Abstract
Several members of the glycoside hydrolase 61 (GH61) family of proteins have recently been shown to dramatically increase the breakdown of lignocellulosic biomass by microbial hydrolytic cellulases. However, purified GH61 proteins have neither demonstrable direct hydrolase activity on various polysaccharide or lignacious components of biomass nor an apparent hydrolase active site. Cellobiose dehydrogenase (CDH) is a secreted flavocytochrome produced by many cellulose-degrading fungi with no well-understood biological function. Here we demonstrate that the binary combination of Thermoascus aurantiacus GH61A (TaGH61A) and Humicola insolens CDH (HiCDH) cleaves cellulose into soluble, oxidized oligosaccharides. TaGH61A-HiCDH activity on cellulose is shown to be nonredundant with the activities of canonical endocellulase and exocellulase enzymes in microcrystalline cellulose cleavage, and while the combination of TaGH61A and HiCDH cleaves highly crystalline bacterial cellulose, it does not cleave soluble cellodextrins. GH61 and CDH proteins are coexpressed and secreted by the thermophilic ascomycete Thielavia terrestris in response to environmental cellulose, and the combined activities of T. terrestris GH61 and T. terrestris CDH are shown to synergize with T. terrestris cellulose hydrolases in the breakdown of cellulose. The action of GH61 and CDH on cellulose may constitute an important, but overlooked, biological oxidoreductive system that functions in microbial lignocellulose degradation and has applications in industrial biomass utilization.
INTRODUCTION
Cellulose is Earth's most abundant biopolymer, and the biological exploitation of this carbohydrate as an energy source plays a critical role in the carbon cycle and the planet's ecology. Industrial production of fuels and chemicals from this plentiful and renewable resource holds the potential to displace petroleum-based processes, promoting energy self-sufficiency and reducing environmental costs (14, 22, 23, 41). However, the recalcitrance of cellulose to hydrolytic depolymerization is a barrier to both microbial and industrial utilization of lignocellulosic biomass (1, 11, 30, 37, 40).
Lignocellulosic biomass is composed largely of three major components: cellulose, a β-1,4-linked glucose polymer arrayed predominantly in crystalline fibrils; hemicellulose, a mixed carbohydrate polymer; and lignin, a complex polyalkylaromatic compound (11). The heterogeneity and recalcitrance of lignocellulose require the synergistic activities of a number of hydrolytic enzymes for efficient conversion to biologically fermentable monosaccharides. The canonical enzymatic cellulose hydrolysis scheme involves cellobiohydrolases (CBH) which processively degrade cellulose chains from the ends, producing cellobiose (EC 3.2.1.91 for CBHs that proceed from the nonreducing ends of cellulose chains; there is no assigned EC number for cellobiohydrolases that release cellobiose from the reducing ends), endo-β-1,4-glucanases (EG; EC 3.2.1.4) which internally clip cellulose, shortening chains, and generating reactive ends for CBH, and β-glucosidases (BG; EC 3.2.1.21) which hydrolyze soluble cellodextrins to glucose, mitigating CBH product inhibition arising from cellobiose (4, 10, 37, 38, 40). Hemicellulases, ligninases, and other secreted accessory proteins, such as swollenin, increase the accessibility of cellulose to these hydrolytic cellulase activities (10, 2, 15, 19, 27, 29, 39).
Another group of proteins broadly expressed in the fungal kingdom and implicated in biomass breakdown is the glycoside hydrolase 61 (GH61) family of proteins. The addition of specific GH61 proteins has been shown to greatly increase the performance of Hypocrea jecorina (Trichoderma reesei) cellulases in lignocellulose hydrolysis (3, 8). Despite extensive investigation, the mechanism of GH61's enhancement of lignocellulose degradation has remained elusive. These proteins were originally classified as glycoside hydrolases, based on the detection of low levels of endoglucanase activity (17, 18, 26). However, subsequent investigations have revealed that, when purified and within clearly defined assay conditions, the GH61 molecules which enhance lignocellulose breakdown by cellulases do not hydrolyze either cellulose substrates, such as microcrystalline cellulose or phosphoric acid-swollen cellulose (PASC), or hemicellulosic substrates, such as xylan (8). The crystal structures of two GH61 proteins show no evidence of a conventional glycoside hydrolase active site. Both GH61 crystal structures possess a bound metal near a group of planar residues, presenting a putative active site of unknown function (8, 16). A structurally homologous site is found on chitin binding protein-21 (CBP21) (34, 35), a bacterial protein which has recently been shown to cleave chitin into reducing-end oxidized chitin oligomers in a reaction dependent on the presence of O2 and reducing agent (36). Based on that result, it was proposed that CBP21 degrades chitin by a chitin oxygenase activity in a reaction mechanism which consumes both O2 and reducing agent as cosubstrates (36). Very recently, CelS2, another bacterial homolog of GH61, was shown to cleave cellulose into a mixed population of reducing-end oxidized and reduced cellodextrin in a reductant-dependent reaction (6). Given the structural similarity of both CBP21 and CelS2 to GH61 (6, 8, 16, 34), it was further proposed that GH61 attacks cellulose by a similar reductant-dependent oxygenase mechanism (6, 36).
Cellobiose dehydrogenase (CDH; EC 1.1.99.18), the only known example of a secreted flavocytochrome, is another fungal enzyme which has no clearly defined biological function despite extensive study over the last 30 years (5, 43). The suggested roles for CDH are diverse, including activating hydrolytic cellulases by relief of cellobiose product inhibition, being a component of the respiratory chain, preventing cellulose or lignin repolymerization after cleavage, acting as an antimicrobial agent, supporting manganese peroxidase, acting in the reduction of toxic quinones, or initiating Fenton-type hydroxyl radical-mediated cleavage of cellulose, hemicellulose, or lignin (5, 9, 43). Of these, the hydroxyl radical cleavage of the cellulose hypothesis is most widely proposed in current literature. What is known is that CDH both binds cellulose and catalyzes the oxidation of cellodextrins, lactose, and maltodextrins to their corresponding lactones using a diverse selection of electron acceptors (9).
Evidence of a linkage between the activities of CDH and GH61 has previously been shown in the patent literature (32, 33). With a range of pure cellulose substrates, the combination of both GH61 and CDH increased cellulose conversion by hydrolytic cellulases and cellulase mixtures. In this study, the nature of GH61-CDH stimulation of hydrolytic cellulase activity is examined, demonstrating that the combination of GH61 and CDH cleaves cellulose and that the oligosaccharides released in this cleavage differ from those produced by the activities of canonical hydrolytic cellulases. Further, we show that the in vitro combination of CDH and GH61 coexpressed by Thielavia terrestris enhances cellulose breakdown by canonical T. terrestris cellulose hydrolases, demonstrating evidence of a microbial oxidoreductive cellulose depolymerizing enzyme system active in parallel with the better-described hydrolytic cellulase enzyme system.
MATERIALS AND METHODS
Substrates and reagents.
Reagents and buffers were commercial products of reagent grade or better. PASC was prepared from Avicel (FMC BioPolymer; PH 101) (44). Bacterial cellulose was prepared as described previously (32). All reagents and substrates were prepared in high-purity Milli-Q (Millipore) water.
Preparation of enzymes.
A cellulase preparation, Celluclast Plus (Novozymes), was used as a control for complete cellulose hydrolysis. The mix contained Hypocrea jecorina cellulases and Aspergillus oryzae Cel3 BG (AoBG). Native H. jecorina Cel7A (HjCBH-I), recombinant H. jecorina Cel5A (HjEG-II), recombinant T. terrestris Cel6A (TtCBH-II), recombinant T. terrestris Cel7E EG (TtEG), recombinant T. terrestris GH61E (TtGH61E), recombinant T. terrestris Cel3A beta-glucosidase (TtBG), recombinant AoBG, recombinant Aspergillus fumigatus Cel3 BG (AfBG), recombinant Humicola insolens CDH (HiCDH), recombinant T. terrestris CDH (TtCDH), and recombinant Thermoascus aurantiacus GH61A (TaGH61A) were expressed and purified as previously reported (7, 8, 20, 32, 33). Protein quantification was performed using bicinchoninic acid (BCA) analysis (Pierce) or the Bio-Rad protein assay (Bio-Rad; for T. terrestris enzymes only).
Instrumentation.
A Powerwave X (Biotek Instruments) or SpectraMax (Molecular Devices) plate reader was used for spectrophotometric measurement using Costar 96-well microplates. Ultra-high-performance liquid chromatography (UPLC) electrospray ionization mass spectrometry (UPLC-ESI-MS) and tandem mass spectrometry (MS/MS) were performed on a Waters Micromass Technologies Q-ToF microhybrid orthogonal quadrupole mass spectrometer, coupled to an Acquity nano-flow HPLC system with a bridged ethyl hybrid (BEH) amide column.
Enzymatic cellulose cleavage assays.
Cellulose conversion assays were carried out at 25 g/liter microcrystalline cellulose (Avicel), 5 g/liter PASC, or 2.3 g/liter bacterial cellulose in Milli-Q (Millipore) water buffered with 50 mM sodium acetate [pH 5], 1 mM MnSO4, at 50°C for 1 to 5 days (unless otherwise noted in figure legends), using protein loadings (depending on substrate, stated in figure legends) of 10 to 50 mg GH61 protein per gram cellulose, 1 to 10 mg CDH per gram cellulose, and 1 to 50 mg BG per gram cellulose. Control incubations excluding either GH61 or CDH alone were performed in the same manner. Triplicates were run in each experiment. Samples were filtered using a 0.45-μm Multiscreen 96-well filter plate (Millipore) prior to further analysis. Glucose and cellobiose concentrations were quantified in the filtrates by HPLC with an Agilent 1100 system equipped with a refractive index detector, using an Aminex HPX-87H analytical column (Bio-Rad), eluting with 5 mM H2SO4 at 0.6 ml/min, at 65°C. The extent of hydrolysis, based on percent cellulose conversion, was calculated from the ratio of measured glucose plus cellobiose in a sample relative to a limit digest by a high loading of cellulase (100 mg Celluclast Plus per g cellulose).
GH61-CDH reactions prepared for UPLC-ESI-MS were similarly performed with 5 g/liter PASC in Milli-Q (Millipore) water buffered with 20 mM ammonium acetate, 0.1 mM MnSO4 at pH 5.0 treated with 50 mg TaGH61A per gram cellulose, and 5 mg HiCDH per gram cellulose and with or without 5 mg AfBG per gram cellulose, at 50°C for 4 days. All reaction mixtures, prior to refractive index detection (RID)-HPLC or UPLC-ESI-MS analysis, were filtered as described and then passed through a 10-kDa-molecular-mass-cutoff membrane using ultrafiltration (Vivacell [Sartorius-Stedim] or VivaSpin [GE Healthcare]).
RESULTS
Influence of TaGH61A and HiCDH on hydrolytic cellulase conversion of cellulosic substrates.
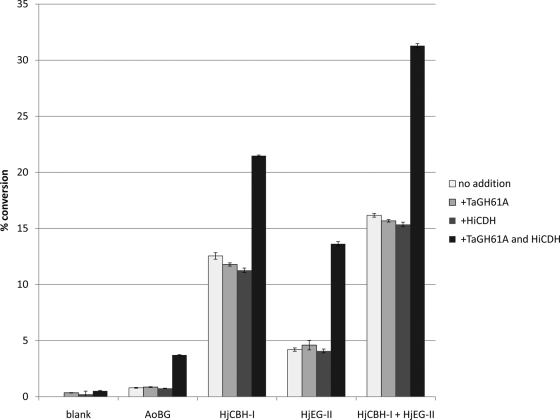
In order to test the effects of TaGH61A and HiCDH on cellulose hydrolase activity, a microcrystalline cellulose conversion assay was conducted using the monocomponent cellulases HjEG-II, HiCBH-I, and AoBG or the combination of HjCBH-I and HjEG-II. The detection of glucose and cellobiose released was used as a metric for conversion. Shown in Fig. 1, the combination of both TaGH61A and HiCDH greatly increased cellulose conversion by all hydrolytic cellulases tested compared to addition of TaGH61A or HiCDH individually. An increase in cellulose conversion to glucose was also observed for AoBG, which is otherwise inactive on insoluble cellulose. This increase in glucose release in the presence of BG may indicate the release of soluble cellodextrin from cellulose, which serves as a BG substrate.
Fig. 1.
Microcrystalline cellulose conversion assay. HiCDH was loaded at 1 mg protein per gram cellulose, TaGH61A was loaded at 5 mg protein per gram cellulose, and HjCBH-I, HjEG-II, and AoBG were loaded at 10 mg protein per gram cellulose. Assay conditions were 50 mM NaOAc at pH 5, 1 mM MnSO4, 50°C, and a 72-h time course. Monosaccharide and disaccharide were quantified by RID-HPLC using an Aminex HPX-87H column, with percent cellulose conversion calculated compared to a limit digest by a high loading of cellulase. Error bars are for triplicate cellulose cleavage reactions, each measured once.
In order to confirm that the combined enzymatic activities of TaGH61A and HiCDH, rather than an activation of one protein by a contaminant in either preparation, were responsible for the increase in microcrystalline cellulose conversion by cellulase, experiments were conducted in which either heat-inactivated TaGH61A and fresh HiCDH or fresh TaGH61A and heat-inactivated HiCDH were added to individual cellulases in microcellulose conversion assays similar to those shown in Fig. 1. No synergistic effect was seen when either HiCDH or TaGH61A was previously heat inactivated prior to the assay (data not shown). Similarly, pretreatment of microcrystalline cellulose with either HiCDH or TaGH61A individually, followed by heat inactivation of the material and a cellulose cleavage assay with addition of cellulase and either TaGH61A or HiCDH, demonstrated that both active TaGH61A and HiCDH must be present simultaneously in order to increase cellulase conversion of cellulose (data not shown).
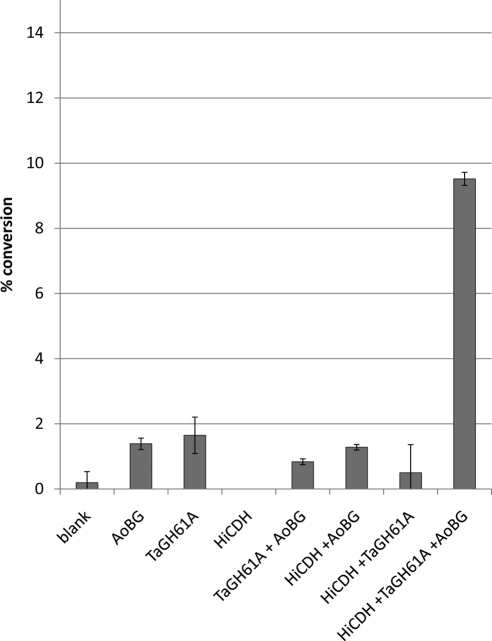
A PASC cleavage assay was conducted in order to test the activity of TaGH61A and HiCDH on a highly amorphous cellulose substrate. AfBG was added to hydrolyze any released cellodextrins, allowing glucose detection as a conversion metric. Shown in Fig. 2, treatment of PASC with AfBG, the binary combination of AfBG and TaGH61A, or the binary combination of AfBG and HiCDH resulted in very low PASC conversion, while the ternary combination of TaGH61A, HiCDH, and AfBG resulted in significant cellulose conversion (30% at 2 days). The addition of radical quenching agent mannitol or dimethyl sulfoxide (DMSO) (25) was only slightly inhibitory to cellulose conversion by the combination of TaGH61A, HiCDH, and AfBG. The activity of this enzyme mixture was strongly inhibited by the addition of the chelating agents EDTA and EGTA but not inhibited by the chelator nitrilotriacetic acid (NTA). The addition of the O2 mimic sodium azide also resulted in the reduction of cellulose conversion by the combination of TaGH61A, HiCDH, and AfBG.
Fig. 2.
PASC conversion assay (5 g/liter PASC digestion). HiCDH and AfBG were loaded at 5 mg protein per gram cellulose, and TaGH61A was loaded at 20 mg protein per gram cellulose. Assay conditions were 50 mM NaOAc at pH 5, 1 mM MnSO4, 50°C, and a 2- to 120-h time course. Monosaccharide release was determined by RID-HPLC using an Aminex HPX-87H column, with percent cellulose conversion calculated compared to a limit digest by a high loading of cellulase. Error bars are for triplicate cellulose cleavage reactions, each measured once.
Shown in Fig. 3, the activity of a combination of TaGH61A, HiCDH, and AoBG was tested on highly crystalline bacterial cellulose. The ternary combination of TaGH61A, HiCDH, and AoBG resulted in significant conversion of bacterial cellulose, while treatment with individual enzyme components and binary combinations resulted in little glucose formation from this substrate.
Fig. 3.
Bacterial cellulose conversion assay (2.3 g/liter bacterial cellulose). HiCDH and AfBG were loaded at 5 mg protein per gram cellulose, and TaGH61A was loaded at 20 mg protein per gram cellulose. Assay conditions were 50 mM NaOAc at pH 5, 1 mM MnSO4, 50°C, and 3-day time course. Monosaccharide release was determined by RID-HPLC using an Aminex HPX-87H column, with percent cellulose conversion calculated compared to a limit digest by a high loading of cellulase. Error bars are for triplicate cellulose cleavage reactions, each measured once.
In similar assays using hemicellulosic substrates (e.g., birch wood xylan), the combination of TaGH61A and HiCDH was tested for their effect on hemicellulose conversion by various hemicellulose hydrolases. For all hemicellulose substrates tested, hemicellulose conversion was not increased by individual addition of TaGH61A, HiCDH, or the combination of TaGH61A and HiCDH (data not shown), indicating cellulose specificity for TaGH61A and HiCDH.
Analysis of cellulose cleavage products of TaGH61A and HiCDH by LC-MS and MS/MS.
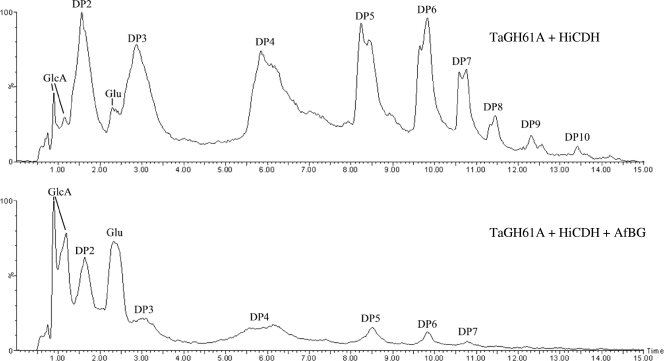
In order to further explore the nature of the TaGH61A and HiCDH action upon cellulose, a mass spectrometric analysis of TaGH61A-and-HiCDH-treated PASC samples was undertaken. Analytes were separated by UPLC with a BEH amide column, and electrospray ionization and detection by a quadropole/time-of-flight mass spectrometer followed. Shown in Fig. 4 is the LC elution profile, using total ion count (TIC) as a bulk readout, for PASC treatment by either the binary combination of TaGH61A and HiCDH or the ternary combination of TaGH61A, HiCDH, and AfBG. This data set shows that the combination of TaGH61A and HiCDH cleaves PASC, resulting in the release of soluble oligosaccharide. Elution times for these oligosaccharides corresponded with those of cellodextrin aldonic acid standards (not shown), which would be expected in the presence of CDH which oxidizes the reducing end of cellodextrins. AfBG-treated samples show the conversion of most oligosaccharides to glucose and some quantity of gluconic acid (GlcA), though a minority of oligosaccharides are resistant to BG cleavage.
Fig. 4.
LC-MS TIC profiles of UPLC separation of samples with a BEH amide column. (Top) PASC, 5 g/liter, treated with 50 mg TaGH61A per gram cellulose and 5 mg HiCDH per gram cellulose in 20 mM ammonium acetate at pH 5, 0.1 mM MnSO4, for 4 days at 50°C. (Bottom) PASC, 5 g/liter, treated with 50 mg TaGH61A per gram cellulose, 5 mg HiCDH per gram cellulose, and 5 mg AfBG per gram cellulose in 20 mM ammonium acetate at pH 5, 0.1 mM MnSO4, for 4 days at 50°C. Note that all cellodextrins in this sample set were observed as cellodextrin acids; e.g., DP4 marks cellotetraonic acid.
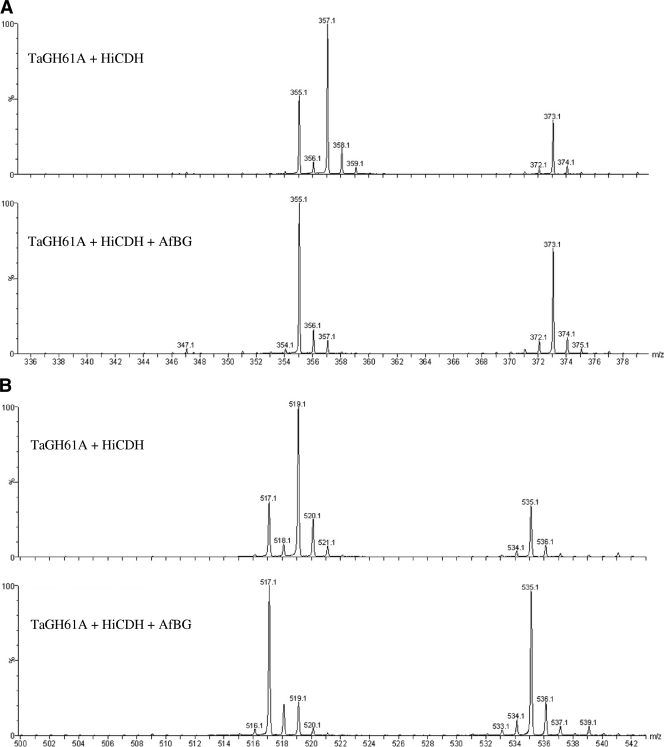
Figure 5A shows the mass spectra for the degree of glucose polymerization 2 (DP2) time window of the elution profile, and Fig. 5B shows the mass spectra for the DP3 region of the elution profile. In the cases of both DP2 and DP3 oligomers, three masses were detected: a +16-Da mass predicted for a reducing-end C-1-oxidized cellodextrin aldonic acid which was BG labile and also +14-Da and +32-Da species which were both BG resistant. This pattern of +14 Da, +16 Da, and +32 Da repeats itself at all oligosaccharide lengths (not shown). BG cleaves the terminal glucose from the nonreducing end of cellodextrin (7), as such a BG-resistant oligosaccharide population may result from modification of a cellodextrin at any point other than the reducing end.
Fig. 5.
(A) MS-1 spectra of cellobionic acid (DP2) region of LC elution profile. (Top) PASC, 5 g/liter, treated with TaGH61A and HiCDH. (Bottom) PASC, 5 g/liter, treated with TaGH61A, HiCDH, and AfBG. (B) MS-1 spectra of cellotrionic acid (DP3) region of LC elution profile. (Top) PASC, 5 g/liter, treated with TaGH61A and HiCDH. (Bottom) PASC, 5 g/liter, treated with TaGH61A, HiCDH, and AfBG.
MS/MS analysis, shown in Fig. S1 in the supplemental material, confirmed that the +16-Da species were reducing-end C-1 oxidized (gluconic acid equivalent at the reducing end) for both DP2 and DP3, based on a comparison to gluconic acid and cellobionic acid standards. We were unable to unambiguously identify the +14-Da and +32-Da species based on this MS/MS datum set, though reducing-end C-1 oxidation coupled with a non-reducing-end modification is consistent with the observed fragmentation pattern and BG resistance of these species.
Cellulose cleaving activity by GH61 and CDH from T. terrestris and synergy with T. terrestris cellulose hydrolases.
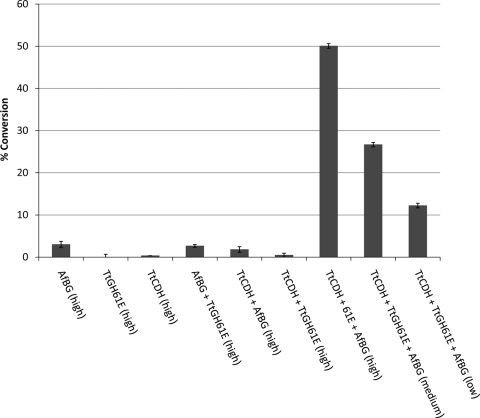
Recombinant TtCDH was tested for activity with recombinant TtGH61E in a PASC cleavage assay, with AfBG present in order to allow monosaccharide quantification. Shown in Fig. 6, the ternary combination of TtGH61E, TtCDH, and AfBG resulted in substantial, dose-dependent conversion of PASC, while the binary combinations of either TtGH61E or TtCDH with AfBG did not result in significant monosaccharide release.
Fig. 6.
PASC conversion to monosaccharide by TtCDH, TtGH61E, and AfBG. Reactions were carried out with 5 g/liter PASC in 100 mM sodium acetate at pH 5.0 with 1 mM CaCl2 for 3 days at 50°C, using single enzymes as well as binary and ternary combinations. Enzyme loadings at highest concentrations (high) were (in mg protein per gram cellulose) 5 mg TtCDH per gram cellulose, 50 mg TtGH61E per gram cellulose, and 5 mg AfBG per gram cellulose. Medium protein loadings (medium) were 2.5 mg TtCDH per gram cellulose, 25 mg TtGH61E per gram cellulose, and 5 mg AfBG per gram cellulose. Low protein loadings (low) were 1 mg TtCDH per gram cellulose, 10 mg TtGH61E per gram cellulose, and 5 mg AfBG per gram cellulose. Error bars are for triplicate cellulose cleavage reactions, each measured once.
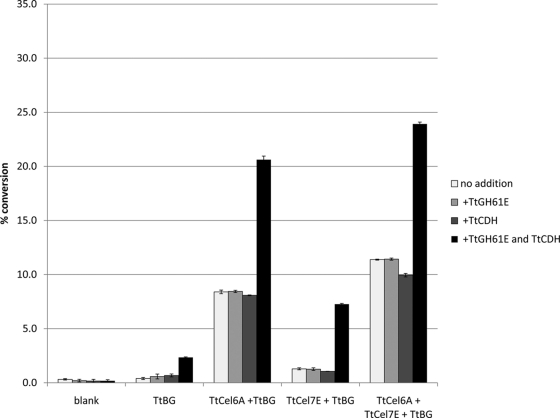
In order to investigate the effect that the interaction of TtGH61E and TtCDH may have on the canonical cellulose hydrolase enzyme system, several purified recombinant T. terrestris cellulose hydrolases, including TtCBH-II, TtEG, TtBG, and combinations thereof, were assayed for microcrystalline cellulose-degrading activity in the presence of TtGH61E, TtCDH, and the combination of TtGH61E and TtCDH. Shown in Fig. 7, the addition of both TtCDH and TtGH61E to TtBG resulted in an increase in the conversion of microcrystalline cellulose, relative to TtBG alone or TtBG with either TtGH61E or TtCDH alone, of ∼2%. The addition of TtGH61E and TtCDH to TtCBH-II, TtEG, or the combination of TtCBH-II and TtEG, all in the presence of TtBG, resulted in increases in microcrystalline cellulose conversion of ∼12%, ∼6%, and ∼14%. Compared to the addition of the binary combination of TtGH61E and TtCDH, the addition of either TtGH61E or TtCDH individually did not result in an increase of microcrystalline cellulose conversion by T. terrestris cellulose hydrolases under these assay conditions. Similar results, that the addition of both TtGH61E and TtCDH enhance cellulose conversion by T. terrestris cellulose hydrolases, were seen in the absence of TtBG (not shown).
Fig. 7.
Microcrystalline cellulose conversion assay by combinations of TtGH61E, TtCDH, TtBG, TtCel6A, and TtCel7E. Reactions were carried out with 25 g/liter microcrystalline cellulose (Avicel) in 100 mM sodium acetate at pH 5.0 with 1 mM CaCl2 for 3 days at 50°C, using single enzymes as well as enzyme combinations. Enzyme loadings were 5 mg TtGH61E per gram cellulose, 0.5 mg TtCDH per gram cellulose, 1 mg TtBG per gram cellulose, 5 mg TtCel6A per gram cellulose, and 0.75 mg TtCel7E per gram cellulose. Error bars are for triplicate cellulose cleavage reactions, each measured once.
DISCUSSION
We have shown that, in the absence of hydrolytic cellulases, the binary combination of TaGH61A and HiCDH catalyzes cellulose cleavage into soluble oligosaccharides and that this activity is highly synergistic with cellulose breakdown by canonical cellulose hydrolases. Soluble cellulose cleavage products of TaGH61A and HiCDH are further shown to have a length that extends from DP2 to DP10 and are mixed species, including reducing-end oxidized species as well as non-reducing-end modified species.
The ascomycete T. terrestris secretes a number of canonical cellulose hydrolases when grown in the presence of cellulose (8, 20); formerly, these cellulases have been presumed to account for the majority of cellulose-degrading activity by this fungus. However, cellulose-induced secretion of GH61 and CDH by T. terrestris results in an additional cellulose-degrading enzyme activity. The combined increases in conversion, which are significantly greater than the simple addition of conversions from TtGH61E and TtCDH with canonical T. terrestris hydrolases, indicate synergy between the canonical cellulose hydrolase components and the cellulose cleavage system of GH61 and CDH. A very recent report also reveals coproduction of CDH and GH61 by the distantly related basidiomycete Phanerochaete chrysosporium in response to cellulose and xylan, indicating that a GH61-CDH-based oxidative carbohydrate depolymerization system may not be uncommon (12).
The synergy of the GH61-CDH activity with canonical cellulose hydrolase activities argues for a lack of redundancy in the substrates and activities of these systems and suggests that the combination of GH61 and CDH may be opening up new sites for hydrolase action. One possibility is that they act by attacking highly crystalline surfaces, upon which canonical cellulose hydrolases show low activity (28, 31). Canonical hydrolytic endocellulases (endoglucanases) show high activity on amorphous, cellodextrin-like strands of cellulose, while canonical hydrolytic exocellulases (cellobiohydrolases) processively degrade crystalline cellulose but require a cellodextrin chain end to initiate attack. GH61 and CDH may be hypothesized to have an endocrystalline cellulose cleaving activity, unknown with canonical cellulose hydrolases, accounting for the observed degree of synergy with canonical endo- and exocellulose hydrolases. This hypothesis is consistent with the persistence of long oligosaccharide products in the presence of both TaGH61A and HiCDH and is supported by the inability of TaGH61A and HiCDH to cleave DP2 to DP5 soluble cellodextrin standards (data not shown). The structure of GH61, which has one flat surface bearing a metal binding site and putative carbohydrate binding tyrosyl residues, but no active site cleft that could accommodate a cellodextrin strand, is consistent with this hypothesis (8, 16). If the crystalline surface is the target of GH61-CDH activity, two potential modes of cellulose attack would be indicated: most likely is a random nicking of the surface that opens new sites for hydrolase attack, but also possible is a semiprocessive attack either parallel with or perpendicular to the grain of the cellulose fibril.
The specific biochemical mechanism by which GH61A and CDH cleave cellulose remains unknown. GH61 homology to the chitin oxygenase CBP-21 (36) and cellulose cleaving CelS2 (6) suggests that GH61 is the species responsible for cellulose cleavage and that CDH is somehow supporting GH61 activity. Indeed, in the absence of CDH and upon addition of ascorbate, TaGH61A has been show to have cellulose cleaving activity (13), though the specific details of the mechanism by which GH61 cleaves cellulose are beyond the scope of this study and are being actively pursued by ongoing studies. Mixed-product species argue for radical involvement, possibly from cleavage by diffusive Fenton-type radical species (e.g., hydroxyl radical). Arguing against diffusive, unshielded, radical species is the observation that hydroxyl radical quenching agents only slightly inhibit the reaction; further, no cross-ring cleavage products are detected, and no cleavage of noncellulose carbohydrates is observed (data not shown). The analysis of TaGH61A and HiCDH cellulose cleavage products is complicated by CDH cellodextrin oxidase activity on possible cleavage products, making it unclear if oligosaccharide products are reducing-end C-1 oxidized prior to, during, or subsequent of cleavage. However, if deprived of C-1-reduced oligosaccharide substrate as an electron donor, CDH activity in this system would be dissimilar to any function shown in the previous literature (5, 9, 43), presenting the possibility that some reduced cellodextrin may be produced in this cleavage reaction, as seen in the products of CelS2 (6), which later serves as a CDH donor. In addition to CBP21, CelS2, and GH61, oxidoreductive carbohydrate cleavage has also been shown in the NAD+-dependent mechanisms of the GH4 and GH109 families (21, 24, 30, 42). GH4 activity is divalent cation dependent, similar to results shown for GH61, CelS2, and CBP21 (6, 8, 36), but it is unclear if further similarities exist within these enzyme systems. Further study will be required to answer these questions and allow full mechanistic elucidation as well as industrial exploitation of this microbial oxidative cellulose depolymerization enzyme system.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul Harris, Kim Borch, Hui Xu, Katja Johansen, Keith McCall, Janne E. Tønder, Leonardo De Maria, Christel Jørgensen, Kim Brown, Jason Quinlan, Kristian Krogh, K. C. McFarland, Brett McBrayer, and Hanshu Ding for technical assistance and discussion and Robert L. Starnes and Claus C. Fuglsang from Novozymes for critical reading of the manuscript.
This material is based upon work supported by the United States Department of Energy under award number DE-FC36-08GO18080. This report was prepared as an account of work sponsored by an agency of the United States government. Neither the United States government nor any agency thereof, nor any of their employees, makes any warranty, express or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States government or any agency thereof. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the United States government or any agency thereof.
All authors are employed by Novozymes, Inc., which is a producer of cellulases for industrial usage. No other conflicts of interest are declared.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Aden A., Foust T. 2009. Technoeconomic analysis of the dilute sulfuric acid and enzymatic hydrolysis process for the conversion of corn stover to ethanol. Cellulose 16:535–545 [Google Scholar]
- 2. Berlin A., et al. 2005. Evaluation of novel fungal cellulase preparations for ability to hydrolyze softwood substrates—evidence for the role of accessory enzymes. Enzyme Microb. Technol. 37:175–184 [Google Scholar]
- 3. Brown K., et al. June 2010. Polypeptides having cellulolytic enhancing activity and nucleic acids encoding same. U.S. patent 7,741,466 [Google Scholar]
- 4. Brumer H. 2010. Carbohydrases. In Horváth I. T. (ed.), The encyclopedia of catalysis, 2nd ed. Wiley, New York, NY: doi:10.1002/0471227617.eoc037 [Google Scholar]
- 5. Cameron M. D., Aust S. D. 2001. Cellobiose dehydrogenase—an extracellular fungal flavocytochrome. Enzyme Microb. Technol. 28:129–138 [DOI] [PubMed] [Google Scholar]
- 6. Forsberg Z., et al. 2011. Cleavage of cellulose by a CBM33 protein. Prot. Sci. doi:10.1002/pro.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harris P., Golightly E. March 2010. Polynucleotides encoding polypeptides having beta-glucosidase activity. U.S. patent 7,670,819 B2 [Google Scholar]
- 8. Harris P. V., et al. 2010. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49:3305–3316 [DOI] [PubMed] [Google Scholar]
- 9. Henriksson G., Johansson G., Petersson G. 2000. A critical review of cellobiose dehydrogenase. J. Biotechnol. 78:93–113 [DOI] [PubMed] [Google Scholar]
- 10. Henrissat B., Rodriguez H., Viet C., Schülein M. 1985. Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Nat. Biotechnol. 3:722–726 [Google Scholar]
- 11. Himmel M. E., et al. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807 [DOI] [PubMed] [Google Scholar]
- 12. Hori C., Igarashi K., Katayama A., Samejima M. 2011. Effects of xylan and starch on secretome of the basidiomycete Phanerochaete chrysosporium grown on cellulose. FEMS Microbiol. Lett. doi:10.1111/j.1574-6968.2011.02307.x. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 13. Johanson K. 2011. Function GH61 enzymes in biomass deconstruction, 5-04, p. 58. Abstr. 33rd Symp. Biotechnol. Fuels Chemicals. [Google Scholar]
- 14. Jørgensen H., Kristensen J. B., Felby C. 2007. Enzymatic conversion of lignocellulose into fermentable sugars: challenges and opportunities. Biofuel Bioprod. Bioref. 1:119–134 [Google Scholar]
- 15. Juhasz T., Szengyel Z., Reczey K., Siika-Aho M., Viikari L. 2005. Characterization of cellulases and hemicellulases produced by Trichoderma reesei on various carbon sources. Process Biochem. 40:3519–3525 [Google Scholar]
- 16. Karkehabadi S., et al. 2008. The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 A resolution. J. Mol. Biol. 383:144–154 [DOI] [PubMed] [Google Scholar]
- 17. Karlsson J., et al. 2001. Homologous expression and characterization of Cel61A (EG IV) of Trichoderma reesei. Eur. J. Biochem. 268:6498–6507 [DOI] [PubMed] [Google Scholar]
- 18. Koseki T., et al. 2008. Biochemical characterization of a glycoside hydrolase family 61 endoglucanase from Aspergillus kawachii. Appl. Microbiol. Biotechnol. 77:1279–1285 [DOI] [PubMed] [Google Scholar]
- 19. Kubicek C. P., Eveleigh D. E., Esterbauer H., Steiner W., Kubicek-Pranz E. M. 1990. Trichoderma reesei cellulases: biochemistry, genetics, physiology and application. Royal Society of Chemistry, Cambridge, United Kingdom [Google Scholar]
- 20. Langston J. 2011. Comparative proteomics of the Thielavia terrestris secretome, and associated proteins for the conversion of lignocellulosic biomass. Ph.D. dissertation, University of California, Davis, CA [Google Scholar]
- 21. Liu Q. P., et al. 2007. Bacterial glycosidases for the production of universal red blood cells. Nat. Biotechnol. 25:454–464 [DOI] [PubMed] [Google Scholar]
- 22. Lynd L. R., et al. 2008. How biotech can transform biofuels. Nat. Biotechnol. 26:169–172 [DOI] [PubMed] [Google Scholar]
- 23. Ragauskas A. J., et al. 2006. The path forward for biofuels and biomaterials. Science 311:484–489 [DOI] [PubMed] [Google Scholar]
- 24. Rajan S. S., et al. 2004. Novel catalytic mechanism of glycoside hydrolysis based on the structure of an NAD+/Mn2+-dependent phospho-a-glucosidase from Bacillus subtilis. Structure 12:1619–1629 [DOI] [PubMed] [Google Scholar]
- 25. Repine J. E., Eaton J. W., Anders M. W., Hoidal J. R., Fox R. B. 1979. Generation of hydroxyl radical by enzymes, chemicals, and human phagocytes in vitro. Detection with the anti-inflammatory agent, dimethyl sulfoxide. J. Clin. Invest. 64:1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saloheimo M., Nakari-Setala T., Tenkanen M., Penttila M. 1997. cDNA cloning of a Trichoderma reesei cellulase and demonstration of endoglucanase activity by expression in yeast. Eur. J. Biochem. 249:584–591 [DOI] [PubMed] [Google Scholar]
- 27. Saloheimo M., et al. 2002. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 269:4202–4211 [DOI] [PubMed] [Google Scholar]
- 28. Samejima M., Junji Sugiyama J., Kiyohiko Igarashi K., Eriksson K. L. 1997. Enzymatic hydrolysis of bacterial cellulose. Carbohydr. Res. 305:281–288 [Google Scholar]
- 29. Selig M. J., Knoshaug E. P., Adney W. S., Himmel M. E., Decker S. R. 2008. Synergistic enhancement of cellobiohydrolase performance on pretreated corn stover by addition of xylanase and esterase activities. Bioresour. Technol. 99:4997–5005 [DOI] [PubMed] [Google Scholar]
- 30. Service R. F. 2010. Is there a road ahead for cellulosic ethanol? Science 329:784–785 [DOI] [PubMed] [Google Scholar]
- 31. Srisodsuk M., Kleman-Leyer K., Keränen S., Kirk T. K., Teeri T. T. 1998. Modes of action on cotton and bacterial cellulose of a homologous endoglucanase-exoglucanase pair from Trichoderma reesei. Eur. J. Biochem. 251:885–892 [DOI] [PubMed] [Google Scholar]
- 32. Sweeney M. D., Vlasenko E., Abbate E. June 2010. Methods for increasing hydrolysis of cellulosic material in the presence of cellobiose dehydrogenase. U.S. patent 20,100,159,536 A1 [Google Scholar]
- 33. Sweeney M. D., Vlasenko E. 2009. Methods for determining cellulolytic enhancing activity of a polypeptide. U.S. patent 20,100,159,494 A1 [Google Scholar]
- 34. Vaaje-Kolstad G., Horn S. J., van Aalten D. M., Synstad B., Eijsink V. G. 2005. The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 280:28492–28497 [DOI] [PubMed] [Google Scholar]
- 35. Vaaje-Kolstad G., Houston D. R., Riemen A. H., Eijsink V. G., van Aalten D. M. 2005. Crystal structure and binding properties of the Serratia marcescens chitin-binding protein CBP21. J. Biol. Chem. 280:11313–11319 [DOI] [PubMed] [Google Scholar]
- 36. Vaaje-Kolstad G., et al. 2010. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330:219–222 [DOI] [PubMed] [Google Scholar]
- 37. Viikari L., Alapuranen M., Puranen T., Vehmaanperä J., Siika-Aho M. 2007. Thermostable enzymes in lignocellulose hydrolysis. Adv. Biochem. Eng. Biotechnol. 108:121–145 [DOI] [PubMed] [Google Scholar]
- 38. Vocadlo D. J., Davies G. J. 2008. Mechanistic insights into glycosidase chemistry. Curr. Opin. Chem. Biol. 12:539–555 [DOI] [PubMed] [Google Scholar]
- 39. Wang M., et al. 2010. High-level expression and efficient purification of bioactive swollenin in Aspergillus oryzae. Appl. Biochem. Biotechnol. 162:2027–2036 [DOI] [PubMed] [Google Scholar]
- 40. Wilson D. B. 2009. Cellulases and biofuels. Curr. Opin. Biotechnol. 20:295–299 [DOI] [PubMed] [Google Scholar]
- 41. Wyman C. E. 2003. Potential synergies and challenges in refining cellulosic biomass to fuels, chemicals, and power. Biotechnol. Prog. 19:254–262 [DOI] [PubMed] [Google Scholar]
- 42. Yip V. L., Withers S. G. 2006. Breakdown of oligosaccharides by the process of elimination. Curr. Opin. Chem. Biol. 10:147–155 [DOI] [PubMed] [Google Scholar]
- 43. Zamocky M., et al. 2006. Cellobiose dehydrogenase—a flavocytochrome from wood-degrading, phytopathogenic and saprotropic fungi. Curr. Protein Pept. Sci. 7:255–280 [DOI] [PubMed] [Google Scholar]
- 44. Zhang Y. H., Cui J., Lynd L. R., Kuang L. R. 2006. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: evidences from supramolecular structures and enzymatic hydrolysis. Biomacromolecules 7:644–648 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.