Abstract
Neuraminidase (NA) inhibitors are among the first line of defense against influenza virus infection. With the increased worldwide use of the drugs, antiviral susceptibility surveillance is increasingly important for effective clinical management and for public health epidemiology. Effective monitoring requires effective resistance detection methods. We have developed and validated a novel genotyping method for rapid detection of established NA inhibitor resistance markers in influenza viruses by single nucleotide polymorphism (SNP) analysis. The multi- or monoplex SNP analysis based on single nucleotide extension assays was developed to detect NA mutations H275Y and I223R/V in pandemic H1N1 viruses, H275Y in seasonal H1N1 viruses, E119V and R292K in seasonal H3N2 viruses, and H275Y and N295S in H5N1 viruses. The SNP analysis demonstrated high sensitivity for low-content NA amplicons (0.1 to 1 ng/μl) and showed 100% accordant results against a panel of defined clinical isolates. The monoplex assays for the H275Y NA mutation allowed precise and accurate quantification of the proportions of wild-type and mutant genotypes in virus mixtures (5% to 10% discrimination), with results comparable to those of pyrosequencing. The SNP analysis revealed the lower growth fitness of an H275Y mutant compared to the wild-type pandemic H1N1 virus by quantitatively genotyping progeny viruses grown in normal human bronchial epithelial cells. This novel method offers high-throughput screening capacity, relatively low costs, and the wide availability of the necessary equipment, and thus it could provide a much-needed approach for genotypic screening of NA inhibitor resistance in influenza viruses.
INTRODUCTION
Influenza viruses are important human pathogens, and antivirals are the only control option in the absence of a specific vaccine. The neuraminidase (NA) inhibitors (oral oseltamivir, inhaled zanamivir) are a class of specific antivirals targeted at influenza viruses, and they have served as the front line of influenza prevention and treatment for more than a decade (2, 24, 35, 40). In 2009, the U.S. Food and Drug Administration granted Emergency Use Authorization for the treatment of selected cases of pandemic H1N1 influenza infection with the investigational intravenous NA inhibitor peramivir (3). However, the emergence of NA inhibitor-resistant variants can substantially reduce the efficacy of chemoprophylaxis and treatment. Monitoring the emergence of NA inhibitor resistance is advisable not only for clinical intervention strategies but also for public health epidemiology.
NA inhibitor resistance caused by NA subtype-specific mutations has emerged at different rates (1, 17, 25). The H275Y NA mutation in the 2009 pandemic H1N1 influenza viruses causes cross-resistance to oseltamivir and peramivir but not to zanamivir (10). A novel I223R NA mutation causing modest multidrug resistance has recently been identified in several cases of 2009 H1N1 infection (36). The overall frequency of oseltamivir resistance in the pandemic H1N1 viruses remained as low as 0.7 to 1.1% (10, 13), emerging mainly in the context of prophylaxis and treatment and rare community transmission (2, 10). The H275Y NA mutation is also the most common oseltamivir resistance marker in the seasonal H1N1 viruses (26, 38). In the seasonal H3N2 influenza viruses, the E119V NA framework mutation confers resistance only to oseltamivir, whereas the R292K mutation in the NA catalytic active site confers cross-resistance to all three NA inhibitors (1, 17). Prior to 2007, emergence of resistant variants in both the seasonal N1 and N2 subtypes was generally low during treatment: <1% in adults and 4% to 8% in children (16, 22). However, oseltamivir-resistant seasonal H1N1 viruses with the H275Y mutation showed a startlingly high prevalence worldwide (from 25% in Europe to almost 100% in the United States) during the 2007 to 2009 seasons in the absence of drug selection pressure (26, 38). In avian H5N1 influenza viruses, the H275Y and N295S NA mutations emerged during oseltamivir treatment in infected humans and confer high and moderate oseltamivir resistance, respectively (15, 18). All of these findings of resistance in different subtypes and precedent global spread of resistant seasonal H1N1 viruses emphasize the necessity for continued monitoring of NA inhibitor susceptibility among circulating influenza viruses.
The NA inhibitor susceptibility of influenza viruses can be evaluated by phenotypic and genotypic methods. Phenotypic analysis includes cell culture-based infectivity reduction assays and biochemical NA inhibition assays (28, 39). However, infectivity reduction assays are not recommended due to unpredictable changes of HA receptor binding in resistant viruses (31, 39). Biochemical NA inhibition assays using different substrates are predominantly used (28, 31, 39), and these assays yield the 50% inhibitory concentration (IC50) values of drugs for NA enzyme activity. Phenotypic assays require propagation of viruses in cell cultures or embryonated chicken eggs, and the assay procedures are complex and labor-intensive. Genotypic methods directly analyze viral NA gene sequences to identify the presence of established NA inhibitor resistance markers. Although genotypic methods require viral subtypes or lineages to be differentiated in advance, they generally offer simple, rapid screening of large quantities of clinical isolates. Thus, the NA inhibitor susceptibility in influenza viruses can be easily determined by genotypic analysis and subsequently confirmed by phenotypic assays.
Rapid, sensitive, accurate, and high-throughput genotypic methods for detection of NA inhibitor resistance markers are especially valuable in antiviral resistance surveillance. The resistance markers in NA gene are most commonly analyzed by the Sanger method of DNA sequencing, but this method requires purification of PCR-amplified NA segments and is time-consuming for a large number of samples. Two other methods, TaqMan real-time PCR-based assays and pyrosequencing, provide rapid, sensitive, and quantitative analysis of resistance markers in influenza viruses (7, 9, 20, 23), but they require costly specific labeled primers or probes. Further, pyrosequencing requires specific equipment that is not widely available. Other PCR-based methods, such as the reverse transcriptase PCR (RT-PCR)/restriction fragment length polymorphism assay, rolling circle amplification, and bead-based PCR, have been proved experimentally feasible (11, 29, 32, 37). However, most are time-consuming, low-throughput, and/or costly, and few allow quantification of the wild-type and mutant genotypes present in a mixed sample.
In our pursuit for a novel alternative method, we found that single nucleotide polymorphism (SNP) genotyping could provide an option, because all known NA inhibitor resistance markers are caused by a single nucleotide mutation in the NA gene. The SNP analysis by the SNaPshot Multiplex System (Applied Biosystems) has been a reliable method in genetics to identify SNPs at known locations in the human genome (4, 12, 19, 21, 27) but had not been applied to influenza viruses. The assay is based on template-directed single nucleotide extension and could be described as “mini-sequencing” of only one nucleotide. An extension probe is designed to anneal to the template in a position that places the mutation site immediately adjacent to the 3′ end of the probe, and the use of dideoxynucleoside triphosphates (ddNTPs) allows the extension of only one nucleotide from the 3′ end of the probe (see Fig. S1 in the supplemental material). Labeling of each ddNTP with a different fluorescent dye allows the differentiation of the genotype at the SNP by the color of the extended probes. This assay can be used in the mono- or multiplex format and can simultaneously detect as many as 10 known polymorphism sites.
In the present study, we describe (i) a novel mono- and multiplex SNP analysis method for the rapid screening of established NA inhibitor resistance-associated mutations in pandemic H1N1, seasonal H1N1 and H3N2, and avian H5N1 influenza viruses and (ii) a quantitative SNP analysis to detect the ratios of wild-type and mutant genotypes in mixed samples. We have successfully validated this method against a panel of defined clinical isolates and have applied it to assess the shifts in oseltamivir IC50s in mixed viral populations and the relative growth fitness of wild-type and resistant viruses.
MATERIALS AND METHODS
Influenza viruses and RT-PCR.
A panel of pandemic H1N1 viruses and seasonal H1N1 and H3N2 viruses which were previously characterized by Sanger sequencing or pyrosequencing was used to validate the new SNP detection method (see Table S1 in the supplemental material). All virus samples except the Denmark and Hong Kong isolates were provided by Alexander I. Klimov (U.S. Centers for Disease Control and Prevention [CDC]). The Denmark isolates were provided by Lars P. Nielsen (Statens Serum Institute, Denmark). The Hong Kong isolate was provided by Malik Peiris (University of Hong Kong). Virus stocks were propagated in Madin-Darby canine kidney (MDCK) cells (ATCC, Manassas, VA). Viral RNAs were extracted by using the RNeasy minikit (Qiagen, Valencia, CA). RT-PCR primer sequences for amplifying NA segments containing the residues of interest for each subtype of influenza virus were obtained in published reports by the CDC (5–7, 30) and were all unlabeled oligonucleotides. The NA segment was amplified by using the Platinum one-step RT-PCR system (Invitrogen, Carlsbad, CA) for 35 cycles according to the manufacturer's instructions. The amplicons were confirmed as a clear, strong band free of nonspecific products on a 1.5% agarose gel before further experiments.
Plasmid construction and site-directed mutagenesis.
Plasmid standards were generated containing the full-length wild-type NA gene of A/California/4/09 pandemic H1N1 virus, A/Brisbane/59/07 seasonal H1N1 virus, and A/Hong Kong/218849/06 seasonal H3N2 virus. The H275Y, I223R, and I223V mutations were introduced into the NA gene of A/California/04/09 virus, H275Y into the NA gene of A/Brisbane/59/07 virus, and E119V and R292K into the NA gene of A/Hong Kong/218849/06 virus by using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA). Wild-type NA, H275Y NA, and N295S NA plasmids of A/Vietnam/1203/04 virus were constructed as described previously (41). No infectious highly pathogenic H5N1 viruses were used in this study due to the required higher biosafety level. All the NA plasmids of the defined genotype were confirmed by Sanger sequencing and were used to generate NA amplicons for positive and negative controls and condition optimization for SNaPshot assays.
Design of extension probes for detection of resistance markers.
Extension probes to detect a nucleotide of interest in the NA genes were designed to anneal with the template adjacent to the site of interest, leaving the next nucleotide to be extended from the 3′ end of the probe to match the site of interest. About 1,000 sequences of 2009 pandemic H1N1 viruses (2009-2010) and contemporary seasonal H1N1 and H3N2 viruses (2006 to 2009), and 255 sequences of avian origin human cases of H5N1 viruses (2000 to 2010) were acquired from the NCBI GenBank. The consensus sequences of each genotype were used for probe design. A few sites with high numbers of frequent changes were modified into degenerate nucleotides (Tables 1 and 2). In general, the extension probes can be forward or reverse, depending on whether the local sequences are prone to secondary structure and, in a multiplex assay, whether the probe overlaps with the others. Probes in multiplex assay were of different lengths, so that they could be separated by size in capillary electrophoresis (CE). Oligonucleotide T can be added to the 5′ end of probes to increase the length without increasing the sequence complexity. Template and probe sequences were assessed for specific complementarity and absence of self-complementarity.
Table 1.
Probes for detection of the H275Y NA mutation in seasonal and pandemic H1N1 viruses by monoplex SNaPshot assay
| Subtype/lineage | NA resistance marker | SNP in the codon of marker | Extension probe |
|||
|---|---|---|---|---|---|---|
| Primera | Sequence | Size (bp) | Signal | |||
| 2009 pandemic H1N1 | H275Y | CAC–TAC | pPdmN1-275-F | CAGTCGAAATGAATGCCCCTAATTAT | 26 | C/T |
| pPdmN1-275-R | ATCAGGATAACAGGAGCATTCCTCATAGT | 29 | G/A | |||
| Human seasonal H1N1 | H275Y | CAT–TAT | pHuN1-275-F | AATCAATAGAGTTRAATGCACCCAATTTT | 29 | C/T |
| pHuN1-275-R | TGTCTGGGTAACAGGARCATTCCTCATAAT | 30 | G/A | |||
“F” and “R” in the primer names indicate forward and reverse directions, respectively.
Table 2.
Probes for detection of NA inhibitor resistance-associated mutations in influenza viruses by multiplex SNaPshot assay
| Subtype/lineage | NA resistance marker | SNP in the codon of marker | Extension probe |
|||
|---|---|---|---|---|---|---|
| Primera | Sequenceb | Size (bp) | Signal | |||
| 2009 pandemic H1N1 | H275Y | CAC–TAC | pPdmN1-275-F | CAGTCGAAATGAATGCCCCTAATTAT | 26 | C/T |
| I222V | ATA–GTA | pPdmN1-222V-F | AATAACAGACACTATCAAGAGTTGGAGAAACAAT | 34 | A/G | |
| I222R | ATA–AGA | pPdmN1-222R-R | TTTTTACACATGCACATTCAGACTCTTGTGTTCTCAAT | 38 | A/C | |
| Human H3N2 | R292K | AGA–AAA | pHuN2-292-R | TTTTCCTATTGGAKCCTTTCCAGTTGTCT | 29 | C/T |
| E119V | GAA–GTA | pHuN2-119F | TTTTTTCCGCTGGTGGGGACATCTGGGTGACAAGAG | 36 | A/T | |
| Human H5N1 | H275Y | CAC–TAC | pH5N1-275-R | TTTTTTTTCAGGATAACAGGAGCAYTCCTCATAGT | 35 | A/G |
| N295S | AAT–AGT | pH5N1-295-R | ATACCCATGGCCYATTTGAKCCATGCCAA | 29 | C/T | |
“F” and “R” in the primer names indicate forward and reverse directions, respectively.
Underlining indicates addition of unspecific oligonucleotide T at the 5′ end in order to increase the size of the probes.
Single nucleotide probe extension assay.
The SNaPshot kit (ABI, Carlsbad, CA) was used for template-directed single nucleotide probe extension as described previously (8, 41), with modifications. First, NA fragments with site of interest were amplified by RT-PCR or PCR from viral RNA samples or NA plasmids. The amplicons were then digested with shrimp alkaline phosphatase (SAP) and exonuclease I (USB Corp) to remove unincorporated primers and dNTPs or were purified with a column kit (Qiagen, Valencia, CA). The digested or purified amplicons served as the DNA templates for further single nucleotide probe extension. The extension reaction mixture consisted of 5 μl SNaPshot reaction mix, 2 μl DNA amplicons, and 0.2 μmol/liter of the extension probes in a 10-μl final volume, and the reaction was conducted as directed by the manufacturer. After the reaction, a unit of SAP was added to remove 5′ phosphoryl groups of unincorporated dideoxynucleotide. Then 1 μl of the reaction product was mixed with deionized formamide and LIZ120 (ABI) size standard and injected into the ABI 3730xl capillary electrophoresis instrument (ABI) according to the manufacturer's protocol. Data were analyzed by using ABI GeneMapper software to determine signal type and signal peak height and peak area for each SNP site.
Concentration-response curves of the SNaPshot assay.
The sensitivity of the monoplex or multiplex assays was evaluated by serial dilution of NA amplicons of defined genotype. For example, 2-fold and 1.5-fold serial dilutions of wild-type or H275Y mutant NA amplicons of the pandemic H1N1 virus (35 to 0.02 ng/μl of an ∼300-bp purified PCR product) were used to generate concentration-response curves. Each diluted sample was tested in triplicate (intra-assay precision) and the serial dilution was repeated at least twice (interassay reproducibility). The average responses (peak height of each fluorescence signal) were plotted against the concentrations of DNA templates on a logarithmic scale. The endpoint detection limit was determined on the basis of the lower plateau of the concentration-response curves.
Quantification of viral genotypes by SNaPshot assay.
The essentially linear region in the concentration-response curves was determined, and the data from the linear portion were plotted separately on a decimal scale. R2 was used to evaluate the goodness of linearity of the selected portion. If R2 was <0.9, the data at the bending points of the curve can be further excluded and R2 will be calculated again for the remaining data; if R2 was >0.9, the linear equation derived from the selected portion will be accepted. Thus, within the linear range, the response (peak height of the signal) was expressed as a function of template concentration: Y = aX + b. For example, for the H275Y forward probe, the response C signal is Yc = acXc + bc, and the T signal is Yt = atXt + bt. For any Y signal within the linear range, X can be resolved by which is the concentration of amplicons of the genotype present in the SNaPshot reaction mixture. If the original template was diluted F-fold, the original concentration should be F × X. When both C and T signal were detected in a mixed sample, the ratio of two genotypes was calculated by the formula when both signals within the linear range were acquired from one assay (same dilution), or from the formula when two signals were acquired from different dilutions of original templates. The first formula was recommended for having no dilution error. Similarly, the ratio of G to A genotypes in a mixed sample detected by H275Y reverse probe can be derived by .
To test the accuracy of the relative quantification of two genotypes by this method, a series of spike-in samples was generated at 11 different ratios of wild-type and H275Y mutant NA amplicon concentrations (e.g., 100%, 90% … 10%, 0% wild-type NA amplicons) and tested by SNaPshot assay and the corresponding formula. Several dilutions of the spike-in samples can be used to acquire fluorescence signals within the linear range determined above. The detected ratios were then compared with the original spike-in ratios to assess the precision and accuracy of relative quantification by SNaPshot assay.
Pyrosequencing.
Pyrosequencing was performed on NA amplicons of pandemic viruses according to the CDC protocol (7). First, fragments of the wild-type and H275Y mutant NA genes of A/California/4/2009 virus were amplified by using a biotinylated reverse primer. Spike-in samples of 11 different ratios of wild-type and H275Y NA amplicons were generated as described above by using the biotinylated amplicons. Pyrosequencing reactions were then performed with the spike-in samples by using sequencing primer pPdmN1-275-F (Table 1) on the Pyromark Q24 system, using the allele quantification module. The same spike-in samples were also subjected to SNaPshot assay by using extension probe pPdmN1-275-F (Table 1). The ratios of mixed samples detected by the two methods were compared.
Detection of resistant subpopulation by NA inhibition assay.
The NA activity and NA inhibition assays were performed as previously described (8, 34). Briefly, the NA activity of wild-type A/Denmark/524/09 virus and H275Y mutant A/Denmark/528/09 virus was measured by a fluorescence-based assay using 100 μM 2′-(4-methylumbelliferyl)-alpha-d-N-acetylneuraminic acid (MUNANA) substrate at 37°C for 30 min. The Synergy 2 microplate reader (BioTek) was used to detect the fluorescence at excitation and emission wavelengths of 360 and 460 nm, respectively. The two viruses were then mixed in 11 ratios on the basis of their relative NA enzyme activity (100%, 90%…10%, 0% of wild-type NA activity in a total of 4,000 fluorescence units of mixed NA activity). The virus mixtures were analyzed by phenotypic NA inhibition assay and genotypic SNaPshot assay to determine the resistant subpopulation. In the NA inhibition assay, the apparent IC50 of oseltamivir in the sample mixtures was determined by plotting the percent inhibition of NA activity as a function of compound concentration calculated by using GraphPad Prism 4 software. In the SNaPshot assay, the forward probe pPdmN1-275-F was used to detect and quantify the subpopulation in the mixture.
Evaluation of viral growth fitness by SNP analysis.
The growth fitness of the oseltamivir-resistant variant A/Denmark/528/09 virus was assessed by competitive growth assay in primary culture of normal human bronchial epithelial (NHBE) cells. The NHBE cells (Lonza, Basel, Switzerland) were cultured on an air-liquid interface in 12-well plates as described previously (33). The H275Y mutant A/Denmark/528/09 virus and wild-type A/Denmark/524/09 virus were premixed at 1:1, 1:4, and 4:1 ratios based on their PFU titers. Four-week-differentiated NHBE cells were infected with the mixed viruses at an estimated multiplicity of infection (MOI) of 0.001 PFU/cell. Briefly, 300 μl of mixed viruses was added to the apical surface of cells and was removed after 1 h of incubation at 37°C. One or one-half milliliter of BEGE medium (Lonza) was then added to the basolateral reservoirs or apical surface, respectively, for further culture. Apical supernatants containing progeny viruses were collected at 12 h and 24 h postinfection (p.i.) and stored at −70°C for further SNaPshot assay or titration in MDCK cells.
Statistical analysis.
The unpaired t test or analysis of variance (ANOVA) was used for all comparisons. All nonlinear and linear regression was performed by using GraphPad Prism 4 software (La Jolla, CA).
RESULTS
Extension probes for detection of resistance markers in NA genes.
We first designed probes for detection of the H275Y NA mutation in pandemic or seasonal H1N1 viruses by monoplex SNaPshot assay. The forward and reverse probes were designed to provide two independent assessments, especially when the ratios of wild-type and mutant genotypes were to be analyzed (see below) (Table 1). To obtain consistent quantitative data, the optimal size of extension probes was 25 to 45 bp. Smaller probes would cause irregular migration of extension products, resulting in broader, asymmetrical peaks or shifted peaks (data not shown). We also designed probes for simultaneous detection of multiple resistance markers in multiplex SNaPshot assays. The following resistance markers identified in clinical isolates were included in the multiplex assays: H275Y and I223V/R in 2009 pandemic H1N1 viruses, E119V and R292K in seasonal H3N2 viruses, and H275Y and N295S in avian H5N1 viruses (Table 2). Shorter PCR amplicons (∼100 to ∼300 bp) and longer probes (>20 bp) were utilized to increase probe binding specificity.
Image profiles obtained from SNaPshot assay.
The results of SNaPshot assays are presented as images with colored peaks (Fig. 1 and 2). In typical image profiles, the specificity of probes is discriminated by their size (location of bands on the x axis) relative to a set of internal size standards. The genotype of each extended probe is differentiated by the color of each peak, which is determined by the fluorescent dye assignment of the ddNTPs incorporated into the probe. The fluorescence intensity of each signal is indicated as the peak height (y axis) or the area under the peak; the peak heights were used for all analyses in this study.
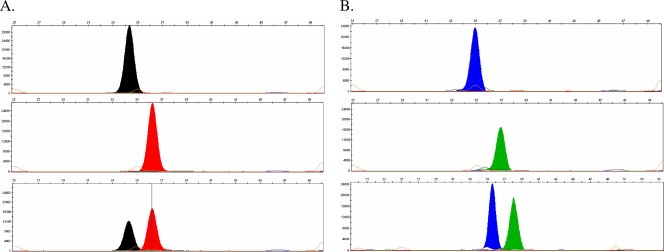
Fig. 1.
Representative images of monoplex SNaPshot assay for H275Y in the pandemic H1N1 virus. (A) The NA275 forward probe (pPdmN1-275-F) detected a single black peak (C genotype) in a homogeneous H275 wild-type sample (upper panel), a single red peak (T genotype) in a homogeneous H275Y mutant sample (middle panel), and dual peaks (C and T genotypes) in a mixed H275 + H275Y sample (1:1 ratio) (lower panel). (B) The NA275 reverse probe (pPdmN1-275-R) detected a single blue peak (G genotypes, upper panel), a single green peak (A genotype, middle panel), and dual peaks (G and A genotype, lower panel) in homogeneous H275 and H275Y samples and a mixed H275 + H275Y sample (1:1 ratio), respectively. In all images, the x axis represents relative capillary mobility determined by a set of sizing standards and the y axis represents relative fluorescence signal intensity.
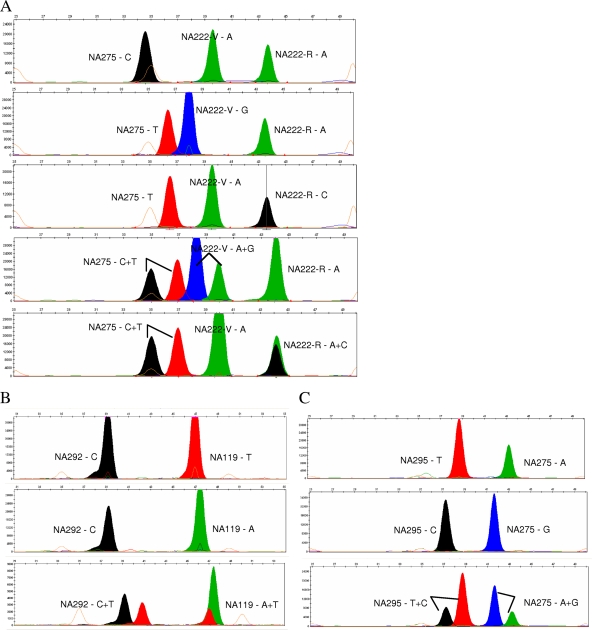
Fig. 2.
Representative examples of detection of resistance markers in influenza A viruses by multiplex SNaPshot assay. Image files of multiplex SNaPshot assays are presented as colored bands at different positions. The probe specificity is determined by its position in capillary electrophoresis based on its size (horizontal axis), and the genotype extended at each probe is determined by its color code. (A) Triplex SNaPshot assay for H275Y, I222V, and I222R markers in pandemic H1N1 viruses detected (from top to bottom) a wild-type H275 + I222 sample, a double mutant H275Y + I223V sample, a double mutant H275Y + I223R sample, and a mixed sample of wild-type and both double mutants. (B) Duplex SNaPshot assay for detection of R292K and E119V/I markers in seasonal H3N2 viruses detected (from top to bottom) a wild-type R292 + E119 sample, a single mutant R292 + E119V sample, and a mixed wild-type, R292K, and E119V sample. (C) Duplex SNaPshot assay for detection of H275Y and N295S markers in H5N1 viruses detected (from top to bottom) a wild-type N295 + H275 sample, a double mutant N295S + H275Y sample, and a mixed wild-type and double mutant sample.
Figure 1 shows the results of H275Y monoplex assays of pandemic H1N1 virus using both a forward and a reverse probe. The forward probe detected the single base C or T in the homogeneous wild-type or H275Y mutant samples and detected both C and T in a mixture of wild-type and H275Y mutant viruses (Fig. 1A). When the reverse probe was applied, the complementary G/A signal were detected instead of the C/T signal at the H275Y site (Fig. 1B). Representative results of the three sets of multiplex assays (for pandemic H1N1, seasonal H3N2, and avian H5N1 viruses) were also demonstrated in homogeneous wild-type or mutant samples and in mixed samples (Fig. 2). A triplex assay was designed to detect not only the H275Y mutation but also the I223V/R mutation in pandemic H1N1 viruses (Fig. 2A), which was recently reported to confer multidrug resistance (36).
Sensitivity of detection of resistant markers by SNaPshot assay.
To test the detection limits of the SNaPshot assay, we generated concentration-response curves for the monoplex assay of the 275 site of pandemic H1N1 viruses using the forward and reverse probes (Fig. 3A and B). All four fluorescence signals from the extended probes showed a typical sigmoid concentration-response pattern: a plateau of minimal response at low concentrations, a plateau of maximal response at high concentrations, and a relatively linear range between the two plateaus. The lower plateaus of all four curves occurred at approximately ∼800 to ∼1,000 relative fluorescence units (RFU), which corresponded to concentrations of ∼0.05 to ∼0.1 ng/μl template DNA (∼300-bp PCR product). The results indicated that the monoplex SNaPshot assay detects low concentrations of templates with high sensitivity. The concentration-response curves of the monoplex assay of the 275 site in seasonal H1N1 viruses showed similar curves with similar sensitivities (data not shown). The detection limit of the multiplex assays was reduced to approximately 0.1 to 1 ng/μl template DNA (data not shown), probably because a longer DNA template had to be used to cover more sites of interest; the increased probability of secondary structure in longer templates may reduce the accessibility of the binding sites to the probes.
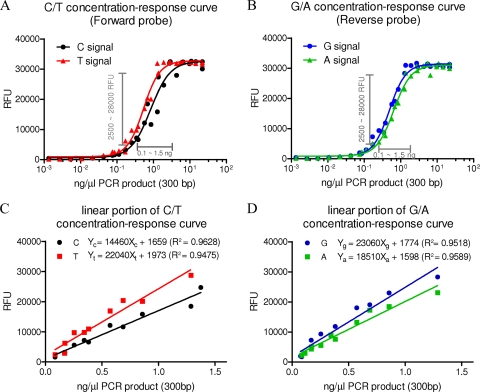
Fig. 3.
Concentration-response curves generated from H275Y monoplex assay. (A and B) Signal responses (in relative fluorescence units [RFU]) of C and T genotypes acquired by using the forward probe (pPdmN1-275-F) (A) and of G and A genotypes acquired by using the reverse probe (pPdmN1-275-R) (B) for the H275Y marker in 2009 pandemic H1N1 viruses; the linear concentration-response range is indicated. (All data points represent the average responses from duplicate determinations.) (C and D) The linear portions of the concentration-response curve are plotted on a decimal scale, and linear curve fitting is derived. R2 indicates the goodness of fit.
The performance of each mono- or multiplex SNaPshot assay for detection of resistance markers in influenza viruses was validated against a panel of pandemic H1N1, seasonal H1N1, and H3N2 virus isolates with defined genotypes (see Table S1 in the supplemental material); the results were 100% concordant with the previously defined genotypes when the template DNA of a concentration above 1 ng/μl was used. Of note, in our experiments, when the concentrations of NA templates were used below the lower detection limit, false-positive or false-negative results together occurred in around 50% of all tested samples (n > 20).
Algorithm for relative quantification of viral genotypes by SNaPshot assay.
Because each concentration-response curve had an essentially linear portion, we used the linear range to quantify the concentration of each genotype on the basis of signal intensity. The portion between 2,500 and 28,000 RFU and the corresponding concentrations of ∼0.1 to ∼1.5 ng/μl DNA in the curves were used for linear curve fitting, and the graphs and equations of the function Y = aX + b for all four signals were acquired (Fig. 3C and D). The equations can be used to calculate the concentration of each genotype present in templates. In most cases when a mixture is detected only, the relative proportion (the ratio of wild-type to resistant genotype) is of most interest; therefore, two equations were converted into a single formula expressing the ratio of two genotypes in units of the two signal responses. For example, to calculate the concentration ratios of wild-type and H275Y mutant genotypes in a mixture from signals within the linear range (2,500 to 28,000 RFU), we used the formula
for results from the forward probe and
for results from the reverse probe.
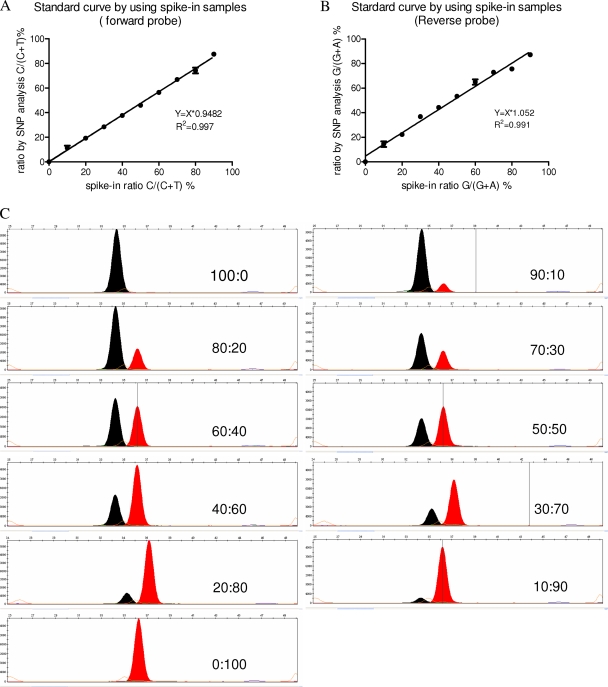
To assess the precision and accuracy of quantification of the genotypes by the SNaPshot assay, we tested a series of standard mixtures with different ratios of wild-type and H275Y mutant templates. Both the forward and reverse probes and their corresponding formulas were used to provide two independent assessments of each mixed sample and were compared to the original spike-in ratios. The results showed excellent correlation between the spike-in ratios and the detected ratios by both probes in the SNaPshot assay (R2 > 0.99) (Fig. 4A and B), suggesting the high accuracy of the method. The intra-assay (n = 3) and interassay (n = 2 or 3) variability was small (standard deviations [SD] = 1.2% and 3.6%, respectively), suggesting great reproducibility of the method. The signal images of the 11 standard mixtures acquired by the forward probe showed that the C signal peak height progressively increased as the proportion of wild-type template increased in the mixture, and vice versa for the T signal peak (Fig. 4C). Additional experiments using samples with lower abundance of either genotype showed that the SNaPshot assay could detect a minor population as small as ∼2% to ∼5% in a mixture, depending on the concentration of template, although multiple dilutions of templates were required to get both signals within the linear range (data not shown).
Fig. 4.
Relative quantification of viral genotypes in a set of mixture standards with different ratios of wild-type and H275Y NA amplicons of pandemic H1N1 virus. (A and B) The ratios of mixture standards were detected by using the forward and reverse probes, respectively (data represent means ± SD of results from triplicate determinations); the correlation between the detected and spike-in ratios was measured by linear curve fitting. (C) Representative images of the mixture standards using the forward probes; the spike-in ratios (wild type:H275Y mutant) are indicated.
It should be noted that the ratios derived from the above formulas technically represented the concentration ratios only of the two genotypes presented in the SNaPshot reaction. The concentration ratios of the templates approximate their molar ratios because the templates of two genotypes are different at only one nucleotide. From RT-PCR amplification to probe extension, the reaction efficiency is assumed to be equivalent for all templates presented in the same reaction; therefore, the final molar ratio of the two genotypes should be the same as the molar ratio of their RNA/DNA copies in the original sample prior to the reaction. Thus, theoretically, the detected concentration ratio of the two genotypes represents the molar ratio of viral RNA copies of the two genotypes. It is also worth noting that for a given probe and protocol, the concentration-response curve for each signal is assay specific, so the curves and corresponding linear equations must be generated separately for each mutation genotype.
Comparison of relative quantification by SNaPshot assay versus pyrosequencing.
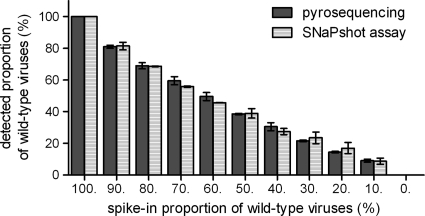
To compare the accuracy of relative quantification of two genotypes by SNaPshot with that by an established method, we performed pyrosequencing and SNaPshot assays of the same serial mixture standards containing different ratios of wild-type and H275Y mutant NA amplicons of H1N1 pandemic virus. There was no significant difference between the results acquired by the two methods (Fig. 5), and the detected ratios by both methods showed good correlation with the spike-in ratios (R2> 0.9) (Fig. 5). Thus, the relative quantification of the two genotypes by the SNaPshot assay had the same accuracy and efficiency as allele quantification by pyrosequencing.
Fig. 5.
The relative quantification of viral genotypes of by SNaPshot assay and by pyrosequencing. A set of mixture standards with different ratios of wild-type and H275Y NA amplicons of pandemic H1N1 viruses was generated. The ratios detected by SNaPshot assay and by pyrosequencing are indicated and compared (data represent means ± SD of results from triplicate determinations).
Detection of resistant subpopulations by SNaPshot assay versus NA inhibition assay.
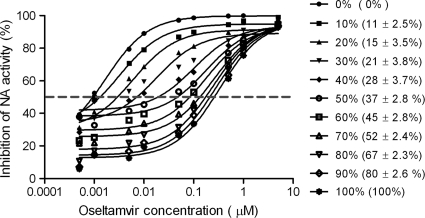
We next compared the SNaPshot assay with the phenotypic NA inhibition assay for detection of oseltamivir-resistant populations in mixed samples. In NA inhibition assays of the viral mixtures, the inhibition curves gradually shifted to higher concentrations as the NA activity of the mutant virus in the mixtures increased, leading to increasing apparent IC50s of the mixtures (Fig. 6). The shifts of the apparent IC50s were most marked when either subpopulation was greater than 20%. When a minor subpopulation was <20% in the mixture, the IC50s did not change sufficiently to detect it. In contrast, SNP analysis was able to differentiate and quantify the minor subpopulations in all mixtures, and the detected ratios of the two genotypes were consistent with the mixed NA activity ratios (Fig. 6). Therefore, genotypic SNP analysis detected minor populations in mixtures with greater efficiency than did the phenotypic NA inhibition assay. The results also showed that the apparent IC50s of the mixtures were dependent on the NA activity contributed by wild-type and resistant viruses as well as on the copies of each virus.
Fig. 6.
Detection of resistant subpopulations by SNaPshot and NA inhibition assays. Apparent IC50s of mixed viral samples were determined by NA inhibition assay (all data points represent the average response of duplicate determinations). Percentages represent the contribution of NA activity from H275Y mutant viruses to a total 4,000 RFU of NA activity in mixed wild-type and H275Y mutant samples. Percentages in parentheses indicate the proportion of H275Y genotype in the same mixed viral samples detected by the SNP analysis (data represent means ± SD of results from triplicate determinations).
Application of SNP analysis to assess the relative growth fitness of resistant virus.
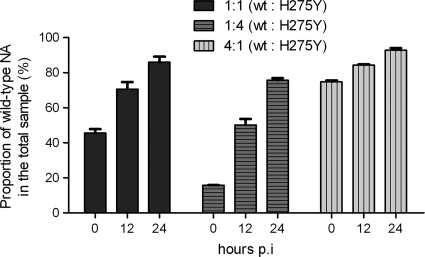
We used the quantitative SNaPshot assay to evaluate the competitive growth fitness of an oseltamivir-resistant pandemic virus. A pair of pandemic H1N1 viruses, one wild type (A/Denmark/524/2009) and one H275Y mutant (A/Denmark/528/2009), were characterized by full-genome sequencing and showed only H275Y NA mutation between two viruses (8). To evaluate the effect of the H275Y mutation on the replication fitness of the virus, we inoculated differentiated NHBE cells with mixed viruses containing three different ratios of wild-type to mutant virus (1:1, 1:4, 4:1 by PFU titer) and used the SNaPshot assay to examine the proportions of each genotype in the progeny viruses. The results showed that the wild-type viruses had outgrown the resistant mutant viruses by 24 h p.i. regardless of their original ratio in the inocula (Fig. 7). This result confirmed the lower replication fitness of the H275Y mutant pandemic virus in host cells.
Fig. 7.
Comparison of viral growth fitness by quantitative SNP analysis. Differentiated NHBE cells were coinoculated with three different ratios of wild-type and H275Y mutant pandemic H1N1 virus (1:1, 1:4, 4:1 by PFU titer). Quantitative SNP analysis was used to detect the ratio of wild-type and H275Y mutant progeny viruses collected at different times postinoculation (data represent means ± SD of results from triplicate determinations).
DISCUSSION
We have described and validated a novel SNP analysis for rapid detection and quantification of NA inhibitor resistance markers in 2009 pandemic H1N1, contemporary seasonal H1N1 and H3N2, and highly pathogenic avian H5N1 influenza viruses. This method allows multiplex assays for detection of multiple resistance markers in different influenza virus subtypes and monoplex assays for relative quantification of H275Y genotypes in H1N1 viruses. Detection and quantification of the resistance markers by the SNP analysis were highly sensitive and reliable for low-content DNA template and low abundance of a genotype in the mixtures. The method was successfully applied to analyze mixed viral populations.
To our knowledge, our study is the first attempt to apply SNP analysis for detection and quantification of drug resistance markers in respiratory viruses. The SNaPshot single-nucleotide extension assay has been a valuable tool for interrogating multiple genomic loci in human and animal genomes (4, 12, 19, 21, 27). However, the quantification capability of the SNaPshot assay was not investigated in these studies because human genome in somatic cells can be only either homozygous or heterozygous. The assay had previously been described only once in the virology literature, to detect two drug resistance markers in the HIV protease gene; this study also explored the quantitative analysis of different genotypes (14). In this study, concentration-response curves of different genotypes differed in their kinetics and plateaus, and the ratios of different genotypes were calculated based on the normalization of signal intensity of G, A, and T relative to C at certain concentrations. In our study, the sigmoid concentration-response curves of different genotypes differed only in their kinetics but had comparable plateaus. The linear portion of each curve was utilized to derive a linear equation for calculation of the concentration of each genotype, and a pair of linear equations was converted for relative quantification of the two genotypes within a wide range of amplicon concentrations and signal responses. The differences between the two studies suggest that the concentration-response curves by SNaPshot assay may be highly assay specific, depending on the probes, templates, and equipment that are used. For quantification purposes, the assay-specific concentration-response curve should be generated individually for each marker. Importantly, because the four fluorescent dyes are fundamentally different in their emission capacity, the intensities of their signals should never be directly compared to calculate the ratios of the genotypes. The fluorescence signals must always be normalized. To do this, we used the equations described; another study used the relative ratios of the different signals (14).
A novel method must always be compared with other established methods. We found that the H275Y monoplex assay could detect a minor subpopulation of ∼2% to ∼5% in a mixture, which was more sensitive than the ∼10% to ∼20% level detected by commonly used Sanger sequencing (empirical detection level) and also provided the advantages of a clean background and easy discrimination. The accuracy and precision of relative quantification by SNP analysis were also comparable to those of pyrosequencing. We also compared the detection of subpopulations by genotypic SNP analysis with that by phenotypic NA inhibition assay. It remains unclear how the apparent IC50s are affected by the proportions of drug-resistant subpopulations in mixtures, which may be dependent on different resistance markers (39). In our mixtures containing an H275Y mutant pandemic virus, the apparent IC50s gradually shifted upward as the proportion of resistant virus increased, but the test could not reliably detect a minor subpopulation of <∼10% to ∼20%. The SNP analysis, which can quantify a minor subpopulation in a mixture, can be used for further validation when the results of NA inhibition assays are ambiguous.
The quantitative analysis of genotypes presented in mixed samples is an appealing feature of the SNP analysis and can provide a useful tool for viral quasispecies analysis. Since the method can be easily adapted for detection of other influenza virus pathogenesis or lineage markers that are caused by single nucleotide mutations, it can be applied to analyze viral quasispecies generated by any mutations of interest in clinical or laboratory samples. We used the quantitative SNP analysis to evaluate the fitness of two viral populations in competitive growth assays and found that the H275Y mutant resistant pandemic virus had lower growth fitness than the wild-type virus in ferrets (8) or NHBE cells. The SNP analysis is also useful to confirm the samples mixed by infectivity titer, functional activity, etc. The confirmation of artificially mixed samples could be important, as viral infectivity titers are not always accurate due to the dilution process of infectivity assays.
The influenza virus SNP analysis is compared with other commonly used genotypic methods in terms of time and reagent costs, labor intensity and equipment availability, etc. (see Table S2 in the supplemental material). The SNP analysis offers several advantages that may encourage its wider laboratory use in the future. First, the method provides easy high-throughput capacity for screening of resistance markers in clinical samples. The SNaPshot assay involves only a single PCR procedure followed by a probe extension reaction. All procedures can be performed in 96-well microtiter plates in a mid- or high-throughput manner. Second, its multiplex feature is helpful for screening multiple targets simultaneously. In the present study, we developed probes only for the resistance markers identified in clinical isolates, but the SNaPshot assay can be readily multiplexed for as many as 10 SNP sites in a single reaction, further increasing its high-throughput potential. Third, SNaPshot assays require no expensive labeled primer or probes and therefore are relatively cost-effective. Finally, SNaPshot assays require only a capillary sequencer, which is widely available and can be automated. The main limitation of the SNP analysis is that it can detect only known resistance markers; this limitation is common to most genotypic methods, such as TaqMan real-time PCR-based assays.
In summary, our influenza virus SNP analysis relies on two key features: (i) the fact that all known NA inhibitor resistance markers result from SNPs in the codons of certain critical residues and (ii) the chemical principle of a template-directed single nucleotide extension assay. With its advantages in costs, screening capacity, and the availability of the necessary equipment, this genotypic method could provide a valuable tool for monitoring NA inhibitor resistance-associated mutations in circulating influenza viruses. Such monitoring is essential for public health, as NA inhibitors remain the primary therapeutic option for influenza infection.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by contract no. HHSN266200700005C from the National Institute of Allergy and Infectious Diseases and by the American Lebanese Syrian Associated Charities (ALSAC).
We thank Sharon Naron for editorial assistance. The NA inhibitor oseltamivir carboxylate was provided by F. Hoffmann La Roche, Ltd. (Basel, Switzerland).
While this study did not utilize corporate funding, Elena A. Govorkova and Robert G. Webster are currently performing a different study funded by F. Hoffmann-La Roche, Ltd., Basel, Switzerland. We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 5 July 2011.
REFERENCES
- 1. Abed Y., Baz M., Boivin G. 2006. Impact of neuraminidase mutations conferring influenza resistance to neuraminidase inhibitors in the N1 and N2 genetic backgrounds. Antivir. Ther. 11:971–976 [PubMed] [Google Scholar]
- 2. Bautista E., et al. 2010. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N. Engl. J. Med. 362:1708–1719 [DOI] [PubMed] [Google Scholar]
- 3. Birnkrant D., Cox E. 2009. The Emergency Use Authorization of peramivir for treatment of 2009 H1N1 influenza. N. Engl. J. Med. 361:2204–2207 [DOI] [PubMed] [Google Scholar]
- 4. Budowle S. A., Gonzalez S., Budowle B., Eisenberg A. J., Grange R. W. 2008. A novel SNaPshot assay to detect the mdx mutation. Muscle Nerve 37:731–735 [DOI] [PubMed] [Google Scholar]
- 5. Deyde V. M., et al. 2009. Detection of molecular markers of antiviral resistance in influenza A (H5N1) viruses using a pyrosequencing method. Antimicrob. Agents Chemother. 53:1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deyde V. M., et al. 2009. Pyrosequencing as a tool to detect molecular markers of resistance to neuraminidase inhibitors in seasonal influenza A viruses. Antiviral Res. 81:16–24 [DOI] [PubMed] [Google Scholar]
- 7. Deyde V. M., et al. 2010. Detection of molecular markers of drug resistance in 2009 pandemic influenza A (H1N1) viruses by pyrosequencing. Antimicrob. Agents Chemother. 54:1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duan S., et al. 2010. Oseltamivir-resistant pandemic H1N1/2009 influenza virus possesses lower transmissibility and fitness in ferrets. PLoS Pathog. 6:e1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duwe S., Schweiger B. 2008. A new and rapid genotypic assay for the detection of neuraminidase inhibitor resistant influenza A viruses of subtype H1N1, H3N2, and H5N1. J. Virol. Methods 153:134–141 [DOI] [PubMed] [Google Scholar]
- 10. Gubareva L. V., et al. 2010. Comprehensive assessment of 2009 pandemic influenza A (H1N1) virus drug susceptibility in vitro. Antivir. Ther. 15:1151–1159 [DOI] [PubMed] [Google Scholar]
- 11. Guo L., et al. 2009. Rapid identification of oseltamivir-resistant influenza A(H1N1) viruses with H274Y mutation by RT-PCR/restriction fragment length polymorphism assay. Antiviral Res. 82:29–33 [DOI] [PubMed] [Google Scholar]
- 12. Hurst C. D., Zuiverloon T. C., Hafner C., Zwarthoff E. C., Knowles M. A. 2009. A SNaPshot assay for the rapid and simple detection of four common hotspot codon mutations in the PIK3CA gene. BMC Res. Notes 2:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hurt A. C., et al. 2011. Oseltamivir-resistant influenza viruses circulating during the first year of the influenza A(H1N1) 2009 pandemic in the Asia-Pacific region, March 2009 to March 2010. Euro Surveill. 16(3):pii=19770 [PubMed] [Google Scholar]
- 14. Ibe S., Fujisaki S., Fujisaki S., Morishita T., Kaneda T. 2006. Quantitative SNP-detection method for estimating HIV-1 replicative fitness: application to protease inhibitor-resistant viruses. Microbiol. Immunol. 50:765–772 [DOI] [PubMed] [Google Scholar]
- 15. Ison M. G., Gubareva L. V., Atmar R. L., Treanor J., Hayden F. G. 2006. Recovery of drug-resistant influenza virus from immunocompromised patients: a case series. J. Infect. Dis. 193:760–764 [DOI] [PubMed] [Google Scholar]
- 16. Kiso M., et al. 2004. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364:759–765 [DOI] [PubMed] [Google Scholar]
- 17. Lackenby A., Thompson C. I., Democratis J. 2008. The potential impact of neuraminidase inhibitor resistant influenza. Curr. Opin. Infect. Dis. 21:626–638 [DOI] [PubMed] [Google Scholar]
- 18. Le Q. M., et al. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437:1108. [DOI] [PubMed] [Google Scholar]
- 19. Li L., et al. 2011. Simultaneous detection of CYP3A5 and MDR1 polymorphisms based on the SNaPshot assay. Clin. Biochem. 44(5–6):418–422 [DOI] [PubMed] [Google Scholar]
- 20. Liu C. M., et al. 2010. Rapid quantification of single-nucleotide mutations in mixed influenza A viral populations using allele-specific mixture analysis. J. Virol. Methods 163:109–115 [DOI] [PubMed] [Google Scholar]
- 21. Lou C., et al. 2011. A SNaPshot assay for genotyping 44 individual identification single nucleotide polymorphisms. Electrophoresis 32:368–378 [DOI] [PubMed] [Google Scholar]
- 22. McKimm-Breschkin J., et al. 2003. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob. Agents Chemother. 47:2264–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mehta T., et al. 2010. Detection of oseltamivir resistance during treatment of 2009 H1N1 influenza virus infection in immunocompromised patients: utility of cycle threshold values of qualitative real-time reverse transcriptase PCR. J. Clin. Microbiol. 48:4326–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moscona A. 2005. Neuraminidase inhibitors for influenza. N. Engl. J. Med. 353:1363–1373 [DOI] [PubMed] [Google Scholar]
- 25. Moscona A. 2005. Oseltamivir resistance—disabling our influenza defenses. N. Engl. J. Med. 353:2633–2636 [DOI] [PubMed] [Google Scholar]
- 26. Moscona A. 2009. Global transmission of oseltamivir-resistant influenza. N. Engl. J. Med. 360:953–956 [DOI] [PubMed] [Google Scholar]
- 27. Murphy K. M., et al. 2003. A single nucleotide primer extension assay to detect the APC I1307K gene variant. J. Mol. Diagn. 5:222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nguyen H. T., Sheu T. G., Mishin V. P., Klimov A. I., Gubareva L. V. 2010. Assessment of pandemic and seasonal influenza A (H1N1) virus susceptibility to neuraminidase inhibitors in three enzyme activity inhibition assays. Antimicrob. Agents Chemother. 54:3671–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nukiwa N., et al. 2010. Simplified screening method for detecting oseltamivir resistant pandemic influenza A (H1N1) 2009 virus by a RT-PCR/restriction fragment length polymorphism assay. J. Virol. Methods 170:165–168 [DOI] [PubMed] [Google Scholar]
- 30. Okomo-Adhiambo M., et al. 2010. Detection of E119V and E119I mutations in influenza A (H3N2) viruses isolated from an immunocompromised patient: challenges in diagnosis of oseltamivir resistance. Antimicrob. Agents Chemother. 54:1834–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Okomo-Adhiambo M., et al. 2010. Host cell selection of influenza neuraminidase variants: implications for drug resistance monitoring in A(H1N1) viruses. Antiviral Res. 85:381–388 [DOI] [PubMed] [Google Scholar]
- 32. Operario D. J., Moser M. J., St. George K. 2010. Highly sensitive and quantitative detection of the H274Y oseltamivir resistance mutation in seasonal A/H1N1 influenza virus. J. Clin. Microbiol. 48:3517–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oshansky C. M., Barber J. P., Crabtree J., Tripp R. A. 2010. Respiratory syncytial virus F and G proteins induce interleukin 1alpha, CC, and CXC chemokine responses by normal human bronchoepithelial cells. J. Infect. Dis. 201:1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Potier M., Mameli L., Belisle M., Dallaire L., Melancon S. B. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal. Biochem. 94:287–296 [DOI] [PubMed] [Google Scholar]
- 35. Roberts N. A., Govorkova E. A. 2009. The activity of neuraminidase inhibitor oseltamivir against all subtypes of influenza viruses, p. 93–118 In Mitrasinovic P. M. (ed.), Global view of the fight against influenza. Nova Science Publishers, Hauppauge, NY [Google Scholar]
- 36. van der Vries E., Stelma F. F., Boucher C. A. 2010. Emergence of a multidrug-resistant pandemic influenza A (H1N1) virus. N. Engl. J. Med. 363:1381–1382 [DOI] [PubMed] [Google Scholar]
- 37. Wang B., et al. 2010. Detection of the rapid emergence of the H275Y mutation associated with oseltamivir resistance in severe pandemic influenza virus A/H1N1 09 infections. Antiviral Res. 87:16–21 [DOI] [PubMed] [Google Scholar]
- 38. Weinstock D. M., Zuccotti G. 2009. The evolution of influenza resistance and treatment. JAMA 301:1066–1069 [DOI] [PubMed] [Google Scholar]
- 39. Wetherall N. T., et al. 2003. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: report of the neuraminidase inhibitor susceptibility network. J. Clin. Microbiol. 41:742–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. WHO 2010. WHO guidelines for pharmacological management of pandemic (H1N1) 2009 influenza and other influenza viruses. WHO, Geneva, Switzerland: http://www.who.int/csr/resources/publications/swineflu/h1n1_use_antivirals_20090820/en/index.html [PubMed] [Google Scholar]
- 41. Yen H. L., et al. 2007. Neuraminidase inhibitor-resistant recombinant A/Vietnam/1203/04 (H5N1) influenza viruses retain their replication efficiency and pathogenicity in vitro and in vivo. J. Virol. 81:12418–12426 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.