Abstract
Previous studies have shown that repression of the Saccharomyces cerevisiae SER3 gene is dependent on transcription of SRG1 from noncoding DNA initiating within the intergenic region 5′ of SER3 and extending across the SER3 promoter region. By a mechanism dependent on the activities of the Swi/Snf chromatin remodeling factor, the HMG-like factor Spt2, and the Spt6 and Spt16 histone chaperones, SRG1 transcription deposits nucleosomes over the SER3 promoter to prevent transcription factors from binding and activating SER3. In this study, we uncover a role for the Paf1 transcription elongation complex in SER3 repression. We find that SER3 repression is primarily dependent on the Paf1 and Ctr9 subunits of this complex, with minor contributions by the Rtf1, Cdc73, and Leo1 subunits. We show that the Paf1 complex localizes to the SRG1 transcribed region under conditions that repress SER3, consistent with it having a direct role in mediating SRG1 transcription-dependent SER3 repression. Importantly, we show that the defect in SER3 repression in strains lacking Paf1 subunits is not a result of reduced SRG1 transcription or reduced levels of known Paf1 complex-dependent histone modifications. Rather, we find that strains lacking subunits of the Paf1 complex exhibit reduced nucleosome occupancy and reduced recruitment of Spt16 and, to a lesser extent, Spt6 at the SER3 promoter. Taken together, our results suggest that Paf1 and Ctr9 repress SER3 by maintaining SRG1 transcription-dependent nucleosome occupancy.
INTRODUCTION
The packaging of eukaryotic DNA into chromatin presents a major obstacle to transcription initiation by preventing access of transcription factors to promoter DNA and also to transcription elongation by physically hindering the passage of RNA polymerase II (RNA Pol II) (44). Eukaryotic cells rely on the activities of three major classes of factors to alter chromatin architecture during transcription. The first class consists of chromatin remodeling factors that use the energy of ATP hydrolysis to alter the position or occupancy of nucleosomes (16). The second class contains histone-modifying enzymes that add or remove covalent modifications, such as acetyl, methyl, and ubiquitin groups, which can alter the dynamic properties of nucleosomes and influence the binding of additional regulatory proteins (9). The third class is comprised of histone-interacting proteins that function as chaperones to mediate the disassembly and reassembly of nucleosomes during transcription by RNA Pol II (23).
Recent studies have provided evidence that the Saccharomyces cerevisiae SER3 gene is controlled by transcription-dependent chromatin dynamics (27, 85). SER3 encodes an enzyme required for serine biosynthesis that is repressed when serine is abundant and is rapidly activated upon serine depletion (50). SER3 repression is mediated by the serine-dependent transcription of SRG1, initiating within intergenic noncoding DNA (ncDNA) 5′ of SER3 and extending across the SER3 promoter region before terminating near the SER3 translational start site (48, 50, 86). SRG1 intergenic transcription acts in cis to promote nucleosome occupancy of the SER3 promoter, thereby occluding this region from the binding of transcription factors. SRG1 transcription-dependent nucleosome occupancy of the SER3 promoter requires the functions of the Swi/Snf chromatin remodeling complex, an HMG-like protein Spt2, and the Spt6 and Spt16 histone chaperones (27, 49, 85). Swi/Snf is recruited to the SRG1 promoter by the Cha4 activator protein in a serine-dependent manner and is thought to mobilize nucleosomes, allowing RNA Pol II to initiate transcription of SRG1 (49, 50). As a consequence of SRG1 transcription, RNA Pol II traverses the SER3 promoter, where it deposits and maintains nucleosomes over this region by a mechanism that is dependent on Spt6, Spt16, and Spt2 (27, 85).
Another factor that is important for facilitating transcription elongation through chromatin is the Paf1 complex (30). The Paf1 complex is conserved across all eukaryotes and has important roles in embryonic development (2, 84, 90), maintenance of stem cell fate (22), and tumorigenesis (11, 45, 54). In S. cerevisiae, the Paf1 complex is comprised of five subunits, Paf1, Ctr9, Rtf1, Cdc73, and Leo1 (42, 55, 76, 79) that colocalize with RNA Pol II across transcribed genes exiting near the polyadenylation sites (38, 42, 53, 67). During elongation, the Paf1 complex has been shown to promote histone modifications (15, 41, 59, 94), alter the phosphorylation state of the RNA Pol II carboxy-terminal domain (CTD) (56, 61), and facilitate proper transcription termination (34, 56, 64, 75, 87). Colocalization of the Paf1 complex with RNA Pol II is dependent on the Bur1/Bur2 and Spt4/Spt5 transcription elongation factors (43, 46, 69, 102). Other factors, such as Spt6, FACT (composed of Spt16 and Pob3), and Ccr4-Not, may also contribute to the recruitment of the Paf1 complex, but their roles are not clearly defined (34, 57, 63).
One of the primary functions of the Paf1 complex is to promote histone modifications associated with active transcription (30). In yeast, the Paf1 complex promotes monoubiquitylation of histone H2B at lysine 123 (K123ub) by the ubiquitin conjugase Rad6 and ubiquitin ligase Bre1 (59, 94). Ubiquitylation of H2B is required for subsequent methylation of histone H3 at lysine 4 (K4me) and lysine 79 (K79me) by the Set1 and Dot1 methyltransferases, respectively (41, 59, 60, 74, 82, 94). These modifications are predominantly dependent on the Rtf1 subunit of the Paf1 complex, specifically involving a central region of the protein termed HMD for the histone modification domain (87, 91). Furthermore, the Paf1 and Ctr9 subunits are required for trimethylation of histone H3 at lysine 36 (K36me3) by the Set2 methyltransferase (14). Together, these modifications control histone acetylation across transcribed genes through the recruitment of histone deacetylases (10, 32, 36, 39, 66, 100).
Other connections of the Paf1 complex to chromatin have also been described. In yeast, Paf1 and Ctr9 were shown to affect the rate of induction of GAL genes by promoting nucleosome eviction from the GAL1-10 promoter during activation (51). The Drosophila melanogaster Paf1 complex has been shown to facilitate recruitment of the Spt6 and FACT histone chaperones during transcription (1). These studies established the importance of the Paf1 complex in transcription through chromatin, but little is known about how the complex mediates this function.
In this study, we examine the role of the Paf1 complex in SRG1 transcription-dependent repression of SER3. We find SER3 repression to be primarily dependent on the Paf1 and Ctr9 subunits and provide genetic evidence suggesting that these factors act in a previously described pathway with the Bur1/Bur2 kinase/cyclin and the Spt4/Spt5 transcription elongation complex. Our results indicate that while the Paf1 complex colocalizes with SRG1 transcription, its absence does not reduce SRG1 transcript levels, and its role in SER3 repression is largely independent of its ability to orchestrate covalent histone modifications. Rather, we find that Paf1 and Ctr9 promote both nucleosome occupancy over actively transcribing SRG1 and normal recruitment of Spt6 and Spt16 to this region. Our results suggest that the Paf1 and Ctr9 subunits of the Paf1 complex repress SER3 by facilitating SRG1 transcription-dependent nucleosome occupancy of the SER3 promoter, possibly by stabilizing the association of Spt6 and Spt16.
MATERIALS AND METHODS
Yeast strains and media.
Saccharomyces cerevisiae strains used in this study were derivatives of a GAL2+ strain of S288C (93) and are listed in Table 1. Strains were created using standard genetic crosses or by transformation (3). Gene replacements of PAF1, RTF1, CTR9, LEO1, CDC73, RAD6, BRE1, BUR2, and HTA2-HTB2 with KanMX or URA3 have been previously described (14, 18, 87). Other alleles that have been previously described include spt4Δ1::URA3 (83), spt4Δ2::HIS3 (4), spt5-194 (92), bur2Δ2::URA3 (15), HTA1-htb1K123R (87), rtf1Δ100::URA3 (80), and the epitope-tagged versions of PAF1, CTR9, LEO1, CDC73, RTF1, SPT6, and RPB3 (35, 79, 91). For most experiments, yeast cells were grown at 30°C in YPD medium containing 1% yeast extract, 2% peptone, and 2% glucose (72). For the complementation experiment (see Fig. 2C), yeast cells were grown at 30°C in synthetic complete medium lacking uracil (sc − ura) (72).
Table 1.
Saccharomyces cerevisiae strains used in this study
| Namea | Genotype |
|---|---|
| FY4 | MATa |
| FY5 | MATα |
| KY399 | MATα rtf1Δ100::URA3 leu2Δ1 ura3-52 trp1Δ63 |
| KY716 | MATα his3Δ200 lys2-128δ ura3-52 spt5-194 |
| KY735 | MATα his4-912δ lys2-128δ leu2Δ1 ura3-52 arg4-12 spt4Δ1::URA3 |
| KY785 | MATahis4-912δ lys2-128δ leu2Δ(0 or 1) trp1Δ63 CTR9-6×MYC::LEU2 LEO1-3×HA::HIS3 SPT5-FLAG |
| KY786 | MATahis4-912δ lys2-128δ leu2Δ(0 or 1) ura3-52 trp1Δ63 CTR9-6×MYC::LEU2 3×HA-CDC73::URA3 SPT5-FLAG |
| KY1349 | MATabur2Δ2::URA3 his4-912δ lys2-128δ suc2::UAS (−1900/−390) ura3-52 trp1Δ63 |
| KY1451 | MATα paf1Δ::KanMX bur2Δ::KanMX |
| KY1700 | MATα paf1Δ::KanMX |
| KY1703 | MATartf1Δ::KanMX |
| KY1706 | MATα cdc73Δ::KanMX |
| KY1712 | MATα rad6Δ::KanMX |
| KY1713 | MATabre1Δ::KanMX |
| KY1721 | MATα 3×HA-PAF1 |
| KY1805 | MATα leo1Δ::KanMX |
| KY2167 | MATα HTA1-htb1K123R (hta2-htb2)Δ::KanMX ura3Δ0 |
| KY2170 | MATactr9Δ::KanMX leu2Δ1 |
| KY2172 | MATα (hta2-htb2)Δ::KanMX ura3Δ0 |
| KY2082 | MATα ura3-52 lys2-128δ leu2Δ1 trp1Δ63 3×HA-RTF1 |
| YJ759 | MATactr9Δ::KanMX leu2Δ0 ura3Δ0 his3Δ0 lys2Δ0 |
| YJ786 | MATα paf1Δ::KanMX ura3Δ0 his3Δ200 lys2Δ0 |
| YJ882 | MATα ura3-52 or ura3Δ0 lys2Δ0 or lys2-128δ his3Δ200 leu2Δ0 or leu2Δ1RPB3-3×HA::LEU2 SPT6-FLAG |
| YJ1013 | MATaura3-52 or ura3Δ0 lys2Δ0 or lys2-128δ leu2Δ0 or leu2Δ1RPB3-3×HA::LEU2 SPT6-FLAG rtf1Δ::KanMX |
| YJ1014 | MATaura3-52 or ura3Δ0 lys2Δ0 or lys2-128δ leu2Δ0 or leu2Δ1 his3Δ200RPB3-3×HA::LEU2 SPT6-FLAG rtf1Δ::KanMX |
| YJ1016 | MATaura3-52 or ura3Δ0 lys2Δ0 or lys2-128δ leu2Δ0 or leu2Δ1 his3Δ200RPB3-3×HA::LEU2 SPT6-FLAG ctr9Δ::KanMX |
| YJ1030 | MATahis4-912δ trp1Δ63 ura3-52 or URA3lys2-128δ leu2Δ1 SPT6-FLAG RPB3-3×HA::LEU2 paf1Δ::URA3 |
| YJ1031 | MATα his4-912δ trp1Δ63 ura3-52 or URA3lys2-128δ leu2Δ1 SPT6-FLAG RPB3-3×HA::LEU2 paf1Δ::URA3 |
| YJ1087 | MATaura3Δ0 pRS416 |
FY and KY strains were kindly provided by Fred Winston and Karen Arndt, respectively.
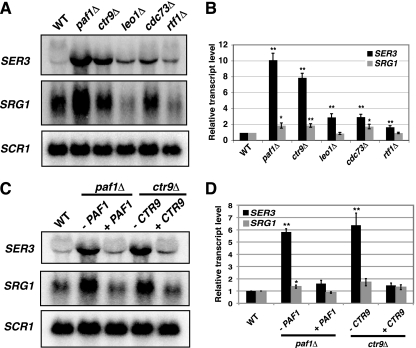
Fig. 2.
SER3 repression is primarily dependent on the Paf1 and Ctr9 subunits. (A) Representative Northern analysis of SER3, SRG1, and SCR1 (loading control) RNA levels in wild-type (FY5), paf1Δ (KY1700), ctr9Δ (KY2170), leo1Δ (KY1805), cdc73Δ (KY1706), and rtf1Δ (KY1703) strains. (B) Quantitation of results from a minimum of seven biological replicates. The values shown are the mean SER3 (black) and SRG1 (gray) transcript levels, normalized to the SCR1 loading control and made relative to the wild-type strains. Error bars indicate standard errors of the means, and asterisks indicate statistical significance compared to the wild type (*, P < 0.05; **, P < 0.01). (C) Representative Northern analysis of SER3, SRG1, and SCR1 (loading control) RNA levels. The wild-type strain (YJ1087) was transformed with a control plasmid (pRS416). The paf1Δ strain (YJ786) was transformed with either pRS416 (−PAF1) or pRS316-HA-PAF1 (+PAF1). The ctr9Δ (YJ759) strain was transformed with either pRS416 (−CTR9) or pAP10 (+CTR9). (D) Quantitation of results from a minimum of three biological replicates. The values shown are the mean SER3 (black) and SRG1 (gray) transcript levels, normalized to the SCR1 loading control and made relative to the wild-type strains. Error bars indicate standard errors of the means, and asterisks indicate statistical significance compared to the wild type (*, P < 0.05; **, P < 0.01).
Plasmids.
pRS316-HA-PAF1 is a URA3-marked, CEN/ARS plasmid carrying a hemagglutinin (HA) epitope-tagged version of PAF1. pAP10 is a URA3-marked, CEN/ARS plasmid carrying CTR9. Both plasmids were kind gifts from K. Arndt (University of Pittsburgh). pRS416 is a URA3-marked CEN/ARS plasmid (7).
Northern analysis.
Total RNA was isolated from cells grown to 1 × 107 to 2 × 107 cells/ml and separated on a 1% formaldehyde-agarose gel as described previously (3). RNA was transferred to a Gene Screen membrane (Perkin-Elmer) and hybridized with radiolabeled probes generated by random-primed labeling of PCR fragments that were amplified from the following genomic sequences: SRG1 (chromosome V [ChrV], nucleotides 322258 to 322559), SER3 (ChrV, nucleotides 324059 to 324307), and SCR1 (ChrV, nucleotides 441741 to 442266), which was used as a control for RNA loading.
Chromatin immunoprecipitation (ChIP).
Cells were grown in YPD at 30°C to a density of 1 × 107 to 2 × 107 cells/ml and then treated with 1% formaldehyde for 20 min. Chromatin was isolated and sonicated as previously described (77) and then incubated with antibodies overnight at 4°C. Anti-FLAG M2 agarose (30 μl; A2220; Sigma) was used to immunoprecipitate Spt6-FLAG. Anti-HA antibody (1 μl; sc-7392; Santa Cruz Biotechnology) was used to immunoprecipitate HA-Paf1, HA-Rtf1, Leo1-HA, HA-Cdc73, and Rpb3-HA. Anti-Myc (1 μl; sc-789; Santa Cruz Biotechnology), anti-Spt16 (1 μl; kind gift from T. Formosa, University of Utah), and anti-histone H3 (5 μl; ab1791; Abcam) antibodies were used to immunoprecipitate Ctr9-MYC, Spt16, and histone H3, respectively. Primary antibody-protein conjugates were isolated by incubating with 30 μl protein A- or protein G-coupled Sepharose beads (GE Healthcare) at 4°C for 2 to 3 h. After purifying DNA through PCR purification columns (Qiagen), the amount of immunoprecipitated (IP) DNA relative to input DNA was determined by quantitative PCR (qPCR) and then normalized to a control region on chromosome V that lacks open reading frames (no ORF), which has been previously described (40).
Nucleosome scanning assay.
Nucleosome scanning assays were performed as previously described (27). Briefly, cells were grown in YPD to 2 × 107 to 3 × 107 cells/ml at 30°C and then treated with 2% formaldehyde followed by 300 mM glycine. A total of 1.2 × 109 cells were spheroplasted with Zymolyase 20T (Seikagaku Biobusiness) and divided into six aliquots, which were then incubated with increasing concentrations of micrococcal nuclease (MNase; nuclease S7; Roche). DNA was extracted, treated with RNase A, and subjected to gel electrophoresis and qPCR to determine the extent of MNase digestion. Well-characterized regions of the GAL1 promoter, one bound by a nucleosome (NB) and another nucleosome-free (NUB), were used as controls (8, 24, 47). The samples in which the concentration of MNase yielded mostly mononucleosome-sized fragments and the NUB/NB ratio was less than 15% were then subjected to further qPCR analyses using primer pairs that amplified ∼100-bp fragments that tile the SER3 locus. The amount of amplification for each SER3 primer pair (SER3-7 to SER3-41) in the digested sample was made relative to the undigested sample and normalized to the GAL1 NB region.
qPCR.
Results of nucleosome scanning and ChIP assays were analyzed with an ABI 7300 or StepOnePlus real-time PCR system and SYBR green reagents (Fermentas). Primer sets that amplified SER3 (SER3-1 to SER3-41) and the no-ORF control have been previously described (27). Primer sets specific to PMA1 5′ ORF, ADH1, GAL1 5′ ORF, and CYC1 have also been described previously (26). Quantitation of real-time PCR results was performed using the Pfaffl method (65).
Western blot analysis.
Whole-cell extracts (WCE) were prepared from cells grown in YPD at 30°C to 1 × 107 to 2 × 107 cells/ml using trichloroacetic acid as previously described (17, 101). Equal volumes of WCE were separated by 10% acrylamide SDS-PAGE, transferred to nitrocellulose (Whatman), and immunoblotted with anti-FLAG antibody (F3165; Sigma) or anti-Spt16 antibody (kind gift from T. Formosa, University of Utah). After incubation with anti-mouse (FLAG) or anti-rabbit (Spt16) horseradish peroxidase (HRP)-conjugated secondary antibody (GE Healthcare), the immunoreactive proteins were visualized by enhanced chemiluminescence detection (Perkin-Elmer) using a Kodak image station 440CF. Blots were then stripped and reprobed with anti-glucose-6-phosphate dehydrogenase (anti-G6PDH) antibody (A9521; Sigma) as a loading control. Quantitation of Spt6-FLAG and Spt16 protein levels was performed by measuring the signal intensities using the Kodak 1D 3.6 software. Spt6-FLAG and Spt16 signals were made relative to the signal from the G6PDH loading control and normalized to wild-type signal, which was set to 1.
RESULTS
Spt4, Spt5, and Bur2 are required to repress SER3 transcription through a pathway involving Paf1.
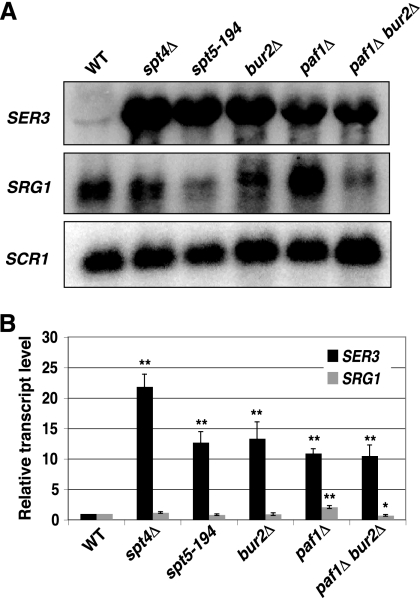
Previously, we showed that transcription of SRG1 intergenic ncDNA across the SER3 promoter region strongly interferes with the initiation of SER3 transcription (48, 50). To identify factors involved in SER3 repression by intergenic transcription, we screened the haploid S. cerevisiae gene deletion collection (Open Biosystems) for gene deletions that derepress a SER3pr-HIS3 reporter gene (J. A. Pruneski, unpublished data). Two of the genes identified in this screen encode the Spt4 and Bur2 transcription elongation factors. Spt4 partners with the essential factor Spt5 to form the yeast homolog of the mammalian DSIF complex (yDSIF) (89). yDSIF associates with Pol II over actively transcribed genes, has numerous physical and genetic interactions with other transcription factors, and facilitates elongation through chromatin (28, 71, 78, 79). Bur2 acts as a cyclin to activate the essential cyclin-dependent kinase Bur1, forming a partial functional homolog of mammalian P-TEFb (96, 99). The Bur1/2 complex plays a variety of roles in transcription through the phosphorylation of substrates, including the CTD of the RNA Pol II subunit Rpb1 (58, 68), the ubiquitin-conjugating enzyme Rad6 (95), and the C-terminal repeat region of Spt5 (46, 102). Phosphorylation by Bur1/2 activates the Spt5 protein, which promotes the recruitment of the Paf1 complex to chromatin (46, 102). To determine if these transcription elongation factors are required for endogenous SER3 repression, we measured SER3 transcript levels in strains lacking functional copies of Spt4, Spt5, Bur2, and the Paf1 subunit of the Paf1 complex (Fig. 1A). In agreement with the results from our genetic screen, both spt4Δ and bur2Δ mutants strongly derepressed SER3, with the spt4Δ mutant being slightly more defective in SER3 repression. A temperature-sensitive mutation of the essential SPT5 gene, spt5-194, not only derepressesd SER3 at a nonpermissive temperature (39°C [data not shown]), as had been previously shown (20), but also at a permissive temperature (30°C) (Fig. 1). Strains lacking Paf1 exhibited increased SER3 transcript levels, similar to those observed in the bur2Δ and spt5-194 strains. This result agrees with previous microarray data that showed increased SER3 levels in a paf1Δ strain (64). Consistent with these factors working in the same pathway, a paf1Δ bur2Δ double mutant derepressed SER3 to a level that was equivalent to either paf1Δ or bur2Δ single mutants (Fig. 1). Importantly, SRG1 transcript levels were not dramatically reduced, indicating that SER3 repression in these mutants cannot be explained solely by reduced SRG1 transcription. Taken together, these results show that Paf1 is required for SER3 repression, likely involving its recruitment to SRG1 by Bur1/2 and Spt4/5, as has been seen at other transcribed regions (46, 102).
Fig. 1.
Spt4, Spt5, Bur2, and Paf1 are required to repress SER3. (A) Representative Northern analysis of SER3, SRG1, and SCR1 (loading control) RNA levels in wild-type (FY4), spt4Δ (KY735), spt5-194 (KY716), bur2Δ (KY1349), paf1Δ (KY1700), and paf1Δ bur2Δ (KY1451) strains. (B) Quantitation of results from a minimum of four biological replicates. The values shown are the mean SER3 (black) and SRG1 (gray) transcript levels, normalized to the SCR1 loading control and made relative to the wild-type strains. Error bars indicate the standard errors from the means, and asterisks indicate statistical significance determined by pairwise comparisons between the wild type and mutant using a two-tailed Student t test (*, P < 0.05; **, P < 0.01).
SER3 repression by the Paf1 complex depends primarily on the Paf1 and Ctr9 subunits.
To further investigate the requirement for Paf1 complex subunits in SER3 repression, we examined SER3 and SRG1 transcript levels from mutant strains that each lacked one of the five subunits of the Paf1 complex (Fig. 2). These experiments revealed strong derepression of SER3 (8- to 10-fold) in paf1Δ and ctr9Δ mutants compared to wild-type strains. Complementation of these strains with plasmid-borne copies of wild-type PAF1 and CTR9, respectively, restored SER3 repression (Fig. 2C and D). More modest defects in SER3 repression (2- to 3-fold increases in SER3 transcript levels) were seen in leo1Δ, cdc73Δ, and rtf1Δ strains (Fig. 2B). Whereas the Paf1 complex has been shown to facilitate transcription elongation (12, 30, 37, 63, 70, 88), it is important to note that the paf1Δ and ctr9Δ strains did not exhibit reduced SRG1 RNA levels. Rather, SRG1 levels were increased 2-fold in these mutants. However, these increases in SRG1 RNA levels were unlikely to impact SER3 levels, as cdc73Δ strains also exhibited higher SRG1 levels but showed only a modest increase in SER3 levels. SRG1 levels showed some variability in leo1Δ and rtf1Δ strains, but when multiple experiments were averaged, there was no difference from wild-type levels (Fig. 2B). We conclude from these data that Paf1 complex-mediated repression of SER3 occurs primarily through the activities of the Paf1 and Ctr9 subunits by a mechanism that does not involve the control of SRG1 transcript levels.
The Paf1 complex localizes to the SER3 promoter when SRG1 is transcribed.
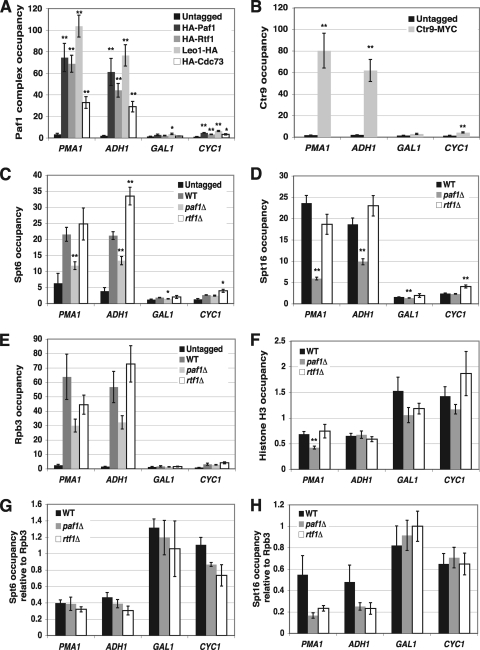
We next performed ChIP experiments to determine if the Paf1 complex is physically associated with the SER3 promoter when SER3 is repressed. Cells expressing previously described epitope-tagged derivatives of Paf1, Rtf1, Leo1, Cdc73, and Ctr9 (79, 91) were grown in serine-rich medium. Chromatin was isolated from these cells after cross-linking with formaldehyde and then subjected to immunoprecipitation with antibodies that recognize the epitope tags. We detected strong occupancy of all Paf1 complex subunits, specifically, at the SER3 promoter (Fig. 3). This is likely a consequence of its colocalization with RNA Pol II over actively transcribed SRG1, similar to what has been previously described for other actively transcribed genes (42, 67, 79). These results suggest that the Paf1 complex may directly contribute to the mechanism by which SRG1 transcription represses SER3 transcription.
Fig. 3.
The Paf1 complex colocalizes with actively transcribed SRG1. (A) ChIP analysis of HA-tagged Paf1 complex subunits at SRG1 (SER3-22 and SER3-19) and the flanking AIM9 (SER3-41) and SER3 (SER3-1) genes from untagged (FY4), 3×HA-PAF1 (KY1721), 3×HA-RTF1 (KY2082), LEO1-3×HA (KY785), and 3×HA-CDC73 (KY786) strains grown in YPD at 30°C. (B) ChIP analysis of Ctr9-Myc from untagged (KY399) and CTR9-6×MYC (KY785) strains grown in YPD at 30°C. The relative occupancies of these factors were calculated using qPCR as described in Materials and Methods. Each value represents the mean ± the standard error of the mean of three biological replicates, and asterisks indicate statistical significance compared to the untagged control (*, P < 0.05; **, P < 0.01). Below the graphs is a schematic of the SRG1/SER3 locus, and the arrows indicate the transcription start sites of SRG1 and SER3. The gray box represents the Cha4 binding site, black boxes indicate TATA sequences, and white boxes are sequences required for SER3 activation. The block arrow indicates SRG1 transcription, and the horizontal black bars mark the location of the DNA fragments amplified by qPCR.
SER3 repression is largely independent of known Paf1 complex-dependent histone modifications.
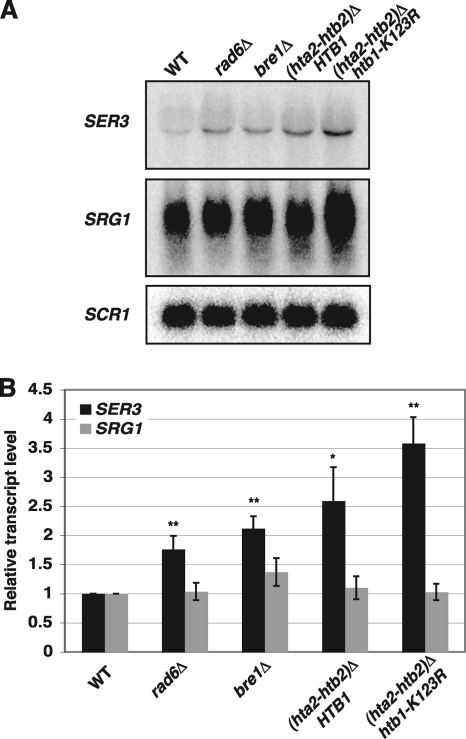
Several studies have indicated a role for the Paf1 complex in mediating transcription-dependent posttranslational modifications of histone proteins. These include H2B K123ub by the Rad6 ubiquitin conjugase and Bre1 ubiquitin ligase and subsequent methylation of H3 K4 and K79 by the Set1 and Dot1 histone methyltransferases, and also the methylation of K36 by Set2 (14, 60, 74, 94). Previously, we showed by Northern analysis that either the deletion of the methyltransferases responsible for methylation of K4, K36, and K79 of histone H3 or the mutation of these lysine residues to alanines had little to no effect on SER3 repression (26, 27). Despite there being no role for the downstream methylation marks, it is possible that the upstream H2B K123ub does regulate SER3 repression. Therefore, we assayed the effects of histone H2B K123ub on SER3 repression. Northern analyses revealed only modest increases in SER3 expression in rad6Δ (2-fold) and bre1Δ (1.5-fold) mutants (Fig. 4). Similarly, a conservative mutation that replaced H2B lysine 123 with arginine also resulted in less than a 2-fold increase in SER3 levels compared to the relevant control strain lacking one copy of the histone H2A and histone H2B genes (compare hta2Δ htb2Δ strains expressing HTB1 or htb1-K123R). These results are consistent with the minor defect in SER3 repression that was observed for cells lacking Rtf1 (Fig. 2), which has been previously shown to be the subunit primarily required for this modification (59, 87, 91, 94). Taken together, the role of the Paf1 complex in SER3 repression seems to be largely independent of its known roles in regulating histone modifications.
Fig. 4.
SER3 repression is largely independent of histone H2B ubiquitylation. (A) Representative Northern analysis of SER3, SRG1, and SCR1 (loading control) RNA levels in wild-type (FY5), rad6Δ (KY1712), bre1Δ (KY1713), (hta2-htb2)Δ (KY2172), and (hta2-htb2)Δ htb1-K123R (KY2167) strains grown in YPD at 30°C. (B) Quantitation of results from a minimum of three biological replicates. The values shown are the mean SER3 (black) and SRG1 (gray) transcript levels, normalized to the SCR1 loading control and made relative to the wild-type strains. Error bars indicate standard errors of the means, and asterisks indicate statistical significance compared to the wild type (*, P < 0.05; **, P < 0.01).
Paf1 and Ctr9 are required for nucleosome occupancy over the SER3 promoter.
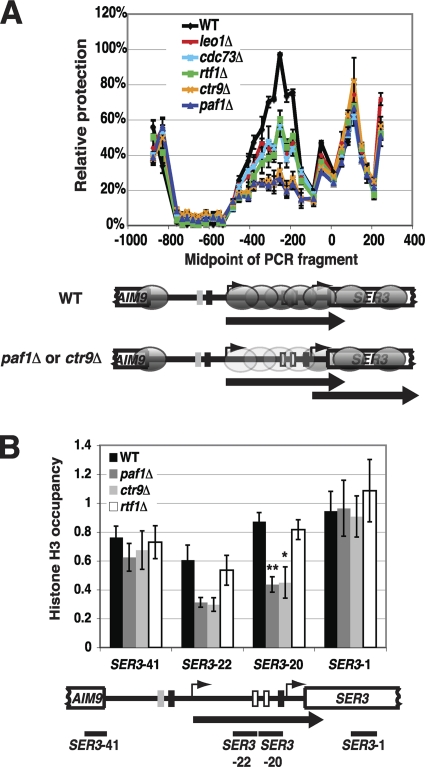
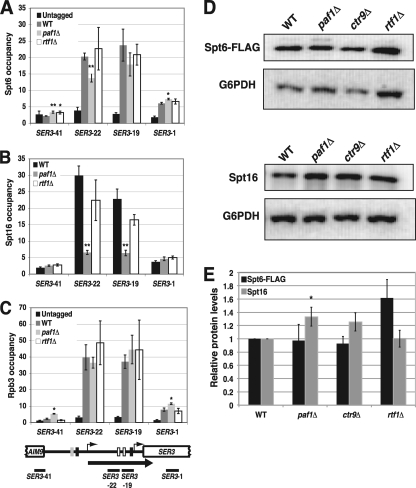
We recently demonstrated a role for chromatin in the repression of SER3 (27). Under conditions in which SRG1 is transcribed and SER3 is repressed, the SER3 promoter is occupied by randomly positioned nucleosomes that prevent transcription factors from binding to this region. When SRG1 is downregulated, the SER3 promoter region becomes nucleosome free, allowing transcription factors to bind and induce SER3 expression (27). These results support a promoter occlusion model whereby intergenic SRG1 transcription deposits and maintains nucleosomes over the SER3 promoter to mediate repression. To test whether the Paf1 complex contributes to SER3 repression by affecting SRG1 transcription-dependent nucleosome occupancy of the SER3 promoter, we first performed nucleosome scanning assays on mutant strains that each lacked one of the five subunits of the Paf1 complex (Fig. 5A). MNase protection across SER3 was determined by qPCR and normalized to the protection of a well-positioned nucleosome in the GAL1 promoter, whose digestion by MNase was unaffected in these mutants (data not shown). In wild-type cells (black line), we observed a broad peak of protection over the SRG1 transcribed unit that overlaps the SER3 promoter, as we have previously reported (27). Strikingly, the effects of these mutants on MNase protection across the SRG1 transcription unit were consistent with their effects on SER3 expression. For paf1Δ and ctr9Δ strains that exhibit strong SER3 derepression, MNase protection across SRG1 was significantly reduced. For leo1Δ, rtf1Δ, and cdc73Δ mutants, which more modestly derepress SER3, we observed more modest decreases in MNase protection across this region.
Fig. 5.
Paf1 and Ctr9 are required for nucleosome occupancy over the SER3 promoter. (A) Nucleosome scanning assays were performed on wild-type (FY4), leo1Δ (KY1805), cdc73Δ (KY1706), rtf1Δ (KY1703), ctr9Δ (KY2170), and paf1Δ (KY1700) strains grown in YPD at 30°C. MNase protection across the SER3 locus was calculated relative to a positioned nucleosome within the GAL1 promoter by using qPCR as described in Materials and Methods. The mean ± standard error of the mean from three biological replicates is plotted at the midpoint for each PCR product. Shown below the graph is a diagram of the SER3 locus, comparing the positions of nucleosomes (gray ovals) extrapolated from the MNase protection data between wild-type and ctr9Δ or paf1Δ strains. The light gray ovals are indicative of reduced nucleosome occupancy compared to the darker ovals (wild-type strains). (B) ChIP analysis of histone H3 from wild-type (FY4), paf1Δ (YJ1030), ctr9Δ (YJ1016), and rtf1Δ (YJ1014) strains grown in YPD at 30°C. The relative occupancies of these factors were calculated as described in Materials and Methods. Each value represents the mean ± the standard error of the mean of at least three biological replicates, and asterisks indicate statistical significance compared to wild type (*, P < 0.05; **, P < 0.01). The P values for the decrease in H3 occupancy over primer set SER3-22 in paf1Δ and ctr9Δ strains were 0.07 and 0.06, respectively. Below the graphs is a schematic of the SRG1/SER3 locus, with the arrow indicating SRG1 transcription and the black bars indicating the location of the DNA fragments amplified by qPCR.
We also performed histone H3 ChIP assays on paf1Δ, ctr9Δ, and rtf1Δ strains (Fig. 5B). Consistent with our nucleosome scanning results, we detected reduced histone H3 occupancy specifically over the SRG1 transcribed region in both paf1Δ and ctr9Δ strains, but not rtf1Δ strains, compared to wild-type strains (Fig. 5B, compare amplicons SER3-20 and SER3-22 to SER3-1 and SER3-41). Taken together, our nucleosome scanning and histone H3 ChIP results indicate that the Paf1 complex, primarily through the activities of the Paf1 and Ctr9 subunits, contributes to SER3 repression by promoting SRG1 transcription-dependent nucleosome occupancy across the SER3 promoter.
Spt16 colocalization with RNA Pol II across SRG1 is reduced in a paf1Δ strain.
We previously showed that the transcription-dependent nucleosome assembly mediating SER3 repression requires the essential histone chaperones Spt6 and Spt16. Impairment of either factor reduces nucleosome occupancy over the SER3 promoter and strongly derepresses SER3, even though SRG1 transcription is maintained (27). Given the importance of these factors in SER3 repression and that a previous study with Drosophila provided evidence to support a role for the Paf1 complex in recruiting Spt6 and FACT to actively transcribed genes (1), we performed ChIP assays to compare the occupancy of Spt6, Spt16, and the Rpb3 subunit of RNA Pol II at actively transcribed SRG1 between wild-type, paf1Δ, and rtf1Δ strains (Fig. 6). Consistent with our Northern blotting data, we found equivalently high levels of Rpb3 associating with actively transcribed SRG1, which overlaps the SER3 promoter (Fig. 6C, amplicons SER3-19 and SER3-22) in all three strains. In wild-type cells, we detected strong Spt6 and Spt16 occupancy that colocalized with Rpb3 across the SRG1 transcription unit (Fig. 6A and B). Whereas the occupancies of these factors were not significantly affected in an rtf1Δ mutant, we observed a 4- to 5-fold reduction in Spt16 occupancy over this region in a paf1Δ mutant, while Spt6 occupancy was more moderately reduced (less than 2-fold). Notably, the decreased association of these factors was not due to a reduction in Spt6 or Spt16 expression levels as determined by Western analysis of whole-cell extracts prepared from wild-type and Paf1 complex mutant strains (Fig. 6D and E). Taken together, these data show that Paf1 is required for Spt16 occupancy and, to a lesser extent, Spt6 occupancy over actively transcribed SRG1, which overlaps the SER3 promoter.
Fig. 6.
Paf1 and Ctr9 are required for Spt6 and Spt16 colocalization to actively transcribed SRG1. ChIP of Spt6-FLAG (A), Spt16 (B), and Rpb3-HA (C) from untagged (FY4) strains and wild-type (YJ882), paf1Δ (YJ1031), and rtf1Δ (YJ1013) strains that all express epitope-tagged versions of Spt6 (SPT6-FLAG) and Rpb3 (RPB3-3×HA). The relative occupancies of these factors were calculated using qPCR as described in Materials and Methods. Each value represents the mean ± the standard error of the mean of three biological replicates, and asterisks indicate statistical significance compared to the wild type (*, P < 0.05; **, P < 0.01). Below the graphs is a schematic of the SRG1/SER3 locus, with the arrow indicating SRG1 transcription and the black bars indicating the location of the DNA fragments amplified by qPCR. (D) Western analysis of Spt6 and Spt16. The wild-type, paf1Δ, ctr9Δ, and rtf1Δ strains shown in panel A were subjected to Western blotting to compare Spt6 (top panel) and Spt16 (bottom panel) protein levels between these strains. Representative immunoblots are shown. These blots were reprobed with an antibody to G6PDH as a loading control. (E) Quantitation of Western analyses from a minimum of four biological replicates. The values shown are the mean Spt6-FLAG (black) and Spt16 (gray) protein levels, normalized to the G6PDH loading control and made relative to the wild-type strains. Error bars indicate standard errors of the means, and asterisks indicate statistical significance compared to the wild type (*, P < 0.05; **, P < 0.01).
Effects of Paf1 complex mutants at other transcribed yeast genes.
Since the Paf1 complex colocalizes with Pol II over actively transcribed genes (38, 42, 53, 67), we performed ChIP assays to determine if the occupancy of histone H3, Spt6, and Spt16 at other transcribed genes is dependent on Paf1, as we have observed for SRG1. The Paf1 complex, Spt6, Spt16, and Rpb3 were present at high levels within the open reading frames of the highly transcribed PMA1 and ADH1 genes compared to background levels of association with the lowly transcribed GAL1 and CYC1 genes (Fig. 7A to E). Similar to our results for SRG1, occupancy of Spt6 and Spt16 was reduced over PMA1 and ADH1 in paf1Δ mutants but not rtf1Δ mutants (Fig. 7C and D). However, in contrast to what we observed at SRG1, Rpb3 occupancy was modestly reduced over PMA1 and ADH1 in paf1Δ strains (Fig. 7E). Since interactions with RNA Pol II may contribute to the recruitment of Spt6 and Spt16 to actively transcribed genes (21, 52, 81), we recalculated Spt6 and Spt16 occupancies relative to Rpb3 occupancy (Fig. 7G and H). Relative to Rpb3 occupancy, a reduction in Spt16 occupancy in paf1Δ cells remained evident over these two highly transcribed genes, although the difference was less significant and was now similar to what we observed for rtf1Δ cells. Additionally, the reduction in Spt6 occupancy that was observed in paf1Δ cells was no longer evident. Interestingly, we found that histone H3 occupancy was significantly reduced at PMA1 in paf1Δ mutants; however, we did not detect any change in histone H3 occupancy at ADH1 (Fig. 7F). Taken together, these data indicate that the requirement of Paf1 and Ctr9 for transcription-dependent nucleosome occupancy and recruitment of Spt6 and Spt16 at SRG1 is not universal for all highly transcribed genes.
Fig. 7.
Effects of the Paf1 complex mutant on histone H3, Spt6, and Spt16 occupancy at other transcribed genes. (A and B) Relative occupancies of HA-Paf1, HA-Rtf1, Leo1-HA, and HA-Cdc73 (A) and of Ctr9-MYC (B) within the coding sequence of two highly transcribed genes, PMA1 and ADH1, and two lowly transcribed genes, GAL1 and CYC1, were determined by qPCR using the ChIP assay shown in Fig. 3. (C to E) Relative occupancies of Spt6-FLAG (C), Spt16 (D), and Rpb3-HA (E) at these four genes were determined by qPCR by using the immunoprecipitated chromatin shown in Fig. 6. (F) Relative occupancy of histone H3 was determined by qPCR using the immunoprecipitated chromatin assayed in Fig. 5B. The occupancies of each of these factors at these genes were normalized to their occupancies at a control region on chromosome V that contained no open reading frames (no ORF). Each value represents the mean ± standard error of the mean for at least three biological replicates, and asterisks indicate statistical significance (*, P < 0.05; **, P < 0.01). (G and H) Spt6-FLAG (G) and Spt16 (H) ChIP data from wild-type, paf1Δ, and rtf1Δ strains (C and D) were normalized to Rpb3-HA ChIP data (E).
DISCUSSION
In this study, we sought to further our understanding of the repression of SER3 by SRG1 intergenic transcription. We uncovered a role for the multifunctional Paf1 transcription elongation complex in SER3 repression. We showed that the Paf1 complex colocalizes with Pol II during SRG1 transcription, and we provided genetic data indicating that the Paf1 complex functions in a previously characterized pathway with Bur1/2 and Spt4/5. Our data indicate that SER3 repression requires the Paf1 and Ctr9 subunits to promote SRG1 transcription-dependent nucleosome occupancy across the SER3 promoter, possibly by facilitating the association of the histone chaperones Spt6 and Spt16.
Our nucleosome scanning and histone ChIP experiments provided evidence that Paf1 and Ctr9 repress SER3 by facilitating SRG1 transcription-dependent nucleosome occupancy of the SER3 promoter (Fig. 5). Previous studies have indicated that a primary function of the Paf1 complex is to establish several histone modifications that are important for chromatin dynamics at actively transcribed genes. However, these marks do not appear to play a major role in SER3 repression by SRG1. First, we have shown that both an rtf1Δ mutant and mutations that prevent Rtf1-dependent monoubiquitylation of histone H2B at K123—either an arginine substitution of lysine 123 or deletion of the RAD6 or BRE1 genes responsible for this mark—only weakly derepress SER3 compared to paf1Δ and ctr9Δ mutants (Fig. 2 and 4). Second, we determined that a parallel set of mutations preventing the subsequent methylation of histone H3 K4 and K79 have no effect on SER3 repression (26, 27). Finally, we had previously shown that SER3 repression is also unaffected by mutations that prevent methylation of histone H3 K36 (26, 27), a modification that is dependent on Paf1, Ctr9, and to a lesser extent Cdc73 (14).
The Paf1 complex has also been shown to play a role in regulating phosphorylation events occurring in the heptapeptide repeats in the CTD of RNA Pol II and transcription termination (34, 56, 61, 64, 75, 87). Although we cannot absolutely rule out a role for these Paf1 complex-dependent activities in regulating SER3 repression, our data suggest that these activities are unlikely to account for the role of the Paf1 complex in maintaining SRG1 transcription-dependent nucleosome occupancy of the SER3 promoter. First, there are distinct differences in the subunit requirements for these activities. Whereas nucleosome occupancy of the SER3 promoter is primarily dependent on Paf1 and Ctr9, additional subunits of this complex are required for the phosphorylation of the serines at position 2 within the heptapeptide repeats (Ser2-P) and proper transcription termination (34, 56, 61, 64, 75, 87). Second, the termination of SRG1 has been mapped to two distinct sites, 75 bp 5′ and 25 bp 3′ of the SER3 translation start site, along with a minor read-through product to the end of SER3 (86). Among Paf1 complex subunit deletions, we did not observe any increase in this read-through product, as might be expected if the Paf1 complex affected SRG1 termination (data not shown). Taken together, our studies of SER3 repression suggest a role for the Paf1 complex in controlling transcription-coupled nucleosome occupancy that is primarily dependent on the Paf1 and Ctr9 subunits and independent of previously characterized activities of this complex.
Interestingly, a possible role for the Paf1 complex in regulating chromatin dynamics during transcription, independent of its roles in promoting histone modifications, was recently proposed by Kim et al. based on an in vitro transcription system where the human Paf1 complex was shown to stimulate elongation through nucleosomes (37). Our studies of SER3 regulation suggest this role may be specific to the Paf1 and Ctr9 subunits, involving a more direct role for these two factors in promoting nucleosome occupancy during transcription. One study previously implicated the Paf1 complex in altering nucleosome occupancy during gene induction. However, rather than promoting nucleosome occupancy, the earlier study showed that Paf1 and Ctr9 were required for efficient histone eviction at the GAL1-10 promoter during the induction of these genes in response to galactose (51). Several studies have also linked efficient induction of GAL1-10 to H2B K123ub by Rad6/Bre1 (29, 33, 97), suggesting that the effect of the Paf1 complex on GAL1-10 induction is likely through its role in promoting histone modifications.
How might Paf1 and Ctr9 promote SRG1 transcription-dependent nucleosome occupancy at the SER3 promoter? One possibility is that Paf1 regulates the histone chaperones Spt6 and/or Spt16, which are required for the maintenance of nucleosomes over the SER3 promoter (27). In yeast, the Paf1 complex has genetic interactions with Spt6 (34) and both physical and genetic interactions with Spt16 (63, 79). The Paf1 complex has also been shown in Drosophila to be required for full recruitment of Spt6 and the FACT subunit SSRP1 during transcription (1). These connections led us to examine whether the recruitment of these factors is affected in Paf1 complex mutants in yeast (Fig. 6). ChIP experiments revealed that Spt16 occupancy over SRG1 is strongly dependent on Paf1 but not Rtf1, which correlates with the effect that each of these factors has on SER3 repression. Western analyses and RNA Pol II ChIP data indicated that this reduction in Spt16 occupancy in paf1Δ cells is not caused by a reduction in Spt16 protein levels or by a reduction in RNA Pol II levels across SRG1 (Fig. 6). Taken together, our results support a role for Paf1 and Ctr9 in promoting FACT occupancy across actively transcribed SRG1. In contrast to Spt16, we found Spt6 occupancy at SRG1 to be only modestly dependent on Paf1. Therefore, while Paf1 and Ctr9 may play a prominent role in Spt16 occupancy at SRG1, other factors are likely to contribute significantly to Spt6 occupancy of this region. Spt6 is known to associate with elongating Pol II through a direct interaction with Pol II CTD containing Ser2-P (21, 81). Although it is unlikely to be part of the Paf1-dependent pathway that represses SER3 as we discussed earlier, this mark may contribute to Spt6 occupancy at SRG1 in a Paf1-independent pathway. Interestingly, Thebault et al. recently reported that Spt6 occupancy at SRG1 is also partially dependent on Spt2, an HMG-like transcription elongation factor (85). Although we cannot rule out the possibility that Spt2 contributes to Spt6 occupancy in a pathway with Paf1, Spt2 may also facilitate Spt6 recruitment independently of Paf1. Overall, our results are consistent with those observed in Drosophila, in which depletion of Paf1, and to a lesser extent Rtf1, led to reduced association of Spt6 and FACT over an actively transcribed gene without affecting Pol II association or global protein levels (1). Interestingly, localization of the Paf1 complex to actively transcribed genes has also been shown to be partially dependent on Spt6 and Spt16 (34, 63). Therefore, it is possible that recruitment of Spt6, Spt16, and the Paf1 complex is interdependent, where the disruption of one of these factors results in reduced associations of the others.
Cells lacking Paf1 not only reduce the association of Spt6 and Spt16 across the SRG1 transcription unit but also reduce nucleosome occupancy over this region that overlaps the SER3 promoter. Because both Spt6 and Spt16 interact with histones (5, 6, 25) and have been implicated in restoring nucleosome occupancy after the passage of RNA Pol II at transcribed genes (5, 13, 31, 35, 52), it is also possible that the loss of these factors over SRG1 is a consequence, rather than a cause, of reduced nucleosomes over this region in the paf1Δ strains. In this case, the Paf1 complex may be required for the transcription-dependent nucleosome reassembly activity of Spt6 and Spt16 rather than recruitment of these factors. Interestingly, Spt2 is also required to promote nucleosome occupancy across the SRG1 transcription unit to repress SER3 transcription (62, 85). In addition, Spt2 has been genetically linked to Paf1 and Ctr9, and its colocalization with RNA Pol II across actively transcribed genes is dependent on Paf1, albeit weakly (62). It will be interesting to decipher the functional interplay between the Paf1 complex, Spt6, FACT, and Spt2 in promoting SRG1 transcription-dependent nucleosome occupancy and SER3 repression.
In addition to the well-characterized role of the Paf1 complex in promoting transcription, whole-genome expression analyses indicate that the Paf1 complex also functions as a negative regulator of transcription (64). Our finding that the Paf1 complex indirectly represses SER3 expression by positively regulating the chromatin dynamics associated with SRG1 intergenic transcription across the SER3 promoter has provided one of the first insights into understanding how this complex negatively regulates transcription. A negative regulatory role for the Paf1 complex has also been recently characterized for the yeast gene ARG1 (18). In this case, Paf1 complex members were found to associate with both the ARG1 promoter and ORF during repressing conditions. Similar to SER3 repression, ARG1 repression is most strongly dependent on the Paf1 and Ctr9 subunits; however, Rtf1 and Rtf1-regulated histone modifications seem to have a greater effect at ARG1 than at SER3 (18). Although the mechanistic role of Paf1 and Ctr9 in ARG1 repression has yet to be defined, it is interesting that transcription antisense to ARG1 has been detected at this locus (19, 98). Additional experiments are required to determine if SER3 and ARG1 repression occur by a common mechanism involving Paf1-mediated chromatin dynamics during transcription of ncDNA.
In addition to SRG1, we assayed the effects of deleting PAF1 on histone H3, Spt6, and Spt16 occupancy over the transcribed regions of two other highly transcribed genes, PMA1 and ADH1 (Fig. 7). Similar to our results for SRG1, we detected reduced Spt6 and Spt16 occupancy levels in cells lacking Paf1. However, in contrast to what we observed at SRG1, RNA Pol II levels for these genes were also slightly reduced in paf1Δ cells. Gene-specific differences in RNA Pol II occupancy have been previously reported for paf1Δ mutants (56). When normalized to RNA Pol II levels, there is no longer a reduction in Spt6 occupancy, while Spt16 occupancy is reduced to similar levels in both paf1Δ and rtf1Δ mutants (Fig. 7G and H). Furthermore, histone H3 occupancy was unaffected at ADH1 and only moderately reduced at PMA1 in paf1Δ cells. These studies suggest that there are likely to be additional factors that influence the role of the Paf1 complex in regulating transcription-coupled nucleosome assembly at specific genes. One factor that may contribute to these gene-specific differences is the properties of the transcribed DNA. SRG1 is transcribed across the promoter region of SER3. In general, promoter regions tend to be comprised of sequences that are refractory to nucleosome formation, whereas the sequences of ORFs generally do not contain these properties (73). This would explain the inherent instability of nucleosomes over the SER3 promoter in the absence of SRG1 transcription. This characteristic of the SRG1 transcription unit makes it unique compared to the transcription of most protein-coding genes and may have allowed us to uncover this new role for the Paf1 complex that may not be readily detectable for other transcribed regions of the genome.
In conclusion, our analysis of SER3 repression by intergenic SRG1 transcription supports a previously uncharacterized role for the Paf1 complex in promoting transcription-dependent nucleosome occupancy. This activity is primarily dependent on the Paf1 and Ctr9 subunits, possibly mediated by their requirement for the recruitment and/or activity of the Spt6 and Spt16 histone chaperones. Additional studies will be necessary to elucidate the precise mechanism by which Paf1 and Ctr9 regulate chromatin dynamics during SRG1 transcription, to determine how broadly these two factors function in a similar manner at other transcribed regions of the genome, and to determine the characteristic of a transcription unit that makes it susceptible to this new activity of the Paf1 complex.
ACKNOWLEDGMENTS
We are grateful to Karen Arndt, Elia Crisucci, Tim Formosa, and Fred Winston for strains and antibodies. We also thank Karen Arndt, Andrea Duina, Brett Tomson, and members of the Martens lab for helpful discussions and critical reading of the manuscript.
This work is supported by funds from the NIH (GM080470) and Pittsburgh Life Sciences Greenhouse to J.A.M.
Footnotes
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Adelman K., et al. 2006. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol. Cell. Biol. 26:250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akanuma T., Koshida S., Kawamura A., Kishimoto Y., Takada S. 2007. Paf1 complex homologues are required for Notch-regulated transcription during somite segmentation. EMBO Rep. 8:858–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ausubel F., et al. 1991. Current protocols in molecular biology. John Wiley and Sons, New York, NY [Google Scholar]
- 4. Basrai M. A., Kingsbury J., Koshland D., Spencer F., Hieter P. 1996. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2838–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belotserkovskaya R., et al. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301:1090–1093 [DOI] [PubMed] [Google Scholar]
- 6. Bortvin A., Winston F. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272:1473–1476 [DOI] [PubMed] [Google Scholar]
- 7. Brachmann C. B., et al. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132 [DOI] [PubMed] [Google Scholar]
- 8. Brickner D. G., et al. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campos E. I., Reinberg D. 2009. Histones: annotating chromatin. Annu. Rev. Genet. 43:559–599 [DOI] [PubMed] [Google Scholar]
- 10. Carrozza M. J., et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592 [DOI] [PubMed] [Google Scholar]
- 11. Chaudhary K., Deb S., Moniaux N., Ponnusamy M. P., Batra S. K. 2007. Human RNA polymerase II-associated factor complex: dysregulation in cancer. Oncogene 26:7499–7507 [DOI] [PubMed] [Google Scholar]
- 12. Chen Y., et al. 2009. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 23:2765–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheung V., et al. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chu Y., Simic R., Warner M. H., Arndt K. M., Prelich G. 2007. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 26:4646–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chu Y., Sutton A., Sternglanz R., Prelich G. 2006. The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol. Cell. Biol. 26:3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clapier C. R., Cairns B. R. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273–304 [DOI] [PubMed] [Google Scholar]
- 17. Cox J. S., Chapman R. E., Walter P. 1997. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8:1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Crisucci E. M., Arndt K. M. 2011. The Paf1 complex represses ARG1 transcription in Saccharomyces cerevisiae by promoting histone modifications. Eukaryot. Cell 10:712–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. David L., et al. 2006. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. U. S. A. 103:5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davis C. A., Ares M., Jr 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 103:3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Diebold M. L., et al. 2010. Noncanonical tandem SH2 enables interaction of elongation factor Spt6 with RNA polymerase II. J. Biol. Chem. 285:38389–38398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ding L., et al. 2009. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 4:403–415 [DOI] [PubMed] [Google Scholar]
- 23. Eitoku M., Sato L., Senda T., Horikoshi M. 2008. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell. Mol. Life Sci. 65:414–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Floer M., et al. 2010. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141:407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Formosa T., et al. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20:3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hainer S. J., Martens J. A. 2011. Identification of histone mutants that are defective for transcription-coupled nucleosome occupancy. Mol. Cell. Biol. 31:3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hainer S. J., Pruneski J. A., Mitchell R. D., Monteverde R. M., Martens J. A. 2011. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 25:29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hartzog G. A., Wada T., Handa H., Winston F. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henry K. W., et al. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jaehning J. A. 2010. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta 1799:379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jamai A., Puglisi A., Strubin M. 2009. Histone chaperone spt16 promotes redeposition of the original H3-H4 histones evicted by elongating RNA polymerase. Mol. Cell 35:377–383 [DOI] [PubMed] [Google Scholar]
- 32. Joshi A. A., Struhl K. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20:971–978 [DOI] [PubMed] [Google Scholar]
- 33. Kao C. F., et al. 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 18:184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaplan C. D., Holland M. J., Winston F. 2005. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 280:913–922 [DOI] [PubMed] [Google Scholar]
- 35. Kaplan C. D., Laprade L., Winston F. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096–1099 [DOI] [PubMed] [Google Scholar]
- 36. Keogh M. C., et al. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593–605 [DOI] [PubMed] [Google Scholar]
- 37. Kim J., Guermah M., Roeder R. G. 2010. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140:491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim M., Ahn S. H., Krogan N. J., Greenblatt J. F., Buratowski S. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23:354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim T., Buratowski S. 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137:259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Komarnitsky P., Cho E. J., Buratowski S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krogan N. J., et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721–729 [DOI] [PubMed] [Google Scholar]
- 42. Krogan N. J., et al. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Laribee R. N., et al. 2005. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr. Biol. 15:1487–1493 [DOI] [PubMed] [Google Scholar]
- 44. Li B., Carey M., Workman J. L. 2007. The role of chromatin during transcription. Cell 128:707–719 [DOI] [PubMed] [Google Scholar]
- 45. Lin L., Zhang J. H., Panicker L. M., Simonds W. F. 2008. The parafibromin tumor suppressor protein inhibits cell proliferation by repression of the c-myc proto-oncogene. Proc. Natl. Acad. Sci. U. S. A. 105:17420–17425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu Y., et al. 2009. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29:4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lohr D. 1984. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic Acids Res. 12:8457–8474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martens J. A., Laprade L., Winston F. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429:571–574 [DOI] [PubMed] [Google Scholar]
- 49. Martens J. A., Winston F. 2002. Evidence that Swi/Snf directly represses transcription in S. cerevisiae. Genes Dev. 16:2231–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martens J. A., Wu P. Y., Winston F. 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19:2695–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marton H. A., Desiderio S. 2008. The Paf1 complex promotes displacement of histones upon rapid induction of transcription by RNA polymerase II. BMC Mol. Biol. 9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mason P. B., Struhl K. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23:8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mayer A., et al. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17:1272–1278 [DOI] [PubMed] [Google Scholar]
- 54. Moniaux N., et al. 2006. The human homologue of the RNA polymerase II-associated factor 1 (hPaf1), localized on the 19q13 amplicon, is associated with tumorigenesis. Oncogene 25:3247–3257 [DOI] [PubMed] [Google Scholar]
- 55. Mueller C. L., Jaehning J. A. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22:1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mueller C. L., Porter S. E., Hoffman M. G., Jaehning J. A. 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14:447–456 [DOI] [PubMed] [Google Scholar]
- 57. Mulder K. W., Brenkman A. B., Inagaki A., van den Broek N. J., Timmers H. T. 2007. Regulation of histone H3K4 tri-methylation and PAF complex recruitment by the Ccr4-Not complex. Nucleic Acids Res. 35:2428–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Murray S., Udupa R., Yao S., Hartzog G., Prelich G. 2001. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol. Cell. Biol. 21:4089–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ng H. H., Dole S., Struhl K. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278:33625–33628 [DOI] [PubMed] [Google Scholar]
- 60. Ng H. H., Xu R. M., Zhang Y., Struhl K. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277:34655–34657 [DOI] [PubMed] [Google Scholar]
- 61. Nordick K., Hoffman M. G., Betz J. L., Jaehning J. A. 2008. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot. Cell 7:1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nourani A., Robert F., Winston F. 2006. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:1496–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pavri R., et al. 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125:703–717 [DOI] [PubMed] [Google Scholar]
- 64. Penheiter K. L., Washburn T. M., Porter S. E., Hoffman M. G., Jaehning J. A. 2005. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell 20:213–223 [DOI] [PubMed] [Google Scholar]
- 65. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pinskaya M., Gourvennec S., Morillon A. 2009. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 28:1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pokholok D. K., Hannett N. M., Young R. A. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799–809 [DOI] [PubMed] [Google Scholar]
- 68. Qiu H., Hu C., Hinnebusch A. G. 2009. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 33:752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Qiu H., Hu C., Wong C. M., Hinnebusch A. G. 2006. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol. Cell. Biol. 26:3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rondon A. G., Gallardo M., Garcia-Rubio M., Aguilera A. 2004. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 5:47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rondon A. G., Garcia-Rubio M., Gonzalez-Barrera S., Aguilera A. 2003. Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J. 22:612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rose M., Winston F., Hieter P. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 73. Segal E., Widom J. 2009. What controls nucleosome positions? Trends Genet. 25:335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shahbazian M. D., Zhang K., Grunstein M. 2005. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell 19:271–277 [DOI] [PubMed] [Google Scholar]
- 75. Sheldon K. E., Mauger D. M., Arndt K. M. 2005. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell 20:225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shi X., et al. 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 17:1160–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shirra M. K., Rogers S. E., Alexander D. E., Arndt K. M. 2005. The Snf1 protein kinase and Sit4 protein phosphatase have opposing functions in regulating TATA-binding protein association with the Saccharomyces cerevisiae INO1 promoter. Genetics 169:1957–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Simic R., et al. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22:1846–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Squazzo S. L., et al. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Stolinski L. A., Eisenmann D. M., Arndt K. M. 1997. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4490–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sun M., Lariviere L., Dengl S., Mayer A., Cramer P. 2010. A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD). J. Biol. Chem. 285:41597–41603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sun Z. W., Allis C. D. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104–108 [DOI] [PubMed] [Google Scholar]
- 83. Swanson M. S., Winston F. 1992. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics 132:325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tenney K., et al. 2006. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 103:11970–11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Thebault P., et al. 2011. Transcription regulation by the noncoding RNA SRG1 requires Spt2-dependent chromatin deposition in the wake of RNA polymerase II. Mol. Cell. Biol. 31:1288–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Thompson D. M., Parker R. 2007. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol. Cell. Biol. 27:92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tomson B. N., Davis C. P., Warner M. H., Arndt K. M. 2011. Identification of a role for histone H2B ubiquitylation in noncoding RNA 3′-end formation through mutational analysis of Rtf1 in Saccharomyces cerevisiae. Genetics 188:273–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tous C., et al. 2011. A novel assay identifies transcript elongation roles for the Nup84 complex and RNA processing factors. EMBO J. 30:1953–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wada T., et al. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang P., et al. 2008. Parafibromin, a component of the human PAF complex, regulates growth factors and is required for embryonic development and survival in adult mice. Mol. Cell. Biol. 28:2930–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Warner M. H., Roinick K. L., Arndt K. M. 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 27:6103–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Winston F., Chaleff D. T., Valent B., Fink G. R. 1984. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107:179–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Winston F., Dollard C., Ricupero-Hovasse S. L. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53–55 [DOI] [PubMed] [Google Scholar]
- 94. Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739–34742 [DOI] [PubMed] [Google Scholar]
- 95. Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. 2005. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol. Cell 20:589–599 [DOI] [PubMed] [Google Scholar]
- 96. Wood A., Shilatifard A. 2006. Bur1/Bur2 and the Ctk complex in yeast: the split personality of mammalian P-TEFb. Cell Cycle 5:1066–1068 [DOI] [PubMed] [Google Scholar]
- 97. Xiao T., et al. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25:637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xu Z., et al. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457:1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yao S., Neiman A., Prelich G. 2000. BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol. Cell. Biol. 20:7080–7087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Youdell M. L., et al. 2008. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 28:4915–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zheng S., Wyrick J. J., Reese J. C. 2010. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol. Cell. Biol. 30:3635–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhou K., Kuo W. H., Fillingham J., Greenblatt J. F. 2009. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. U. S. A. 106:6956–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]