribosomal ribonucleic acid. J Biol Chem. 1984 Jan 10;259(1):224–230. [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douthwaite S., Christensen A., Garrett R. A. Higher order structure in the 3'-minor domain of small subunit ribosomal RNAs from a gram negative bacterium, a gram positive bacterium and a eukaryote. J Mol Biol. 1983 Sep 5;169(1):249–279. doi: 10.1016/s0022-2836(83)80183-1. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Stiegler P., Carbon P., Ungewickell E., Garrett R. A. The topography of the 5' end of 16-S RNA in the presence and absence of ribosomal proteins S4 and S20. Eur J Biochem. 1980 Feb;103(3):439–446. doi: 10.1111/j.1432-1033.1980.tb05967.x. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Glotz C., Brimacombe R. An experimentally-derived model for the secondary structure of the 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1980 Jun 11;8(11):2377–2395. doi: 10.1093/nar/8.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotz C., Zwieb C., Brimacombe R., Edwards K., Kössel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981 Jul 24;9(14):3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Lanter J. M., Woese C. R. Sequence of the 16S Ribosomal RNA from Halobacterium volcanii, an Archaebacterium. Science. 1983 Aug 12;221(4611):656–659. doi: 10.1126/science.221.4611.656. [DOI] [PubMed] [Google Scholar]
- Hughes D. G., Maden B. E. The pseudouridine contents of the ribosomal ribonucleic acids of three vertebrate species. Numerical correspondence between pseudouridine residues and 2'-O-methyl groups is not always conserved. Biochem J. 1978 Jun 1;171(3):781–786. doi: 10.1042/bj1710781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. S., Salim M., Maden B. E. Extensive homologies between the methylated nucleotide sequences in several vertebrate ribosomal ribonucleic acids. Biochem J. 1978 Mar 1;169(3):531–542. doi: 10.1042/bj1690531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klootwijk J., Planta R. J. Analysis of the methylation sites in yeast ribosomal RNA. Eur J Biochem. 1973 Nov 15;39(2):325–333. doi: 10.1111/j.1432-1033.1973.tb03130.x. [DOI] [PubMed] [Google Scholar]
- Köchel H. G., Küntzel H. Nucleotide sequence of the Aspergillus nidulans mitochondrial gene coding for the small ribosomal subunit RNA: homology to E. coli 16S rRNA. Nucleic Acids Res. 1981 Nov 11;9(21):5689–5696. doi: 10.1093/nar/9.21.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Forbes J. M., Stewart M. A., Eason R. 18S coding sequences in amplified ribosomal DNA from Xenopus laevis oocytes are highly homogeneous, unmethylated, and lack major open reading frames. EMBO J. 1982;1(5):597–601. doi: 10.1002/j.1460-2075.1982.tb01214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Reeder R. H. Partial mapping of methylated sequences in Xenopus laevis ribosomal RNA by preparative hybridization to cloned fragments of ribosomal DNA. Nucleic Acids Res. 1979 Mar;6(3):817–830. doi: 10.1093/nar/6.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Salim M. The methylated nucleotide sequences in HELA cell ribosomal RNA and its precursors. J Mol Biol. 1974 Sep 5;88(1):133–152. doi: 10.1016/0022-2836(74)90299-x. [DOI] [PubMed] [Google Scholar]
- Maly P., Brimacombe R. Refined secondary structure models for the 16S and 23S ribosomal RNA of Escherichia coli. Nucleic Acids Res. 1983 Nov 11;11(21):7263–7286. doi: 10.1093/nar/11.21.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man'kin A. S., Kopylov A. M., Rubtsov P. M., Skriabin K. G. Model' vtorichnoi struktury 18 S rRNK ribosom éukariot. Dokl Akad Nauk SSSR. 1981;256(4):1006–1010. [PubMed] [Google Scholar]
- Mankin A. S., Kopylov A. M., Bogdanov A. A. Modification of 18 S rRNA in the 40 S ribosomal subunit of yeast with dimethyl sulfate. FEBS Lett. 1981 Nov 2;134(1):11–14. doi: 10.1016/0014-5793(81)80539-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., McCarroll R., Sogin M. L. Secondary structure of the Dictyostelium discoideum small subunit ribosomal RNA. Nucleic Acids Res. 1983 Nov 25;11(22):8037–8049. doi: 10.1093/nar/11.22.8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A., Brimacombe R. Application of a rapid gel method to the sequencing of fragments of 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1978 Jan;5(1):241–256. doi: 10.1093/nar/5.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A., Brimacombe R. Experimental determination of interacting sequences in ribosomal RNA. Nature. 1979 Sep 27;281(5729):271–276. doi: 10.1038/281271a0. [DOI] [PubMed] [Google Scholar]
- Rubtsov P. M., Musakhanov M. M., Zakharyev V. M., Krayev A. S., Skryabin K. G., Bayev A. A. The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Dec 11;8(23):5779–5794. doi: 10.1093/nar/8.23.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence encoding the 5' end of Xenopus laevis 18S rRNA. Nucleic Acids Res. 1980 Jul 11;8(13):2871–2884. doi: 10.1093/nar/8.13.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Séquence nucléotidique du gène de l'ARN ribosomique 15S mitochondrial de la levure. C R Seances Acad Sci D. 1980 Dec 8;291(12):933–936. [PubMed] [Google Scholar]
- Stiegler P., Carbon P., Ebel J. P., Ehresmann C. A general secondary-structure model for procaryotic and eucaryotic RNAs from the small ribosomal subunits. Eur J Biochem. 1981 Dec;120(3):487–495. doi: 10.1111/j.1432-1033.1981.tb05727.x. [DOI] [PubMed] [Google Scholar]
- Székely M., Sanger F. Use of polynucleotide kinase in fingerprinting non-radioactive nucleic acids. J Mol Biol. 1969 Aug 14;43(3):607–617. doi: 10.1016/0022-2836(69)90362-3. [DOI] [PubMed] [Google Scholar]
- Thomas G., Gordon J., Rogg H. N4-Acetylcytidine. A previously unidentified labile component of the small subunit of eukaryotic ribosomes. J Biol Chem. 1978 Feb 25;253(4):1101–1105. [PubMed] [Google Scholar]
- Torczynski R., Bollon A. P., Fuke M. The complete nucleotide sequence of the rat 18S ribosomal RNA gene and comparison with the respective yeast and frog genes. Nucleic Acids Res. 1983 Jul 25;11(14):4879–4890. doi: 10.1093/nar/11.14.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., Glotz C., Brimacombe R. Secondary structure comparisons between small subunit ribosomal RNA molecules from six different species. Nucleic Acids Res. 1981 Aug 11;9(15):3621–3640. doi: 10.1093/nar/9.15.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]




