Abstract
We recently demonstrated that CDR1 overexpression in azole-resistant isolates of Candida albicans is due to its enhanced transcriptional activation and increased mRNA stability. In this study, we provide the first evidence of transcriptional regulation of CDR1 by Ncb2, the β subunit of NC2, a heterodimeric regulator of transcription. Conditional NCB2 null mutants displayed decreased susceptibility toward azole and an enhanced transcription of CDR1. Interestingly, Ncb2 associated with the CDR1 promoter under both repression and activation; however, an increase in recruitment was observed under both transient and constitutive activation states. By chromatin immunoprecipitation (ChIP) assay, we showed the preferential recruitment of Ncb2 to the core TATA region under activation (azole-resistant isolate), while under repression (azole-susceptible isolate) it was present at the TATA upstream region. Further, ChIP analysis revealed that Ncb2 binding was not restricted to the CDR1 gene; instead, it was observed on the promoters of genes coregulated with CDR1 by the transcription activator Tac1. The tac1Δ null mutants, which fail to show the drug-induced transient activation of CDR1, also showed no increase in Ncb2 recruitment at the promoter. Taken together, our results show that Ncb2, in conjunction with Tac1, is involved in the transcriptional activation of CDR1, opening up new therapeutic possibilities to combat multidrug resistance (MDR) in C. albicans.
INTRODUCTION
Candida albicans is an opportunistic human pathogen which mostly infects immunocompromised patients. It has gained considerable importance due to its ability to acquire resistance to a wide range of drugs during the treatment of its infection. Among various mechanisms contributing to acquired resistance against antifungal agents, drug efflux represents an important strategy adopted by the pathogen (33, 37, 39, 40, 43, 45). Thus, most of the clinical, multidrug-resistant (MDR) isolates of C. albicans are shown to overexpress CDR1, CDR2, or MDR1 drug efflux pump-encoding genes. Cdr1 and Cdr2 belong to the ATP-binding cassette (ABC) superfamily of transporters and use energy driven from ATP hydrolysis to functionally transport drugs outside the cells. Mdr1, on the other hand, is a proton/drug antiporter which belongs to the superfamily of major facilitators (MFS). Notably, major multidrug transporters of Candida spp. belonging to different superfamilies, such as ABC or MFS, display promiscuity toward substrate specificity and use different mechanisms of drug extrusion (32). The regulatory circuits which control the gene expression of either ABC or MFS efflux proteins also are different (25, 29, 38).
The CDR1 gene encodes a major drug efflux protein which is regulated both transcriptionally and posttranscriptionally. The regulation of CDR1 has been extensively studied by us and other groups (4, 5, 6, 8, 18, 21, 35). It has been shown that the CDR1 promoter harbors various consensus cis elements (Sp1, AP-1, and Y-box) as well as specific cis-elements, such as as BRE, NRE (for Negative Regulatory Element), DRE, and SRE in the 5′ flanking region (8, 11, 18, 35). Trans-acting factors regulating CDR1 also have been identified. For example, Ndt80, a homolog of a meiosis-specific transcription factor (TF) in Saccharomyces cerevisiae, has been identified as a potential activator of CDR1 (4). A regulator, Tac1, also has been identified and binds to DRE in both CDR1 and CDR2 promoters (6). Interestingly, transcription factors belonging to the zinc cluster family, such as Fcr1, and the global repressor Tup1 act as negative regulators of CDR1 expression (30, 42). A recent genome-wide location profiling by chromatin immunoprecipitation on a chip (ChIP-chip) showed that another TF of the zinc cluster family, Upc2, which regulates ERG genes, also targets CDR1 (47).
The regulation of the expression of a gene is an intricate process and is tightly controlled by the interplay between various positive and negative factors. In addition to gene-specific transcription factors, there are a variety of accessory factors which act in a global manner. These include chromatin-modifying proteins that positively or negatively affect the formation of active transcription initiation complexes. Negative cofactor 2 (NC2) is an example of one such global regulator of RNA polymerase II (Pol II) which executes both negative and positive roles in transcription (2, 12, 19, 22, 34, 44, 46). NC2 is a heterodimeric complex of two subunits, NC2α and NC2β, and was originally recognized to have a TATA-binding protein (TBP)-binding activity in human nuclear extracts that repressed RNA Pol II transcription (15, 16, 27, 28). It was later identified in S. cerevisiae as an essential factor, and the α- and β-subunit-encoding genes were termed ScBUR6 and ScNCB2, respectively (9, 14, 20, 22, 34). In the present study, we have shown the importance of the β-subunit of NC2 (Ncb2) in the basal and activated transcription of CDR1 in azole-susceptible (AS) and azole-resistant (AR) isolates of C. albicans. We also provide evidence that Ncb2 is involved in the regulation of selective genes, particularly those targeted by transcription activator Tac1.
MATERIALS AND METHODS
Strains and growth conditions.
The Candida albicans strains used in this study are listed in Table S1 in the supplemental material. Strains were grown in YEPD (1% yeast extract, 2% peptone, and 2% dextrose) medium for routine purposes at 30°C or in synthetic defined (SD) dropout medium (0.67% yeast nitrogen base without amino acids and 2% glucose). A 0.2% aliquot of the amino acid dropout mix omitting the required amino acids was added to SD medium. The composition of dropout mix is provided in Table S3 in the supplemental material. For studies with conditional null mutant strain ncb2Δ/PMET3-NCB2, C. albicans cells were grown in YNB (yeast nitrogen base without amino acids) medium supplemented with 0.5 mM methionine and cysteine (M/C) or left unsupplemented. Escherichia coli strains DH5α, DH10β, and BL21(DE3) were used for routine cloning, subcloning, and expression of cloned genes. All E. coli strains were maintained in LB medium. Ampicillin (100 μg/ml) and kanamycin (50 μg/ml) were added to the growth medium when required.
Purification of recombinant proteins.
E. coli DH10β strains harboring plasmid constructs pMALc-2-X-NCB2, pMALc-2-X-BUR6, and pMALc-2-X-TBP were grown at 37°C in LB medium containing 100 μg/ml ampicillin and induced for the expression of the recombinant proteins using 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 2 h. MBP (maltose-binding protein)-tagged proteins were purified using amylose agarose (New England BioLabs) essentially by following the protocol supplied by the vendor. The eluted proteins were dialyzed against dialysis buffer containing 50 mM Tris-Cl (pH 7.5), 1 mM EDTA (pH 8.0), 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 10% glycerol. Similarly for the purification of His-tagged Ncb2, E. coli strain BL21(DE3) harboring plasmid construct pET28a-NCB2 was grown at 37°C in LB medium containing 50 μg/ml kanamycin and induced for protein expression. His-tagged Ncb2 was purified using nickel-nitrilotriacetic acid (Ni-NTA) Sepharose (Genotech Inc.) essentially by following the manufacturer's protocol. The eluted protein fraction was dialyzed against dialysis buffer as mentioned above. Protein concentrations were determined by the Bradford method (Bio-Rad, Hercules, CA).
Antibody production and Western blot analysis.
Purified, recombinant His-tagged Ncb2 protein was injected into rabbits for antibody production. About 200 μg of the purified protein was mixed with Freund's complete adjuvant in a 1:1 (vol/vol) ratio, sonicated for micelle formation, and then injected subcutaneously into New Zealand White rabbits for primary immunization. This was followed by two booster doses of 100 μg protein in Freund's incomplete adjuvant at 3-week intervals. After the second booster dose blood was collected for antiserum. The specificity of the antibody was checked and was used to detect Ncb2 in Western blots at a dilution of 1:10,000. Anti-MBP rabbit polyclonal antibody was purchased from New England BioLabs. Western blot analysis was carried out using standard procedures (36).
EMSA.
For electrophoretic mobility shift assay (EMSA), 0.1 to 0.2 μg of the purified proteins or 15 to 30 μg of C. albicans fractionated cell extract was incubated with [α-32P]dATP-labeled CDR1 promoter fragment probe (320 bp) or 17 bp of NRE containing oligonucleotide in a 20-μl reaction mixture of 50 mM Tris-Cl (pH 7.4), 6% glycerol, 50 mM NaCl, 30 μg/ml bovine serum albumin (BSA), 5 mM Mg2+, 1 mM dithiothreitol (DTT), and the indicated amount of double-stranded poly(dI/dC) carrier DNA (wherever necessary, carrier DNA was omitted from the reaction mixture) at room temperature for 15 min. DNA-protein complexes were resolved at 120 V in a preelectrophoresed 5% native polyacrylamide gel in 1× Tris-borate-EDTA (TBE) buffer (89 mM Tris-borate and 2.5 mM EDTA, pH 8.3) at room temperature. Following the completion of the run, the gel was dried and autoradiographed.
ChIP assay.
Chromatin immunoprecipitation was performed by following the method of Deluen et al. (7) with minor modifications. Briefly, C. albicans cells were grown in YEPD at 30°C until the optical density at 600 nm (OD600) reached 1.0 and then were fixed with 1% formaldehyde for 20 min at room temperature, followed by the addition of glycine to a final concentration of 125 mM. The cells were collected and washed twice with cold Tris-buffered saline (TBS; 20 mM Tris-Cl, pH 7.5, containing 150 mM NaCl). They were resuspended in Zymolyase buffer (50 mM Tris, pH 7.4, 10 mM MgCl2, 1 M sorbitol, and 30 mM DTT) containing Zymolyase 20T (Seikagaku Corporation, Japan) at a concentration of 20 μg/ml and incubated for 2 h at 30°C for spheroplasting. Spheroplasts were washed twice with cold TBS containing 1 M sorbitol, resuspended in ChIP lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8.0, 1% Triton X-100, 0.1% sodium deoxycholate, and 1 mM PMSF), and incubated on ice for 30 min. SDS was added at a final concentration of 0.5%, and the cell suspension was sonicated (seven cycles of a 20-s pulse at a power of 7.0 with 2-min cooling intervals between cycles). Following sonication, the lysate was centrifuged at 15,000 rpm for 30 min and supernatant was collected as soluble total chromatin (STC). The size of fragmented chromatin was examined by agarose gel electrophoresis (∼250 to 500 bp). A sample of 10% STC, referred to as input DNA, was stored at −20°C until further use. Before immunoprecipitation, STC was diluted five times in ChIP lysis buffer and precleared with preimmune serum and 20 μl of a packed volume of protein-A-Sepharose (Bangalore Genei, India) for 2 h. Precleared STC was incubated with preimmune serum or specific anti-Ncb2 antibody overnight at 4°C. The next day, 20 μl of a packed volume of protein A-Sepharose beads preequilibrated with ChIP lysis buffer was added to each aliquot and incubated further for 1 h. The Sepharose bead-bound immune complexes were collected by centrifugation and washed serially twice each with the ChIP lysis buffer, TSE 150 (Tris-Cl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM EDTA, pH 8.0, and 1 mM PMSF), TSE 500 (Tris-Cl, pH 8.0, 500 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM EDTA, pH 8.0, and 1 mM PMSF), buffer III (10 mM Tris-Cl, pH 8.0, 1 mM EDTA, pH 8.0, 250 mM LiCl, 1% NP-40, and 1% sodium deoxycholate), and finally with TE (10 mM Tris-Cl, pH 8, and 1 mM EDTA, pH 8.0). The bound immunoprecipitated complexes were eluted at 65°C in buffer IV (50 mM Tris-Cl, pH 7.5, 10 mM EDTA, pH 8.0, and 1% SDS). The immunoprecipitated as well as input DNA were treated with proteinase K (Sigma Chemical Co., St. Louis, MO), followed by overnight incubation at 65°C. The DNA was extracted two times with phenol-chloroform and precipitated by ethanol with glycogen as a carrier. The input DNA and immunoprecipitated DNA were dissolved in 100 and 20 μl of TE, respectively. For the reproducibility of the data, PCR was performed on IP DNA obtained from immunoprecipitations carried out on at least three different chromatin preparations, and representative data are shown. In PCR analysis, amplifications were carried out for 25 to 28 cycles.
Coimmunoprecipitation.
For coimmunoprecipitation, equal amounts (2 μg) of His-Ncb2, MBP-Bur6, and MBP-TBP were mixed in IP buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 2 mM EDTA, pH 8.0, 1% Triton X-100, and 1 mM PMSF). The mixture was precleared by adding preimmune serum and a 15-μl packed volume of protein A-Sepharose beads. The precleared supernatant was collected and incubated overnight at 4°C with either preimmune serum or anti-Ncb2 antibody. To rule out the possibility of the interaction of Bur6 and TBP with Ncb2 through MBP, MBP alone was used as a negative control. The next day a 20-μl packed volume of protein-A-Sepharose beads was added to each of the tubes, followed by 1 h of incubation. Following brief centrifugation at 500 × g, supernatant was discarded and the beads were washed three times with IP buffer. Bound immune complexes were released by boiling the beads in SDS sample buffer and subjected to SDS-PAGE, followed by Western blotting using anti-MBP antibody.
Spot test.
Stock solutions of fluconazole (1 mg/ml) and ketoconazole (1 mg/ml) were prepared in water and methanol, respectively. Cells were grown overnight on YNB plates and resuspended in 0.9% normal saline to an OD600 of 0.1. In spot assays, 5 μl of 5-fold serial dilutions of each yeast culture (A600 = 0.1) were spotted onto YNB plates supplemented with 0.5 mM methionine and 0.5 mM cysteine (M/C) or left unsupplemented in the absence (control) and presence of the drugs. Growth differences were recorded following the incubation of the plates for 2 days at 30°C.
Conditional NCB2 null mutant construction.
C. albicans strain SN78 (His−, Leu−, Ura−) was used to generate an NCB2 disruptant strain (31). To disrupt the first allele of NCB2, a stretch of DNA, including the NCB2 open reading frame (ORF) and 0.450 kb of its upstream and downstream regions, was amplified from the C. albicans SC5314 genome using primer pair 5′FLKFP (CGGGATCCATAGTTCATCTGATGCCAGTGC) and 3′FLKRP (CCCAAGCTTCAATTCCTAAGATCACAACAGTCC). This fragment was cloned between BamHI and HindIII sites of pUC18 vector, resulting in plasmid pUC-NC. Primers 5′FLKRP (CGACGCGTGGCTATGGTATAATGTTCTATTTAAAGC) and 3′FLK FP (TCCCCGCGGGCTATACTTTCAGAAGCAGC) were used to amplify the entire construct, except the NCB2 ORF region, by inverse PCR. C. dubliniensis HIS1 was amplified from plasmid pSN52 using UNIFLKFP (CGACGCGTACCAGTGTGATGGATATCTGC) and UNIFLKRP (TCCCCGCGGAGCTCGGATCCACTAGTAACG) and was ligated with the inverse PCR product. The ligated product was transformed into E. coli DH5α to obtain a construct containing disruption cassette I or pDC1. The plasmid pDC1 was digested with BamHI and HindIII to release the fragment containing the NCB2 disruption cassette and was used to generate heterozygous strain CaSN78F1 (NCB2/ncb2Δ) by transformation. C. albicans transformation was carried out by the lithium acetate method. The selection of transformants was performed on His− plates. Gene disruption was verified by PCR using primer pair 5′FLKFP and 3′FLKRP, which produced a longer and shorter amplification product in the heterozygous strain. Before disrupting the second allele, NCB2 under MET3 promoter control was integrated into the RP10 locus of CaSN78F1. NCB2 was amplified using primer pair NCB2 I(CGGGATCCATGTCGGAATACTC GGGCTCC) and NCB2RPPst (CGCTGCAGCTAATTAGGTTCAGGTTTGG) and cloned between BamHI and PstI sites of integrative vector CaEXP (3). The resulting plasmid pEXP-NCB2COM was linearized by StuI and was used to transform the heterozygous strain. Transformants were selected on histidine and uracil dropout medium. The resulting strain was termed CaSN78F1COM. The second allele disruption was done in a similar way, except that C. maltosa LEU2 was amplified from plasmid pSN40 using primer pair UNIFLKFP and UNIFLKRP. It was ligated with the inverse PCR product obtained from plasmid pUC-NC as described earlier. The ligated product was transformed into E. coli DH5α to obtain plasmid-containing disruption cassette II or pDC2. The disruption cassette was amplified from plasmid pDC2 using primer pair 5′FLKFP and 3′FLKRP, and 2 μg of amplified product was used for transformation into CaSN78F1COM. Transformants were selected on histidine, uracil, and leucine dropout medium. The screening of NCB2 knockout strains then was carried out by PCR. The resulting NCB2 conditional knockout strain (ncb2Δ/ncb2Δ RP10/rp10::PMET3-NCB2) was termed ncb2Δ/PMET3-NCB2.
CRIP assay.
The chromatin restriction digestion-coupled immunoprecipitation (CRIP) assay was performed essentially as described for the ChIP assay, with the following changes. STC was fragmented to a size of 250 to 500 bp. Fragmented STC was subjected to BamHI digestion. Prior to digestion, chromatin was precipitated at room temperature by adding 1 volume of isopropanol, and the pellet obtained after centrifugation at 8,000 rpm for 10 min at room temperature was washed twice with 70% alcohol at room temperature. The dried pellet was dissolved in 1× BamHI restriction enzyme buffer, and 200 U of BamHI was added. Digestion was performed for 8 h at 37°C, and an aliquot from the reaction mix was analyzed by electrophoresis on a 1.2% agarose gel. After digestion, chromatin was precipitated by ethanol and dissolved in ChIP lysis buffer. ChIP then was performed as described earlier.
RT-PCR.
Total RNA was isolated using an RNeasy Minikit (Qiagen). Reverse transcription-PCR (RT-PCR) was done essentially as mentioned in the RevertAidTM H Minus kit (MBI, Fermentas). Briefly, total isolated RNA was treated with DNase for 30 min at 37°C. This reaction was terminated by the addition of 1 μl of 25 mM EDTA, pH 8.0, and incubation at 65°C for 10 min. One μg of DNA-free total RNA was primed with oligo(dT)18 for first-strand cDNA synthesis at 42°C for 60 min. Reverse transcription was terminated by heating at 70°C for 5 min. Two μl of cDNA product was used in PCR (50 μl) primed with specific forward and reverse primers. For second-strand synthesis, 25 cycles of amplification were carried out. ACT1 was used as an endogenous control. For every RT-PCR analysis, cDNA was synthesized from RNA obtained from at least three different preparations. The amplified products were analyzed by agarose gel electrophoresis and quantified by densitometry.
RESULTS
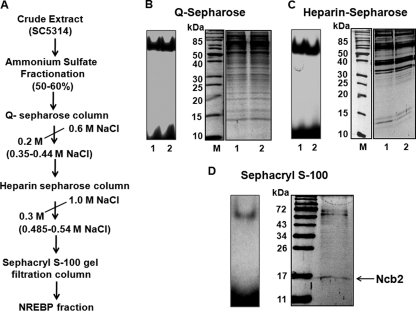
We had demonstrated earlier that an approximately 55-kDa protein purified by a DNA affinity procedure binds to the NRE of the CDR1 proximal promoter (11). The NRE-binding protein (NREBP) was identified as translation elongation factor 1 (Tef1) by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) (N.A. Gaur, unpublished data). However, our subsequent studies suggested that it was a major contaminant, and the NRE-binding activity probably was due to the presence of minor contaminants present in the purified Tef1 protein fraction (data not shown). To purify protein(s) binding to the NRE in the present study, we have employed classical protein purification methods. After multistep protein purification involving ammonium sulfate fractionation, Q-Sepharose, heparin-Sepharose, and Sephacryl S-100 gel filtration chromatography, the final purified fraction displaying NRE-binding activity was found to consist of a mixture of at least three polypeptides of approximately 17, 60, and 70 kDa (Fig. 1A to D). MALDI-TOF analysis of these proteins did not identify the two higher-molecular-mass polypeptides; however, the 17-kDa polypeptide was identified as negative cofactor B2, or Ncb2 (accession no. Q5A016_CANAL; see Fig. S1A to C in the supplemental material). In the Candida genome database (http://www.candidagenome.org), the NCB2 gene is described as an uncharacterized, predicted ORF (orf19.5825). However, in S. cerevisiae, the NCB2 gene encodes a protein which is a part of the basal transcription machinery and is involved in the negative regulation of gene expression by RNA polymerase II (9, 22). Since the negative regulation (repressor function) of the identified protein was in agreement with our expected role of NREBP, we explored its possible role in CDR1 regulation.
Fig. 1.
Copurification of Ncb2 with NRE-binding proteins. (A) Schematic representation of purification procedure. (B) Total cell extract from strain SC5314 was fractionated by ammonium sulfate, and the 50 to 60% fraction enriched in NRE-binding activity was loaded on a Q-Sepharose column. All fractions eluted were tested for NRE-binding activity by EMSA and were analyzed by 12% SDS-PAGE, followed by Coomassie staining. The left panel shows a representative picture of the NRE-binding activity of Q-Sepharose column eluates, and their respective protein profiles are shown in the right panel. (C) The left panel shows the NRE-binding activity of heparin sulfate column eluates. A silver-stained 12% acrylamide gel showing the protein profiles of the respective fractions is shown on the right. (D) Fractions from a heparin-agarose column were subjected to Sephacryl S-100 gel filtration chromatography, and the fractions obtained were analyzed by EMSA. The NRE-binding fractions eluted immediately after the void volume was pooled and was subjected to 12% SDS-PAGE and analyzed by silver staining.
Ncb2 binds to proximal CDR1 promoter in vivo.
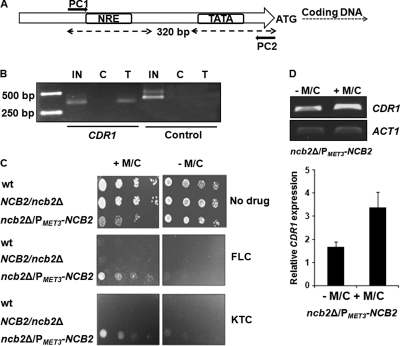
To investigate the role of Ncb2 in CDR1 transcription regulation, NCB2 was cloned and expressed in bacterial expression systems with two different tags at the amino terminus, hexa-His and maltose-binding protein (MBP), as shown in Fig. S2A to C in the supplemental material. However, recombinant Ncb2 did not bind to the NRE of the CDR1 promoter in subsequent electrophoretic mobility shift assays (EMSA) (data not shown). Our inability to detect the NRE-binding activity of recombinant Ncb2 in vitro prompted us to test the functional association of native Ncb2 with the CDR1 promoter in vivo. Polyclonal antibodies were raised against the hexa-His-tagged protein in rabbit as described in Materials and Methods. The specificity of the anti-Ncb2 antibody was confirmed (see Fig. S2D and E in the supplemental material), and chromatin immunoprecipitation (ChIP) was performed on C. albicans laboratory strain SC5314 by following published procedures with minor modifications as described in Materials and Methods (7). The presence of CDR1 promoter DNA in chromatin immunoprecipitated by anti-Ncb2 antibody was analyzed by PCR using CDR1 primer pair PC1 and PC2 for amplifying a 320-bp promoter region (Fig. 2A). As a negative control, a primer pair that amplifies the C-terminal region of the SNQ2 open reading frame was included in the analysis. The CDR1 promoter region was found to be enriched in chromatin immunoprecipitated by anti-Ncb2 antibody, while negligible signal was detected in the case of chromatin immunoprecipitated by control preimmune serum (Fig. 2B).
Fig. 2.
Ncb2 binds to the CDR1 promoter in vivo and represses basal gene expression. (A) Schematic representation of the CDR1 promoter region examined in the ChIP assay. Arrows indicate the position of the primers used. (B) ChIP analysis of Ncb2 association with the CDR1 promoter in C. albicans strain SC5314. IN is input DNA. C (control) and T (test) are immunoprecipitations carried out on cross-linked chromatin using preimmune serum and anti-Ncb2 antibody, respectively. As a negative control, a primer pair that amplifies the C-terminal region of the SNQ2 open reading frame was included in the analysis. (C) Drug susceptibility profiles of the wild-type (wt), heterozygous (NCB2/ncb2Δ), and conditional null mutant (ncb2Δ/ncb2Δ RP10/rp10::PMET3-NCB2) strains. In the figure the conditional null mutant is termed ncb2Δ/PMET3-NCB2. In the spot assay, 5 μl of 5-fold serial dilutions of each yeast culture (A600 = 0.1) was spotted onto drug-containing YNB plates supplemented with 0.5 mM methionine and cysteine (M/C) or left unsupplemented. Growth differences were evaluated by using drug-free controls following the incubation of the plates for 48 h at 30°C. (D) The upper panel shows the RT-PCR analysis of CDR1 transcript levels in the conditional null mutant strain. A culture of ncb2Δ/PMET3-NCB2 cells saturated overnight and grown in YNB was diluted to an OD600 of 0.1 in fresh YNB medium supplemented with 0.5 mM methionine and cysteine or left unsupplemented, and samples for RNA extraction were collected after 10 h of growth. A bar diagram of integrated density values (IDV) of CDR1 amplification normalized with respect to control gene (ACT1) amplification in ncb2Δ/PMET3-NCB2 (supplemented with M/C or left unsupplemented) is shown in the lower panel. Error bars correspond to standard deviations from three replicate experiments.
NCB2 conditional null mutants show enhanced azole resistance and increased CDR1 expression.
To explore the role of Ncb2 in CDR1 transcription regulation, we decided to check CDR1 transcript levels in NCB2 null mutants. In this context, we wanted to disrupt the NCB2 gene from the C. albicans genome. We used auxotrophic strain CaSN78 (His−, Leu−, and Ura−) for the generation of NCB2 null mutants. The strain used was susceptible to drugs with a basal level of CDR1 expression (data not shown). Heterozygous NCB2/ncb2Δ was generated with ease, but the construction of the homozygous ncb2Δ/ncb2Δ strain repeatedly failed, suggesting that similarly to NCB2 of S. cerevisiae, C. albicans NCB2 (CaNCB2) is an essential gene. To overcome the problem, a conditional knockout strain of NCB2 was constructed. Consequently, a copy of the NCB2 gene was placed under a regulatable MET3 promoter and integrated at an ectopic locus (RP10) of the C. albicans genome using selection marker URA3 before disrupting the second allele (see Fig. S3A and B in the supplemental material). The Ncb2 protein levels were analyzed in the conditional mutant strain (ncb2Δ/ncb2Δ RP10/rp10::PMET3-NCB2) under MET3 promoter-repressing conditions (0.5 mM methionine and cysteine) as shown in Fig. S3C. We observed low Ncb2 expression when the MET3 promoter was repressed. We analyzed the drug susceptibility patterns of wild-type and conditional mutant cells with fluconazole (FLC) and ketoconazole (KTC). Interestingly, under MET3 promoter-repressing conditions, the conditional mutant cells displayed enhanced resistance to both drugs compared to that of wild-type CaSN78 and the heterozygous mutant strain (Fig. 2C).
To test if the observed decrease in the susceptibility of the conditional mutant of NCB2 to azole also was associated with the increased expression of CDR1, we measured CDR1 transcript levels in the conditional mutant strains in the absence and presence of M/C by semiquantitative RT-PCR. As shown in Fig. 2D, a 2-fold increase in CDR1 expression was observed in the conditional mutant of Ncb2.
Recombinant Ncb2 in complex with TBP and Bur6 binds to the CDR1 proximal promoter in vitro.
Since we observed that Ncb2 associated with the CDR1 promoter in vivo but the purified recombinant Ncb2 was unable to bind DNA (data not shown), we argued that it could indirectly bind in association with other interacting proteins. In S. cerevisiae, Ncb2 has been shown to interact with NC2α to form NC2 complex, which then exerts its repressor function by binding to TBP. The α-subunit of S. cerevisiae NC2 is encoded by the BUR6 gene. A search for the BUR6 gene in the Candida genome database (CGD) resulted in the identification of orf19.2736, which is listed as an uncharacterized, predicted ORF of 428 bp in length. Similarly, in CGD, orf19.1837 (TBP1) is annotated as the functional homologue of S. cerevisiae and human TBP. We wanted to test if Ncb2 of Candida could interact with BUR6 and TBP1 gene products and form a functional CaNC2-TBP-DNA complex. Therefore, both ORFs were cloned. The putative BUR6 full-length gene (orf19.2736) and TBP1 gene (orf19.1837) were amplified by PCR from SC5314 genomic DNA using gene-specific primers (see Table S2 in the supplemental material). Following PCR, the desired products (0.430 and 0.717 kb, respectively) were cloned into pMAL-c-2X expression vector between BamHI and HindIII sites. Both of the MBP-tagged proteins were expressed and purified to apparent homogeneity as described in Materials and Methods (see Fig. S4 in the supplemental material).
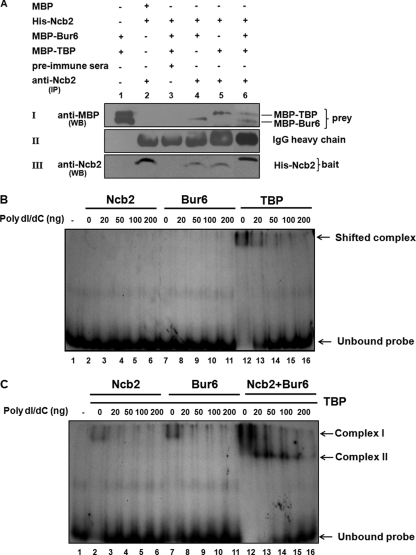
To investigate whether Ncb2, Bur6, and TBP physically interact with each other to form an NC2-TBP complex, we immunoprecipitated Ncb2 by using anti-Ncb2 antibody from a mixture of recombinants, His-Ncb2, MBP-Bur6, and MBP-TBP, expressed in Escherichia coli. The immunoprecipitates obtained were separated by 10% SDS-PAGE, and Western blotting was performed using anti-MBP antibody to check the coimmunoprecipitation of MBP-Bur6 and MBP-TBP (Fig. 3A). As depicted in blot I, both Bur6 and TBP can be immunoprecipitated with anti-Ncb2 antibody (lanes 4 to 6) but not with preimmune serum (lane 3). MBP alone failed to immunoprecipitate with anti-Ncb2 antibody, showing the specificity of the interaction between Ncb2-Bur6 and Ncb2-TBP. Thus, it became apparent that NCB2, BUR6, and TBP1 gene products interact and form NC2-TBP complex in C. albicans. The DNA-binding activity of the recombinant NC2-TBP complex was tested by EMSA. About 0.1 to 0.2 μg of purified recombinant Bur6, Ncb2, and TBP was used in EMSAs either alone or in various combinations. A 320-bp CDR1 proximal promoter fragment containing the TATA box and the NRE was amplified from SC5314 genomic DNA, digested with BglII, labeled with [32P]ATP using Klenow fragment, and subjected to EMSA. As seen in Fig. 3B, neither Ncb2 nor Bur6 alone was able to bind to DNA. TBP, however, did display DNA-binding activity when the reaction was performed in the absence of any carrier DNA (Fig. 3B). We tested the effect of Ncb2, Bur6, and Ncb2-Bur6 on TBP-DNA binding. As depicted in Fig. 3C, Ncb2 did not have a significant effect on TBP-DNA binding, while Bur6 influenced TBP-DNA binding to a significant extent. When Ncb2 and Bur6 were used together with TBP, TBP-DNA binding was dramatically enhanced and a very strong complex was observed (Fig. 3C, complex I). Notably, in the presence of poly(dI/dC), the mobility of this complex changed to the stronger and faster-migrating complex II. This difference in the mobility of complex II could be explained if one considers that the complexes formed in the absence of poly(dI/dC) are very large and partly nonspecific and thus are not well resolved. The formation of NC2-TBP-DNA complex even in the presence of increased amounts of poly(dI/dC) shows that compared to TBP-DNA interaction, complex II is more stable (Fig. 3C).
Fig. 3.
Bur6, Ncb2, and TBP of C. albicans interact together and bind to the CDR1 promoter fragment in vitro. (A) Coimmunoprecipitation of MBP-Bur6 and MBP-TBP with anti-Ncb2 antibody. Protein combinations used in the experiment are shown at the top of the panels. For blot I, lane 1, MBP-Bur6 and MBP-TBP were subjected to Western blotting (not immunoprecipitated) using anti-MBP antibody. For blot I, lanes 2, 4, 5, and 6, samples first were immunoprecipitated using anti-Ncb2 antibody and then subjected to Western blotting using anti-MBP antibody. For blot I, lane 3, the indicated protein mixture was immunoprecipitated using control immunoglobulin IgG (preimmune) and then subjected to Western blotting using anti-MBP antibody. Blot II, lane 1, blank; lanes 2 to 6, IgG heavy chains of the respective antibodies used in immunoprecipitation reactions. Blot III, lane 1, blank; lanes 2 to 6, Western blot of immunoprecipitated bait protein using anti-Ncb2 antibody. IP, immunoprecipitation; WB, Western blotting. (B) Autoradiogram showing the DNA binding of purified recombinant Ncb2, Bur6, and TBP. (C) Autoradiogram showing the effect of Ncb2, Bur6, and Ncb2-Bur6 on TBP-DNA binding. Protein combinations used in the assay are indicated at the top of the figure.
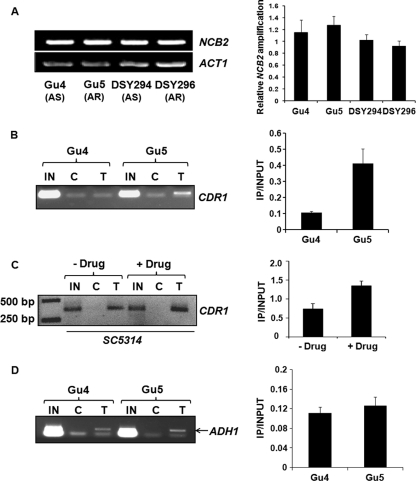
Higher CDR1 promoter occupancy by Ncb2 in AR isolates.
Our results thus far suggested that Ncb2 acts as a repressor of CDR1 expression (Fig. 2C and D). We tested whether there was any variation in the transcript levels of NCB2 between AS and AR isolates of C. albicans. When semiquantitative RT-PCR was performed on RNA isolated from a genetically matched pair of AS and AR clinical isolates of C. albicans, Gu4 and DSY294 as well as Gu5 and DSY296, respectively, no significant alteration in the transcript levels of NCB2 was observed (Fig. 4A). We then explored if the occupancy of the CDR1 promoter by Ncb2 varies between AS and AR isolates. We conducted a ChIP assay in one of the genetically matched pair of isolates, Gu4 and Gu5. Contrary to our expectation, we observed higher CDR1 promoter occupancy by Ncb2 in AR strain Gu5 than in AS strain Gu4. The recruitment of Ncb2 at the CDR1 promoter was approximately 4-fold greater in Gu5 than in Gu4 (Fig. 4B). Similar results were seen in the case of matched-pair isolates DSY294 and DSY296, where AR isolate DSY296 showed more Ncb2 presence at the CDR1 promoter than AS isolate DSY294 (see Fig. S5 in the supplemental material). Importantly, the ChIP assay performed on chromatin prepared from C. albicans SC5314 cells induced for transient CDR1 overexpression by fluphenazine also showed about 1.8-fold enrichment of Ncb2 at the CDR1 promoter in induced cells compared to the enrichment observed in uninduced cells (Fig. 4C). We were curious to check the pattern of the Ncb2 occupancy of a neutral gene promoter in AS and AR isolates. We checked the occupancy of the ADH1 gene promoter by Ncb2 in Gu4 and Gu5 and observed that Ncb2 binding to the ADH1 promoter did not vary significantly between the two strains (Fig. 4D).
Fig. 4.
Enrichment of Ncb2 at activated CDR1 promoters. (A) RT-PCR analysis of NCB2 transcript levels in two genetically matched AS and AR C. albicans clinical isolates. On the left is the NCB2 amplification of reverse-transcribed DNA prepared from the indicated isolates. A bar diagram showing integrated density values (IDV) of NCB2 amplification normalized with respect to constitutive gene (ACT1) amplification in C. albicans clinical isolates is shown in the right panel. Error bars represent standard deviations of results from three different RNA preparations. (B) ChIP analysis of Ncb2 association with the CDR1 promoter in AS and AR isolates. Chromatin immunoprecipitation was performed on chromatin isolated from AS and AR isolates Gu4 and Gu5, respectively. IN is input DNA (10%), and C and T stand for immunoprecipitations carried out on cross-linked chromatin with preimmune serum and anti-Ncb2 antibody, respectively. ChIP was performed in triplicate, and a representative figure is shown. The bar diagram on the right shows an approximately 4-fold enrichment of Ncb2 at the CDR1 promoter in the Gu5 isolate. The enrichment value is represented as a ratio of immunoprecipitation (IP) versus input. Error bars represent standard deviations of values generated from three different experiments. (C) Increased association of Ncb2 with the CDR1 promoter in cells transiently induced for CDR1 expression by fluphenazine compared to that of uninduced cells. The figure is a reverse of the original photo for a better representation. The panel to the right is a bar diagram showing approximately 1.8-fold enrichment of Ncb2 at the fluphenazine-induced CDR1 promoter. Error bars denote standard deviations from three replicate experiments. (D) ChIP analysis of Ncb2 association with the ADH1 promoter in Gu4 and Gu5 isolates. The specific band above the primer dimer is shown by an arrow in the figure.
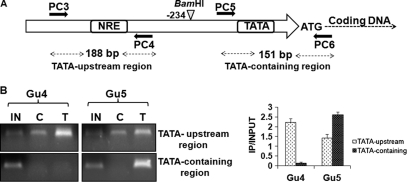
Ncb2 localizes to different sites of the CDR1 promoter in AS and AR isolates.
Our consistent observation that Ncb2 did not dissociate from activated CDR1 promoter but rather showed an increase in promoter occupancy went against the repressor role predicted by our earlier results (Fig. 2C and D). Our subsequent chromatin restriction digestion-coupled immunoprecipitation (CRIP) with AS and AR isolates resolved this discrepancy (41). CRIP was performed with Gu4 and Gu5 isolates, wherein before the immunoprecipitation of chromatin, CDR1 promoter DNA was separated into TATA-containing core promoter and TATA upstream fragments by utilizing a unique BamHI site upstream of TATA. For this the fragmented, soluble total chromatin was digested by BamHI restriction endonuclease as described in detail in Materials and Methods (see Fig. S6 in the supplemental material). Subsequently, the digested chromatin was pulled down by anti-Ncb2 antibody and ChIP was performed as described earlier. The presence of TATA-containing and TATA upstream promoter fragments in immunoprecipitated chromatin was tested by PCR with two primer pairs: PC5 with PC6 and PC3 with PC4 (Fig. 5A). Interestingly, it was observed that in AS strain Gu4, Ncb2 presence was evident at the TATA upstream promoter fragment while there was almost none at the TATA-containing fragment. A reverse scenario in AR strain Gu5 was observed, where anti-Ncb2 antibody strongly immunoprecipitated specifically the TATA-containing DNA fragment of the CDR1 promoter (Fig. 5B).
Fig. 5.
Ncb2 positional occupancy on the CDR1 promoter is different during repression and activation. (A) Schematic representation of the position of primer sets used in a modified chromatin restriction digestion-coupled immunoprecipitation (CRIP) assay. (B) CRIP assay showing the enrichment of Ncb2 at the core promoter region containing the TATA box element in AR isolate Gu5. The enrichment of Ncb2 at the TATA upstream promoter (NRE-containing) fragments is seen in AS isolate Gu4. IN, C, and T represent input DNA (5%) and immunoprecipitates obtained with control preimmune serum and specific anti-Ncb2 antibody, respectively. A bar diagram showing the difference in enrichment of Ncb2 at TATA upstream and TATA-containing fragments of the CDR1 promoter between AS and AR isolates is shown on the right.
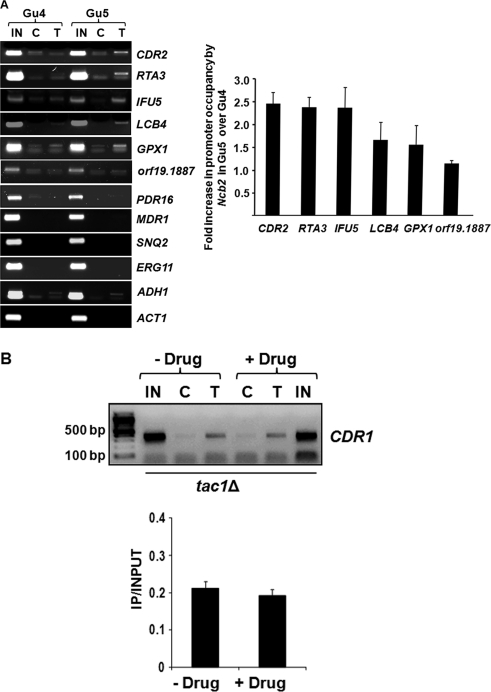
Ncb2 plays a role in regulation of Tac1-activated genes.
Although NC2 is a global regulator of gene expression, data from several microarray experiments and genome-wide ChIP analyses also have suggested gene-specific roles of the transcription factor (12, 26). In this context, we evaluated the possible role of Ncb2 in regulating other MDR genes, particularly those coregulated with CDR1 by the common transcription regulator Tac1. ChIP assay was performed in AS and AR matched-pair isolates Gu4 and Gu5, and Ncb2 association with the promoters of some of the Tac1-regulated genes besides CDR1 (CDR2, RTA3, IFU5, LCB4, GPX1, orf19.1887, and PDR16) was tested. We also included Upc2 (ERG11 and SNQ2)- and Mrr1 (MDR1)-regulated genes and ADH1 and ACT1 as controls. Interestingly, we found that Ncb2 occupancy at the promoters of most of the Tac1-regulated genes followed the same pattern. For example, similarly to CDR1, an increased recruitment of Ncb2 at the respective promoter DNA of CDR2, RTA3, IFU5, LCB4, and GPX1 was seen in AR isolate Gu5 compared to that of AS isolate Gu4. Interestingly, Ncb2 was not seen to be associated with MDR1, SNQ2, and ERG11 promoters. Among control genes included in the analysis, Ncb2 was seen to be bound to the ADH1 promoter to the same extent in AS and AR strains, while it was not seen on the ACT1 promoter in either of the strains (Fig. 6A).
Fig. 6.
Ncb2 plays a role in the regulation of Tac1-activated genes. (A) ChIP analysis of Ncb2 association with the promoters of a selected set of genes of C. albicans in AS and AR isolates. The fold enrichment in Ncb2 recruitment at the promoters of genes regulated by Tac1 in Gu5 above the level of enrichment for Gu4 is represented as a bar diagram on the right. Error bars denote standard deviations for three replicate experiments. (B) ChIP analysis of Ncb2 association with the CDR1 promoter in tac1Δ cells with or without fluphenazine treatment. IN, C, and T are the same as those described in the legend to Fig. 4. The figure is a reverse of the original photo for a better representation. A bar diagram of the figure is shown in the lower panel. Error bars denote standard deviations from three replicate experiments.
To further explore the possible correlation between Tac1 and Ncb2 in CDR1 gene activation, CDR1 promoter occupancy by Ncb2 was examined in a tac1Δ strain. The mutant strain was induced for CDR1 expression by treating cells with fluphenazine, and Ncb2 occupancy was observed in treated and untreated cells. Unlike wild-type cells (Fig. 4C), tac1Δ cells that do not show the transient induction of CDR1 expression also failed to show an increase in Ncb2 recruitment upon drug treatment (Fig. 6B).
DISCUSSION
Resistance to azole antifungals in clinical C. albicans isolates often is caused by the increased expression of genes encoding multidrug efflux pumps, the ATP-binding cassette (ABC) transporters CDR1 and CDR2. The transcriptional regulation of CDR1 is a complicated network which includes its transient induced expression by various stresses and activation by transcription activators (4, 6, 21, 30, 42, 47). Using genetically matched AS and AR isolates of C. albicans, we recently demonstrated that CDR1 overexpression in AR isolates is due to an enhanced transcriptional activation and increased mRNA stability, which is due primarily to the reduced interaction of trans-regulatory factor(s) and the hyperadenylation of its transcript (11, 23, 24).
In the present study, we provide the first evidence of another level of CDR1 regulation mediated by a general repressor of transcription, Ncb2. Ncb2 is one of the two subunits of the NC2 complex, and the most prevalent mechanism of transcription repression by NC2 is through targeting TBP and inhibiting PIC (preinitiation complex) formation. During the purification of the cognate transcription factor of the previously identified cis-element NRE in the CDR1 proximal promoter (10, 11), Ncb2 copurified along with two other proteins as an activity that bound to the NRE in vitro (Fig. 1). Given the importance of the protein in the negative regulation of promoters transcribed by RNA polymerase II, its possible role in CDR1 regulation was examined. Although we were not able to observe the DNA-binding activity of the recombinant, purified Ncb2 protein with the NRE in subsequent gel shift assays, we could demonstrate its binding at the CDR1 promoter by ChIP analysis (Fig. 2B). There might be several reasons for the inability of recombinant Ncb2 to bind to the NRE, such as it might require additional factors for DNA binding or certain posttranslational modifications may play a role in its binding. The role of Ncb2 protein as a repressor of CDR1 was evident from studies with its conditional null mutant. Interestingly, in conditional mutants under the repression of the MET3 promoter transcribing NCB2, we observed the derepression of CDR1 transcription with a simultaneous decrease in drug susceptibilities (Fig. 2C and D). This increase in transcription and in drug resistance, although consistent, was not dramatic. This implies that although CDR1 expression is derepressed in conditional mutants, it is not hyperactivated. This also reaffirms the point that CDR1 is regulated by multiple factors.
In addition to the well-characterized role of NC2 as a repressor, many studies have indicated that it also activates transcription at some other promoters (2, 12, 22, 34, 44). Both an increase and a decrease in transcription was observed in S. cerevisiae cells expressing a mutant form of Bur6, as well as in cells that have altered Ncb2 activity depending upon the gene studied (12, 22, 34). Another study of S. cerevisiae showed that the association of Bur6 with promoters generally correlates well with the transcriptional activity (12). In our ChIP assays involving AS and AR isolate pairs, we found an increased recruitment for Ncb2 at the CDR1 promoter in AR isolates which was inconsistent with its repressor role (Fig. 4B). This raised the question of how Ncb2 as a repressor is also involved in the activated transcription of CDR1. Since the transcription factor was present at the promoter of CDR1 in both basal (in the AS isolate) and activated (in the AR isolate) expression, we questioned whether there can be any difference in positional occupancy in both situations.
The presence of human NC2 at sites other than core promoter DNA has been observed. For example, in a study involving human promoters, the α-subunit of the NC2 complex was present at promoters as far as 600 bp away from the transcription start site (1). It also has been shown experimentally that the human NC2-TBP complex can bind to TATA as well as to non-TATA-containing short oligonucleotides in vitro (13). That NC2 can impart movement to TBP on a 239-bp adenovirus major late promoter DNA, causing it to escape and return to TATA-binding mode, also has been demonstrated by authors who have suggested that this movement of TBP-NC2 complex on promoter explains the repressor and activator functions of NC2 (41).
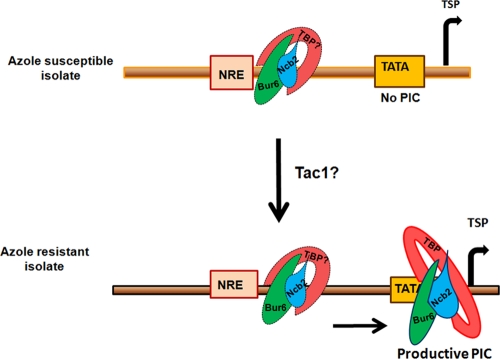
Our CRIP assay, where we had segregated the TATA box region of CDR1 from the rest of the promoter to map the position of Ncb2 in AS and AR isolates, indeed demonstrated that at the repressed CDR1 promoter (in the AS isolate) Ncb2 is localized to the TATA upstream region, while at the activated CDR1 promoter (in the AR isolate) it primarily occupies the core promoter region containing the TATA box (Fig. 5B). Taken together, we provide evidence that in the AS isolate the CDR1 promoter is occupied by Ncb2 (NC2), probably in complex with TBP at nonproductive sites, thus keeping the transcription machinery in a paused state. However, we could not experimentally map the exact position of the transcription factor on the promoter, especially during the basal expression of the gene in AS strains. Recently, the preferential binding of TBP with single-stranded pyrimidine-rich sequence over a double-stranded canonical TATA box sequence [5′-TATA (A/T)A(A/T)(G/A)-3′] has been shown (17). The sequence analysis of the CDR1 proximal promoter revealed the presence of unusually high numbers of polypyrimidine (T) stretches between the TATA box and NRE site. We speculate that these polypyrimidine stretches are the preferred site(s) for stable DNA-TBP-Ncb2 complex formation. A hypothetical model depicting possible positional occupancy by Ncb2 during the repression and activation of CDR1 in AS and AR isolates is shown in Fig. 7. Our data suggest that Ncb2 not only acts as a repressor of basal transcription but also participates in activated CDR1 transcription, and that the role played in activated transcription is mediated by transcription activator Tac1 became apparent when we used a tac1Δ homozygous null mutant strain and examined Ncb2 recruitment at the CDR1 promoter under basal and promoter-inducing conditions. As it is well known that Tac1 is required for the transient induction of CDR1, we modeled CDR1 induction by using fluphenazine (5). Notably, unlike the wild-type strain SC5314, tac1Δ mutants which do not transiently induce CDR1 also did not show an increase in Ncb2 recruitment at the promoter of CDR1 upon fluphenazine treatment (Fig. 4C and 6B). Taking these results together, we suggest that the local concentration of Ncb2 at the TATA region is necessary during the activation of CDR1 transcription, which probably is achieved by the gene-specific transcription factor Tac1. Furthermore, in our ChIP assays with AS and AR isolates, we found that most of the Tac1-activated genes showed increased promoter recruitment of Ncb2 upon activation (AR strain Gu5) (Fig. 6A). This is an interesting observation, showing the interplay between an activator, Tac1, and a repressor, Ncb2, in eliciting the activated transcription of CDR1 in AR isolates, and it merits further investigation.
Fig. 7.
Hypothetical model depicting possible positional occupancy by Ncb2 at the CDR1 promoter during the repression and activation of the gene in AS and AR isolates. Other factors associated with the preinitiation complex (PIC) are not shown. TSP is the transcription start point.
In conclusion, we have identified a basal repressor of CDR1 transcription while searching for the transcription regulator binding to NRE. Although the repression operating through NREBP still remains elusive, it does represent another level at which this important MDR gene is regulated. Given the universal nature of NC2 as a transcription repressor and the manner in which it keeps the CDR1 promoter paused in AS isolates, its multifunctional role at the promoters of highly inducible genes in other eukaryotic systems cannot be ruled out.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Department of Biotechnology of India (BT/PR9100/Med/29/03/2007 and DST-PURSE to R.P. and G.M.). S.S. acknowledges the Council of Scientific and Industrial Research, India, for providing junior and senior research fellowships.
We thank Peter Sudbery, Dominique Sanglard, Joachim Morschhäuser, and A. D. Johnson for providing plasmids and strains. We greatly appreciate S. Krishnamurthy and Sneh Lata Bhadoriya for their valuable input during the preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 19 August 2011.
REFERENCES
- 1. Albert T. K., et al. 2007. Global distribution of negative cofactor 2 subunit-{alpha} on human promoters. Proc. Natl. Acad. Sci. U. S. A. 104:10000–10005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cang Y., Prelich G. 2002. Direct stimulation of transcription by negative cofactor 2 (NC2) through TATA-binding protein (TBP). Proc. Natl. Acad. Sci. U. S. A. 99:12727–12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Care R. S., Trevethick J., Binley K. M., Sudbery P. E. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792–798 [DOI] [PubMed] [Google Scholar]
- 4. Chen C. G., Yang Y. L., Shih H. I., Su C. L., Lo H. J. 2004. CaNdt80is involved in drug resistance in Candida albicans by regulating CDR1. Antimicrob. Agents Chemother. 48:4505–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coste A., et al. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coste A. T., Karababa M., Ischer F., Bille J., Sanglard D. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deluen C., et al. 2002. The Ccr4-Not complex and yTAF1 [yTaf(II)130p/yTaf(II)145p) show physical and functional interactions. Mol. Cell. Biol. 22:6735–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Micheli M., Bille J., Schueller C., Sanglard D. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans. Mol. Microbiol. 43:1197–1214 [DOI] [PubMed] [Google Scholar]
- 9. Gadbois E. L., Chao D. M., Reese J. C., Green M. R., Young R. A. 1997. Functional antagonism between RNA polymerase II holoenzyme and global negative regulator NC2 in vivo. Proc. Natl. Acad. Sci. U. S. A. 94:3145–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gaur N. A., et al. 2005. Expression of the CDR1 efflux pump in clinical Candida albicans isolates is controlled by a negative regulatory element. Biochem. Biophys. Res. Commun. 332:206–214 [DOI] [PubMed] [Google Scholar]
- 11. Gaur N. A., et al. 2004. Identification of a negative regulatory element which regulates basal transcription of a multidrug resistance gene CDR1 of Candida albicans. FEMS Yeast Res. 4:389–399 [DOI] [PubMed] [Google Scholar]
- 12. Geisberg J. V., Holstege F. C., Young R. A., Struhl K. 2001. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol. Cell. Biol. 21:2736–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gilfillan S., Stelzer G., Piaia E., Hofmann M. G., Meisterernst M. 2005. Efficient binding of NC2. TATA-binding protein to DNA in the absence of TATA. J. Biol. Chem. 280:6222–6230 [DOI] [PubMed] [Google Scholar]
- 14. Goppelt A., Meisterernst M. 1996. Characterization of the basal inhibitor of class II transcription NC2 from Saccharomyces cerevisiae. Nucleic Acids Res. 24:4450–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goppelt A., Stelzer G., Lottspeich F., Meisterernst M. 1996. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 15:3105–3116 [PMC free article] [PubMed] [Google Scholar]
- 16. Inostroza J. A., Mermelstein F. H., Ha I., Lane W. S., Reinberg D. 1992. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell 70:477–489 [DOI] [PubMed] [Google Scholar]
- 17. Irani R. J., et al. 2010. TATA binding protein binds tightly to single stranded DNA: a new model for eukaryotic transcription. Nat. Preceedings doi:10.1038/npre.2010.4139.1 [Google Scholar]
- 18. Karnani N., et al. 2004. SRE1 and SRE2 are two specific steroid responsive modules of Candida drug resistance gene 1 (CDR1) promoter. Yeast 21:219–239 [DOI] [PubMed] [Google Scholar]
- 19. Kim J., Parvin J. D., Shykind B. M., Sharp P. A. 1996. A negative cofactor containing Dr1/p19 modulates transcription with TFIIA in a promoter-specific fashion. J. Biol. Chem. 271:18405–18412 [DOI] [PubMed] [Google Scholar]
- 20. Kim S., Na J. G., Hampsey M., Reinberg D. 1997. The Dr1/DRAP heterodimer is a global repressor of transcription in vivo. Proc. Natl. Acad. Sci. U. S. A. 94:820–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krishnamurthy S., Gupta V., Prasad R., Panwar S. L., Prasad R. 1998. Expression of CDR1, a multidrug resistance gene of Candida albicans: in vitro transcriptional activation by heat shock, drugs and human steroid hormones. FEMS Microbiol. Lett. 160:191–197 [DOI] [PubMed] [Google Scholar]
- 22. Lemaire M., Xie J., Meisterernst M., Collart M. A. 2000. The NC2 repressor is dispensable in yeast mutated for the Sin4p component of the holoenzyme and plays roles similar to Mot1p in vivo. Mol. Microbiol. 36:163–173 [DOI] [PubMed] [Google Scholar]
- 23. Manoharlal R., Gaur N. A., Panwar S. L., Morschhauser J., Prasad R. 2008. Transcriptional activation and increased mRNA stability contribute to overexpression of CDR1 in azole-resistant Candida albicans. Antimicrob. Agents Chemother. 52:1481–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manoharlal R., Gorantala J., Sharma M., Sanglard D., Prasad R. 2010. PAP1 [poly(A) polymerase 1] homozygosity and hyperadenylation are major determinants of increased mRNA stability of CDR1 in azole resistant clinical isolates of Candida albicans. Microbiology 156:313–326 [DOI] [PubMed] [Google Scholar]
- 25. Marie C., White T. C. 2009. Genetic basis of antifungal drug resistance. Curr. Fungal Infect. Rep. 3:163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Masson P., Leimgruber E., Creton S., Collart M. A. 2008. The dual control of TFIIB recruitment by NC2 is gene specific. Nucleic Acids Res. 36:539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meisterernst M., Roeder R. G. 1991. Family of proteins that interact with TFIID and regulate promoter activity. Cell 67:557–567 [DOI] [PubMed] [Google Scholar]
- 28. Mermelstein F., et al. 1996. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 10:1033–1048 [DOI] [PubMed] [Google Scholar]
- 29. Morschhäuser J. 2010. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet. Biol. 47:94–106 [DOI] [PubMed] [Google Scholar]
- 30. Murad A. M., et al. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981–993 [DOI] [PubMed] [Google Scholar]
- 31. Noble S. M., Johnson A. D. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4:298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Prasad R., Gaur N. A., Gaur M., Komath S. 2006. Efflux pumps in drug resistance of Candida. Curr. Drug Targets Infect. Disorders 6:69–83 [DOI] [PubMed] [Google Scholar]
- 33. Prasad R., Panwar S. L., Krishnamurthy S. 2000. Principles and clinical applications, p. 601–623 In Calderone R. A., Cihlar R. L. (ed.), Fungal pathogenesis. Marcel Dekker, Inc., New York, NY [Google Scholar]
- 34. Prelich G. 1997. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol. Cell. Biol. 17:2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Puri N., et al. 1999. CDR1, a multidrug resistance gene from Candida albicans, contains multiple regulatory domains in its promoter and the distal AP-1element mediates its induction by miconazole. FEMS Microbiol. Lett. 180:213–219 [DOI] [PubMed] [Google Scholar]
- 36. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Sanglard D., Odds F. C. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73–85 [DOI] [PubMed] [Google Scholar]
- 38. Sanglard D., Coste A., Ferrari S. 2009. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 9:1029–1050 [DOI] [PubMed] [Google Scholar]
- 39. Sanglard D., Ischer F., Monod M., Bille J. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405–416 [DOI] [PubMed] [Google Scholar]
- 40. Sanglard D., et al. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schluesche P., Stelzer G., Piaia E., Lamb D. C., Meisterernst M. 2007. NC2 mobilizes TBP on core promoter TATA boxes. Nat. Struct. Mol. Biol. 14:1196–1201 [DOI] [PubMed] [Google Scholar]
- 42. Talibi D., Raymond M. 1999. Isolation of a putative Candida albicans transcriptional regulator involved in pleiotropic drug resistance by functional complementation of a pdr1 pdr3 mutation in Saccharomyces cerevisiae. J. Bacteriol. 181:231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. White T. C., Marr K. A., Bowden R. A. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Willy P. J., Kobayashi R., Kadonaga J. T. 2000. A basal transcription factor that activates or represses transcription. Science 290:982–985 [DOI] [PubMed] [Google Scholar]
- 45. Wirsching S., Michel S., Kohler G., Morschhäuser J. 2000. Activation of the multidrug resistance gene MDR1 in fluconazole-resistant, clinical Candida albicans strains is caused by mutations in a trans-regulatory factor. J. Bacteriol. 182:400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xie J., Collart M., Lemaire M., Stelzer G., Meisterernst M. 2000. A single point mutation in TFIIA suppresses NC2 requirement in vivo. EMBO J. 19:672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Znaidi S., et al. 2008. Genome-wide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot. Cell 7:836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.