Abstract
Capreomycin and the structurally similar compound viomycin are cyclic peptide antibiotics which are particularly active against Mycobacterium tuberculosis, including multidrug resistant strains. Both antibiotics bind across the ribosomal interface involving 23S rRNA helix 69 (H69) and 16S rRNA helix 44 (h44). The binding site of tuberactinomycins in h44 partially overlaps with that of aminoglycosides, and they share with these drugs the side effect of irreversible hearing loss. Here we studied the drug target interaction on ribosomes modified by site-directed mutagenesis. We identified rRNA residues in h44 as the main determinants of phylogenetic selectivity, predict compensatory evolution to impact future resistance development, and propose mechanisms involved in tuberactinomycin ototoxicity, which may enable the development of improved, less-toxic derivatives.
INTRODUCTION
Tuberculosis has been declared a global public health emergency by the World Health Organization. In 2000, worldwide there were about 9 million new cases and 2 million deaths due to tuberculosis (10). The emergence of multidrug-resistant (MDR) tuberculosis has further complicated treatment and control of the disease (1). Capreomycin and viomycin belong to the tuberactinomycins, an important class of antibiotics with activities against multidrug-resistant tuberculosis (12).
Tuberactinomycins are cyclic peptide antibiotics (for chemical structures, see Fig. S1 in the supplemental material) which target bacterial protein synthesis by binding to the well-conserved intersubunit bridge B2a, formed by interaction between helix 69 (H69) of the 23S rRNA and helix 44 (h44) of the 16S rRNA (Fig. 1A to C) (34). Inhibition of translocation during peptide elongation is the main mechanism of drug action mediating the compound's antibacterial activity (24, 26). Mutational alterations of nucleotides in 23S or 16S rRNA affect drug binding (8, 13, 18, 20, 37). In addition, the loss of 2′-O-methylation of C1920 (rRNA nucleotides are numbered according to those for Escherichia coli throughout the paper) in H69 and that of C1409 in h44 by TlyA reduces susceptibility to capreomycin (17, 21). Interestingly, Thermus thermophilus TlyA modifies only C1920 in H69 of 23S rRNA, but not C1409 in h44 of 16S rRNA. Inactivation of tlyA in T. thermophilus does not affect its sensitivity to capreomycin (25), suggesting that modification of C1409 is the relevant determinant of increased drug susceptibility due to 2′-O-methylation.
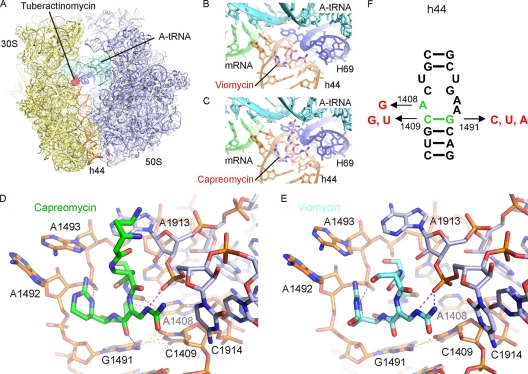
Fig. 1.
Binding site of tuberactinomycins viomycin and capreomycin. (A) Overview of the binding site of the tuberactinomycins (red) at the interface between the small (30S) (yellow) and large (50S) (blue) ribosomal subunits (37). A-tRNA (cyan), P-tRNA (green), and h44 (orange) are in color for reference. (B and C) Enlargement of panel A to show binding site of viomycin (B) and capreomycin (C) to h44 (orange) of the 30S and H69 (blue) of the 50S, with the mRNA and A-tRNA in green and cyan, respectively. (D and E) Interaction of the tuberactinomycins within the ribosomal binding site. Capreomycin (D) (green) and viomycin (E) (cyan) form hydrogen bond interactions (magenta) with h44 of the 16S rRNA (orange) and H69 of the 23S rRNA (blue) (37). Intramolecular hydrogen bonds between rRNA nucleotides are yellow. (F) Secondary structure of decoding site rRNA sequences in the small ribosomal subunit. Nucleotides shown in green represent residues that were exchanged to residues shown in red. rRNA nucleotides are numbered according to the bacterial nomenclature; i.e., to homologous E. coli 16S rRNA positions.
Much of the unwanted side effects of ribosomal antibiotics reflects limitations in selectivity (2, 3), as has been demonstrated for chloramphenicol and linezolid (23, 38). Together with the structurally unrelated class of aminoglycoside antibiotics, the tuberactinomycins share ototoxicity (irreversible loss of hearing) as a common and unique side effect (7, 35).
Tuberactinomycins affect bacterial protein synthesis apparently by stabilizing peptidyl-tRNA in the pretranslocation complex, preventing translocation of the ribosome along the mRNA (11, 19, 24, 26). The recent crystal structure of the 70S ribosome from T. thermophilus in complex with viomycin or capreomycin at a 3.3- to 3.5-Å resolution has delineated important details on drug binding and mechanisms of translocation inhibition (Fig. 1A to C) (34). Based on the structural information available, we set out to characterize the drug target interaction in molecular detail. Specifically, we investigated structure-function relationships by assessing the compounds' determinants of specificity, selectivity, and resistance development.
MATERIALS AND METHODS
Bacterial strains and DNA techniques.
Mycobacterium smegmatis strains mc2 155 Sms ΔrrnB and mc2 155 Sms ΔrrnB ΔrrnA attB::rrnB were used for all experiments. Strategies used to generate the isogenic mutants in this study have been described in detail elsewhere, including (i) transformation with plasmids carrying the rRNA alteration of interest and subsequent integration into the chromosomal rrnA operon by means of homologous recombination (27, 28) and (ii) gene replacement by plasmid exchange (15). Successful gene replacement was controlled by sequence analysis. For a detailed list of plasmids and strains used, see Tables S1 and S2 in the supplemental material.
Clinical strains of Mycobacterium tuberculosis were obtained from the Diagnostic Department, Institute of Medical Microbiology, University of Zurich. Drug-resistant derivatives of Mycobacterium bovis BCG were generated by selective plating.
Isolation and purification of ribosomes.
Ribosomes were purified from bacterial cell pellets by sucrose gradient (10 to 40%) centrifugation as described previously (6). Ribosome concentrations of 70S were determined by absorption measurements on the basis of 23 pmol ribosomes per A260 unit. Integrity and functional activity of purified 70S ribosomes were determined by analytical ultracentrifugation and by assessing their capacities to form initiation complexes (6).
Cell-free luciferase translation assays.
Purified 70S hybrid ribosomes and in vitro-transcribed mRNA were used in translation reactions. Firefly luciferase mRNA was produced using T7 RNA polymerase in vitro. A typical translation reaction mixture contained 0.25 μM 70S ribosomes, 500 ng mRNA, 40% (vol/vol) M. smegmatis S100 extract, 200 μM amino acid mixture, 0.3 μl 50× protease inhibitor cocktail (Roche) and RNasin (40 units; Promega), 0.4 mg/ml tRNAs, and 6 μl commercial S30 premix without amino acids (Promega). The total volume of the reaction mixture was 15 μl. The reaction mixture was incubated at 37°C and stopped on ice. Finally, 100 μl luciferase assay substrate (Promega) was added and the bioluminescence was measured in a luminometer (FLx800; Bio-Tek Instruments).
Misreading was assessed in a gain-of-function assay. We introduced Arg245 (CGC near-cognate) into firefly luciferase to replace the functionally important amino acid His245 (CAC codon). As a negative control we introduced the Arg245 noncognate AGA codon, as ribosomal misreading affects decoding of near-cognate codons but not that of noncognate codons (30). Firefly (F-luc) and Renilla luciferase (R-luc) mRNAs were produced using T7 polymerase in vitro on templates of modified plasmids pGL4.14 (F-luc) and pGL4.75 (R-luc) (both Promega). Mutant 245 F-luc and wild-type (wt) F-luc mRNA was used in in vitro translation reactions, along with R-luc mRNA serving as an internal control. We quantified misreading by calculating mutant firefly/Renilla luciferase compared to wild-type firefly/Renilla luciferase activity. A typical translation reaction mixture contained 0.25 μM 70S ribosomes, 4 μg firefly (F-luc) mRNA, 0.4 μg Renilla luciferase (R-luc) mRNA, 40% (vol/vol) M. smegmatis S100 extract, 200 μM amino acid mixture, 0.6 μl 50× protease inhibitor cocktail (Roche) and RNasin (80 units; Promega), 0.4 mg/ml tRNAs, and 12 μl commercial S30 premix without amino acids (Promega). The total volume of the reaction mixture was 30 μl. The reaction mixture was incubated at 37°C and stopped on ice. Luciferase activity was measured using a dual-luciferase reporter system (Promega). After incubation, 100 μl F-luc substrate was added and the firefly luciferase bioluminescence was measured. Finally, 100 μl R-luc substrate was added and the Renilla luciferase bioluminescence was measured (FLx800; Bio-Tek Instruments).
Antibiotics.
Capreomycin was obtained from Sigma (catalogue no. C4142) and viomycin was obtained from Tocris Bioscience (catalogue no. 3787).
MICs and fitness assays.
Broth microdilution tests were performed in a microtiter plate format in triplicates for the mutants indicated as described previously (15).
Drug susceptibilities of M. tuberculosis and M. bovis BCG were assessed using the MGIT 960 instrumentation equipped with the TBeXiST software (Becton Dickinson) as described previously (33).
The cost of a resistance mutation was determined by direct competition against the isogenic drug-susceptible parental strain as described previously (14, 32).
Figure preparation.
Figure 1A, B, D, and E was prepared using PyMOL (W. L. DeLano, PyMOL molecular graphics system user's manual, DeLano Scientific, San Carlos, CA) from the atomic coordinates in Protein Data Bank (PDB) accession numbers 3KNL and 3KNM (capreomycin) and 3KNH and 3KNI (viomycin) (37).
RESULTS
Determinants of antibacterial activity and fitness of drug resistance mutations.
To study the relative contribution of individual drug-nucleotide contacts to antibacterial activity, we assessed MICs of recombinant Mycobacterium smegmatis constructed previously (32) with point mutations in the h44 drug-binding pocket introduced by site-directed mutagenesis (Table 1). The point mutations sample viable mutational alterations of 16S rRNA key residues participating in drug binding, i.e., A1408G, C1409U/G, and G1491A/C/U (see Fig. 1F; note that A1408C/U and C1409A mutational alterations confer lethality). 1408A→G has limited effects, as it in part decreases susceptibility to capreomycin (4- to 8-fold increase in MIC compared to that of the wild-type) but not susceptibility to viomycin. In contrast, mutational alterations of base pair C1409-G1491 significantly affect drug susceptibility, in particular alterations which result in a purine-pyrimidine (G1491C/U) or in a pyrimidine-purine (C1409G) switch (relative resistance, 100- to 200-fold).
Table 1.
Activity of tuberactinomycins against mutant bacterial cells
| rRNA | MIC (μg/ml) of: |
|
|---|---|---|
| Viomycin | Capreomycin | |
| wt | 4–8 | 4–8 |
| A1408G | 4–8 | 32–64 |
| C1409U | 16 | 64–128 |
| C1409G | 512–1,024 | ≥1,024 |
| G1491A | 128–256 | 512 |
| G1491C | 256–512 | 1,024 |
| G1491U | 256–512 | ≥1,024 |
| C1409U-G1491A | 16–32 | 64 |
| C1409G-G1491C | 16–32 | 64–128 |
| C1409A-G1491U | 128 | 256–512 |
Mutations C1409U and G1491U are associated with capreomycin resistance in Mycobacterium tuberculosis (13, 18, 20) and confer a significant fitness cost. More recently, it has been observed that a secondary site G1491A alteration restoring base pair interaction between the 1409U mutant and residue 1491 evolves in M. tuberculosis to ameliorate the cost of the 1409U resistance mutation (32). We studied the three possible secondary site mutations in M. smegmatis which would restore 1409-1491 base pairing for any of the resistance mutations affecting residue 1409 or 1491, i.e., interactions 1409G-1491C, 1409U-1491A, and 1409A-1491U (Fig. 2). Restoration of 1409-1491 base pairing in part ameliorated fitness (32) but was still associated with a significant degree of tuberactinomycin resistance (Table 1). Of note, the 1409A-1491U interaction retains a high level of drug resistance similar to that of the 1491U resistance alteration alone, yet the 1409A alteration restores the fitness of the 1491U resistance mutation (cost per generation [cpg], 11.7% ± 5.9%) to wild-type levels (cpg of 1409A-1491U, 0.1% ± 2.9%).
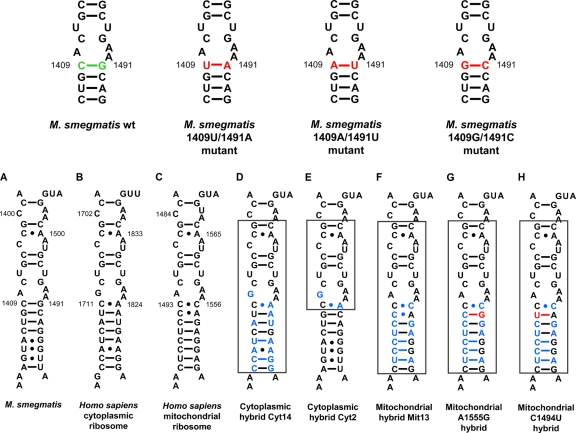
Fig. 2.
(Top) Compensatory mutations in decoding site of bacterial small ribosomal subunit rRNA. Nucleotides shown in green represent residues that were exchanged to residues shown in red. (Bottom) Secondary-structure comparison of decoding site rRNA sequences in the small ribosomal subunit. (A) Decoding region of 16S rRNA helix 44 in wild-type ribosomes of M. smegmatis; rRNA nucleotides are numbered according to the bacterial nomenclature; i.e., to homologous E. coli 16S rRNA positions. (B) Homologous 18S rRNA sequence in human ribosomes; rRNA residues are numbered according to the human cytoplasmic ribosome nomenclature. (C) Homologous 12S rRNA sequence in human mitochondrial ribosomes; rRNA residues are numbered according to the mitochondrial nomenclature. (D to H) Decoding site rRNA of human-bacterial hybrid ribosomes. The transplanted helix is boxed, and nucleotide positions shown in blue represent residues that are specific for human rRNA. Mutations that are associated with hypersusceptibility to aminoglycoside antibiotics, mitochondrial DNA position 1555A→G (G), and 1494C→U (H) are highlighted in red.
Mechanisms involved in selectivity and toxicity.
The mechanism of action of capreomycin and viomycin involves both ribosomal subunits and thus is unique among the ribosome-targeting antibiotics. To study drug selectivity, we used hybrid ribosomes (14, 15) with the bacterial h44 replaced by the corresponding cytosolic homolog, resulting in bacterial ribosomes with a humanized drug-binding pocket (Fig. 2, cyt14). In comparison to native eukaryotic cytosolic ribosomes (rabbit reticulocytes), this should allow the relative contribution of H69 in 23S rRNA to drug selectivity to be determined. Ribosomal drug susceptibility (inhibition of protein synthesis) was assessed in cell-free luciferase translation assays. Compared to bacterial ribosomes, rabbit reticulocytes demonstrated a 3-log10 difference in drug susceptibility. The 50% inhibitory concentration (IC50) values of bacterial ribosomes with a humanized h44 drug-binding pocket were in the same range as those of rabbit reticulocytes (Table 2). To define the relevant polymorphic residues relevant for phylogenetic specificity more precisely, we further constructed ribosomes with the humanized part of h44 further shortened by mutagenesis to result in bacterial ribosomes with the human 1408G and the human 1491A homolog (cyt2). Drug susceptibility of cyt2 ribosomes was comparable to that of cyt14 ribosomes and rabbit ribosomes (Table 2). From these results, we conclude that the phylogenetic variability within h44, more precisely 16S rRNA residues 1408 and 1491, is sufficient to account for the drug's ability to discriminate between bacterial and eukaryotic cytosolic ribosomes.
Table 2.
Ribosomal susceptibility to tuberactinomycin-induced inhibition of protein synthesis
| Type of ribosomes | IC50 (μM) ofa: |
|
|---|---|---|
| Viomycin | Capreomycin | |
| M. smegmatis bacterial | 0.03 ± 0.0057 | 0.03 ± 0.017 |
| Rabbit reticulocyte | 80 ± 12.6 | 50 ± 4.5 |
| Cytohybrid (cyt14) | 16 ± 0.6 | 52 ± 11.7 |
| 1408G/1491A (cyt2) | 53 ± 4.0 | 110 ± 5.8 |
| Mitohybrid (mit13) | 45 ± 5.8 | 75 ± 10.4 |
| A1555G mutant mitohybrid | 11 ± 2.1 | 13 ± 1.1 |
| C1494U mutant mitohybrid | 12 ± 1.7 | 23 ± 4.0 |
Drug concentrations required to inhibit synthesis of active luciferase to 50% (IC50).
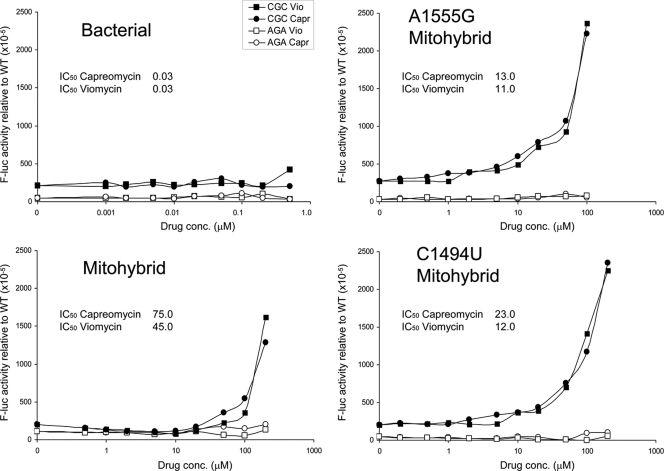
Mitochondrial mistranslation has been identified as an important mechanism contributing to aminoglycoside ototoxicity (14, 16). In addition, aminoglycoside ototoxicity occurs in a genetically inherited form, with drug hypersusceptibility linked to alterations A1555G or C1494U in mitochondrial rRNA (29, 39). Both mutations locate to the mitoribosomes' decoding the A site and replace a C-A non-Watson-Crick interaction with canonical C-G or U-A base pairing, respectively (Fig. 2). Having provided evidence that the large ribosomal subunit contributes little to drug selectivity, we used the hybrid ribosome approach to study the activity of tuberactinomycins against the wild-type mitochondrial h44 drug-binding pocket versus the A1555G and C1494U mutants. In comparison to wild-type mitohybrid ribosomes, the mutant hybrids were found to be significantly more susceptible to tuberactinomycins (Student's t test; P < 0.0001) (Table 2). To study misreading, we established a sensitive gain-of-function assay. We replaced the functionally important amino acid His245 (CAC codon) in the active site of luciferase with the near-cognate CGC codon Arg245. Replacement of His by Arg245 completely eliminates enzymatic activity of the firefly protein, with activity restored by misreading. As a negative control we introduced the Arg245 noncognate AGA codon, as ribosomal misreading affects decoding of near-cognate codons but not that of noncognate codons (30). In contrast to bacterial ribosomes (9, 19), the tuberactinomycin compounds readily induced misreading of the near-cognate CGC codon on mitohybrid ribosomes (Fig. 3). This drug-mediated gain of enzymatic function upon mistranslation of the Arg245 mutant luciferase mRNA was further aggravated in the A1555G and C1494U mutants.
Fig. 3.
Effect of capreomycin and viomycin on misreading during translation elongation. Drug-induced misreading was quantified using the Arg245 (CGC) near-cognate mutant of firefly luciferase (filled circles, capreomycin [Capr]; filled squares, viomycin [Vio]). The Arg245 (AGA) noncognate mutant of firefly luciferase was used as a misreading control (open circles, capreomycin [Capr]; open squares; viomycin [Vio]). Bacterial ribosomes, mitohybrid ribosomes, mutant 1555G mitohybrid ribosomes, and mutant C1494U mitohybrid ribosomes are indicated. Misreading was calculated by comparing mutant firefly/Renilla luciferase activity to wild-type His245 (CAC) firefly/Renilla luciferase activity. The IC50 inhibitory values (drug concentrations which inhibit protein synthesis to 50%) are indicated; for a comparison, see Table 2.
DISCUSSION
Mutations in 16S rRNA positions 1408, 1409, and 1491 are associated with various degrees of resistance to capreomycin and viomycin in M. tuberculosis, while simultaneously conferring in part cross-resistance to the aminoglycosides amikacin and kanamycin (13, 18, 20, 22, 36). Our results refine the role of individual 16S nucleotides in drug binding and resistance (for a comparison to clinical resistant isolates of M. tuberculosis complex and strains generated by selective plating, see Table S3 in the supplemental material). In addition, by modeling compensatory evolution in M. smegmatis we can tentatively predict resistance development in M. tuberculosis. Our results lead us to suggest that the more widespread use of capreomycin may lead to compensatory evolution, in particular by selection for the C1409A-G1491U mutant, allowing for resistance which comes at little cost and which will spread rapidly, as has been observed for streptomycin resistance in M. tuberculosis (4, 5, 31).
Both capreomycin and viomycin bind to the 70S in a cleft formed between H69 of 23S rRNA and h44 of 16S rRNA (Fig. 1). The macrocycle, capreomycidine ring, and ureidodehydroalanine form numerous hydrogen bonds to the ribose phosphate backbone of A1913, C1914 (both 23S rRNA), and A1493 (16S rRNA); side groups of the macrocycle also form contacts to G1491 or A1408 (both 16S rRNA). Despite the close contacts of the tuberactinomycin antibiotics to the phosphate backbone of 23S rRNA residues A1913 and C1914, the polymorphism within h44 of the 16S rRNA is sufficient to account for drug selectivity, leading us to suggest that H69 in the large ribosomal subunit apparently contributes little toward the drug's ability to discriminate between bacterial and eukaryotic cytosolic ribosomes. This is consistent with the high degree of conservation of H69 with A1913 and C1914 being identical between bacteria and eukaryotes. Compared to the effect on viomycin, the effect on capreomycin susceptibility by the 1408A→G mutation is small but significant (relative resistance, 8-fold). This finding is explained by the crystal structure, which suggests two (albeit weak) H bonds between A1408 and capreomycin, but no direct interaction of residue 1408 with viomycin (Fig. 1D and E). As a result of the A1408G substitution, the direct interaction of residue 1408 with capreomycin would be limited to a single H bond. There is one direct contact (H bond) between capreomycin and residue 1491, while viomycin forms no H bonds with base pair 1409-1491. The strong effect of single C1409 or G1491 alterations on tuberactinomycin activity most likely reflects the fact that the base pair forms an important binding surface for the macrocycle ring (Fig. 1D and E), and mutations that break the base pair would induce structural rearrangements that affect drug-ribosome interaction. Restoration of canonical 1409-1491 Watson-Crick base pairing, e.g., 1409G-1491C, recovers the structure of h44 and in part relieves it from the high-level resistance phenotype associated with single C1409 or G1491 alterations.
The functional relevance of rRNA residues in codon-anticodon interaction and stabilization is reflected in the conservation of the small subunit's decoding A site throughout evolution. However, critical variations have evolved between different phylogenetic domains, most prominently in h44 of the 16S rRNA. The binding site for tuberactinomycins in part overlaps that for aminoglycosides in h44 of the small ribosomal subunit. Both classes of antibiotics affect translocation of the tRNA on the ribosome, either by stabilizing the tRNA in the pretranslocation state or by interfering with the conformational changes of the ribosome/tRNA required for translocation (24, 26). Compared to aminoglycosides, the tuberactinomycin antibiotics induce little, if any, misreading on bacterial ribosomes (Fig. 3) (19). Nevertheless, it was observed in the early 1980s that tuberactinomycin antibiotics may have the potential to provoke misreading on bacterial ribosomes when using an in vitro artificial system composed of synthetic homopolynucleotides and noncognate (rather than near-cognate) tRNA as the sole species of aminoacylated tRNA. However, when the system was complemented with additional aminoacylated tRNAs, a strong inhibition of translocation was the prevalent effect with a concurrent loss of misreading (19).
Both tuberactinomycins and aminoglycosides share irreversible hearing loss as a main unwanted effect (7). Similar to aminoglycosides, we suggest that the unwanted side effects of tuberactinomycins reflect limitations in compound selectivity, despite the observation that the tuberactinomycins show a several-hundred-fold-higher activity on bacterial than on eukaryotic ribosomes (Table 2). In support of a common mechanism in aminoglycoside and tuberactinomycin ototoxicity, we find that tuberactinomycins demonstrate key features involved in aminoglycoside ototoxicity: misreading induction in mitohybrid ribosomes and increased susceptibility of mutant mitohybrids with the A1555G and C1494U deafness alleles to drug action. However, the structural determinants of selectivity, i.e., the ability to differentiate between prokaryotic and eukaryotic ribosomes, are notably different. While for the clinically used 6′NH2 4,6-aminoglycosides selectivity to a large extent rests upon 16S rRNA residue 1408 (14, 16), selectivity of tuberactinomycin antibiotics involves mainly 16S rRNA residue 1491. We suggest that the structures available together with the determinants of selectivity identified will enable the creation of new compounds by targeted drug design. The adjacent location of these two different classes of antibiotics should provide a means to combine their selectivity in an additive fashion without decreasing their potency as antibacterial agent, e.g., by chemically linking modified portions of tuberactinomycins and ring I of the 6′NH2 4,6-aminoglycosides. These compounds will be particularly valuable for treatment of tuberculosis, as chemotherapy of tuberculosis requires several months of drug treatment, putting the patients at high risk for ototoxicity.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Janusic and C. Ritter for expert technical assistance and F. Mitterecker and S. Salas for help in manuscript preparation.
This study was supported in part by grants from the University of Zurich and the European Community (PAR, FP7-HEALTH-2009-241476).
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 18 July 2011.
REFERENCES
- 1. Blumberg H. M., et al. 2003. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167:603–662 [DOI] [PubMed] [Google Scholar]
- 2. Böttger E. C. 2007. Antimicrobial agents targeting the ribosome: the issue of selectivity and toxicity—lessons to be learned. Cell. Mol. Life Sci. 64:791–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Böttger E. C., Springer B., Prammananan T., Kidan Y., Sander P. P. 2001. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2:318–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Böttger E. C., Springer B. 2008. Tuberculosis: drug resistance, fitness, and strategies for global control. Eur. J. Pediatr. 167:141–148 [DOI] [PubMed] [Google Scholar]
- 5. Böttger E. C., Springer B., Pletschette M., Sander P. 1998. Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat. Med. 4:1343–1344 [DOI] [PubMed] [Google Scholar]
- 6. Bruell C. M., et al. 2008. Conservation of bacterial protein synthesis machinery: initiation and elongation in Mycobacterium smegmatis. Biochemistry 47:8828–8839 [DOI] [PubMed] [Google Scholar]
- 7. Bryskier A. 2005. Antimicrobial agents: antibacterials and antifungals. ASM Press, Washington, DC [Google Scholar]
- 8. Choi E. C., et al. 1979. Viomycin resistance: alterations of either ribosomal subunit affect the binding of the antibiotic to the pair subunit and the entire ribosome becomes resistant to the drug. Biochem. Biophys. Res. Commun. 87:904–910 [DOI] [PubMed] [Google Scholar]
- 9. Davies J., Gorini L., Davis B. D. 1965. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1:93–106 [PubMed] [Google Scholar]
- 10. Dye C., Espinal M. A., Watt C. J., Mbiaga C., Williams B. G. 2002. Worldwide incidence of multidrug-resistant tuberculosis. J. Infect. Dis. 185:1197–1202 [DOI] [PubMed] [Google Scholar]
- 11. Ermolenko D. N., et al. 2007. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat. Struct. Mol. Biol. 14:493–497 [DOI] [PubMed] [Google Scholar]
- 12. Farmer P., Kim J. Y. 1998. Community based approaches to the control of multidrug resistant tuberculosis: introducing “DOTS-plus.” BMJ 317:671–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feuerriegel S., et al. 2009. Sequence analyses of just four genes to detect extensively drug-resistant Mycobacterium tuberculosis strains in multidrug-resistant tuberculosis patients undergoing treatment. Antimicrob. Agents Chemother. 53:3353–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hobbie S. N., et al. 2008. Mitochondrial deafness alleles confer misreading of the genetic code. Proc. Natl. Acad. Sci. U. S. A. 105:3244–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hobbie S. N., et al. 2007. Engineering the rRNA decoding site of eukaryotic cytosolic ribosomes in bacteria. Nucleic Acids Res. 35:6086–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hobbie S. N., et al. 2008. Genetic analysis of interactions with eukaryotic rRNA identify the mitoribosome as target in aminoglycoside ototoxicity. Proc. Natl. Acad. Sci. U. S. A. 105:20888–20893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johansen S. K., Maus C. E., Plikaytis B. B., Douthwaite S. 2006. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol. Cell 23:173–182 [DOI] [PubMed] [Google Scholar]
- 18. Jugheli L., et al. 2009. High level of cross-resistance between kanamycin, amikacin, and capreomycin among Mycobacterium tuberculosis isolates from Georgia and a close relation with mutations in the rrs gene. Antimicrob. Agents Chemother. 53:5064–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marrero P., Cabanas M. J., Modolell J. 1980. Induction of translational errors (misreading) by tuberactinomycins and capreomycins. Biochem. Biophys. Res. Commun. 97:1047–1052 [DOI] [PubMed] [Google Scholar]
- 20. Maus C. E., Plikaytis B. B., Shinnick T. M. 2005. Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:3192–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maus C. E., Plikaytis B. B., Shinnick T. M. 2005. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McClatchy J. K., Kanes W., Davidson P. T., Moulding T. S. 1977. Cross-resistance in M. tuberculosis to kanamycin, capreomycin and viomycin. Tubercle 58:29–34 [DOI] [PubMed] [Google Scholar]
- 23. McKee E. E., Ferguson M., Bentley A. T., Marks T. A. 2006. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob. Agents Chemother. 50:2042–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Modolell J., Vazquez D. 1977. The inhibition of ribosomal translocation by viomycin. Eur. J. Biochem. 81:491–497 [DOI] [PubMed] [Google Scholar]
- 25. Monshupanee T., Gregory S. T., Douthwaite S., Chungjatupornchai W., Dahlberg A. E. 2008. Mutations in conserved helix 69 of 23S rRNA of Thermus thermophilus that affect capreomycin resistance but not posttranscriptional modifications. J. Bacteriol. 190:7754–7761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peske F., Savelsbergh A., Katunin V. I., Rodnina M. V., Wintermeyer W. 2004. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 343:1183–1194 [DOI] [PubMed] [Google Scholar]
- 27. Pfister P., et al. 2005. Mutagenesis of 16S rRNA C1409–G1491 base-pair differentiates between 6′OH and 6′NH3+ aminoglycosides. J. Mol. Biol. 346:467–475 [DOI] [PubMed] [Google Scholar]
- 28. Prammananan T., Sander P., Springer B., Böttger E. C. 1999. RecA-mediated gene conversion and aminoglycoside resistance in strains heterozygous for rRNA. Antimicrob. Agents Chemother. 43:447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prezant T. R., et al. 1993. Mitochondrial rRNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 4:289–294 [DOI] [PubMed] [Google Scholar]
- 30. Salas-Marco J., Bedwell D. M. 2005. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J. Mol. Biol. 348:801–815 [DOI] [PubMed] [Google Scholar]
- 31. Sander P., et al. 2002. Fitness cost of chromosomal drug resistance-conferring mutations. Antimicrob. Agents Chemother. 46:1204–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shcherbakov D., et al. 2010. Directed mutagenesis of Mycobacterium smegmatis 16S rRNA to reconstruct the in-vivo evolution of aminoglycoside resistance in Mycobacterium tuberculosis. Mol. Microbiol. 77:830–840 [DOI] [PubMed] [Google Scholar]
- 33. Springer B., Lucke K., Calligaris-Maibach R., Ritter C., Böttger E. C. 2009. Quantitative drug susceptibility testing of Mycobacterium tuberculosis by use of MGIT 960 and EpiCenter instrumentation. J. Clin. Microbiol. 47:1773–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stanley R. E., Blaha G., Grodzicki R. L., Strickler M. D., Steitz T. A. 2010. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat. Struct. Mol. Biol. 17:289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sutton W. B., Gordee R. S., Wick W. E., Stanfield L. 1966. In vitro and in vivo laboratory studies on the antituberculous activity of capreomycin. Ann. N. Y. Acad. Sci. 135:947–959 [DOI] [PubMed] [Google Scholar]
- 36. Tsukamura M. 1969. Cross-resistance relationships between capreomycin, kanamycin, and viomycin resistances in tubercle bacilli from patients. Am. Rev. Respir. Dis. 99:780–782 [DOI] [PubMed] [Google Scholar]
- 37. Yamada T., Mizugichi Y., Nierhaus K. H., Wittmann H. G. 1978. Resistance to viomycin conferred by RNA of either ribosomal subunit. Nature 275:460–461 [DOI] [PubMed] [Google Scholar]
- 38. Yunis A. A. 1989. Chloramphenicol toxicity: 25 years of research. Am. J. Med. 87:44N–48N [PubMed] [Google Scholar]
- 39. Zhao H., et al. 2004. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am. J. Hum. Genet. 74:139–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.