Abstract
Bacteria are abundant in the atmosphere, where they often represent a major portion of the organic aerosols. Potential pathogens of plants and livestock are commonly dispersed through the atmosphere, and airborne bacteria can have important effects on human health as pathogens or triggers of allergic asthma and seasonal allergies. Despite their importance, the diversity and biogeography of airborne microorganisms remain poorly understood. We used high-throughput pyrosequencing to analyze bacterial communities present in the aerosol fraction containing fine particulate matter of ≤2.5 μm from 96 near-surface atmospheric samples collected from cities throughout the midwestern United States and found that the communities are surprisingly diverse and strongly affected by the season. We also directly compared the airborne communities to those found in hundreds of samples representing potential source environments. We show that, in addition to the more predictable sources (soils and leaf surfaces), fecal material, most likely dog feces, often represents an unexpected source of bacteria in the atmosphere at more urbanized locations during the winter. Airborne bacteria are clearly an important, but understudied, component of air quality that needs to be better integrated into efforts to measure and model pollutants in the atmosphere.
INTRODUCTION
Scientists have long known that bacteria are ubiquitous in the atmosphere. Bacterial concentrations typically range from 104 to 106 cells m−3 (28), though concentrations may be far higher in proximity to point sources such as compost facilities, feedlots, and wastewater treatment facilities (1, 24, 41). Recent evidence suggests that, even in some relatively unpolluted locations, bacteria or portions of bacteria may represent a major component of the organic aerosols residing in the atmosphere (22, 44). Airborne bacteria can have important effects on human health and the productivity of managed and natural ecosystems. For example, bacteria can cause allergic asthma and seasonal allergies, diseases which are increasingly prevalent in developed nations (14, 35), with allergic asthma currently affecting millions of people in the United States (2). Likewise, important plant and livestock pathogens are dispersed through the atmosphere (20, 38) and there is some evidence that bacteria are capable of influencing atmospheric processes by initiating cloud condensation and ice nucleation events, potentially altering precipitation patterns (10, 34). Despite their abundance and likely importance, we have a limited understanding of the quantities and types of bacteria found in the atmosphere (45). With recent advances in high-throughput sequencing, we can now describe the dynamics of airborne bacterial populations (5, 7) and determine likely sources of bacteria in the atmosphere (6), building a more comprehensive understanding of those bacteria found in the atmosphere and the control of their populations.
With the present study, we describe bacterial abundances and community composition in the near-surface atmosphere at four locations in the Great Lakes region of the United States during the winter and summer seasons. PM2.5 samples (particulate matter with an aerodynamic diameter of less than 2.5 μm) were collected at all locations, as our goal was to capture respirable fine particles, including bacteria, that residents in these areas would be exposed to via inhalation when outside their homes. The bacterial populations in the 108 samples collected were analyzed using a 16S rRNA gene-based barcoded pyrosequencing procedure described previously (5, 6, 18). To complement these analyses of relative bacterial abundances in each sample, absolute bacterial abundances were determined via flow cytometry.
MATERIALS AND METHODS
Sample collection.
Air samples were collected at four locations in the midwestern United States, i.e., one small town with a population of 5,200 (Mayville, WI) and three cities with metropolitan populations of >2 million people (Chicago, Cleveland, and Detroit) (see Fig. S1 and Table S1 in the supplemental material for additional details). The air sampled in each city was collected at a single centralized location (see Table S1 for the geographic coordinates of each sampling site). The sampling campaigns spanned approximately 6 weeks in mid-summer 2007 and 6 weeks in mid-winter 2007. Approximately 20 to 30 samples were collected per location, and each collection period lasted 3 days. The site in Mayville was in a residential area surrounded by cornfields. The other three sites were located in urban corridors, with the Chicago and Detroit sites being in mixed industrial-residential areas and the Cleveland site being located downtown and adjacent to a major interstate. Samples were collected on quartz fiber filters (Whatman, Maidstone, England) mounted inside identical high-volume aerosol samplers at each site. Samplers were installed at existing air-monitoring sites, and therefore samples were collected at an ∼1.5-m height in Mayville and Detroit and an ∼4-m height in Chicago and Cleveland. The samplers draw ambient air at nominally 1.13 m3/min, leading to ∼5,000 m3 of air collected per filter/sample. These samplers employed a two-filter assembly to allow the collection of PM2.5. The top slotted filter collected coarse particles. The bottom filter (20.3 by 25.4 cm), which was the one analyzed in this work, collected PM2.5. The filters were prebaked in an oven at 550°C for 12 h prior to sampling. Baked filters were stored in a sealed box until loaded into the filter holder. The filter holder was cleaned with isopropanol before a new filter was loaded. Blank samples were collected by letting a filter sit inside the unit, while it was not operating, for 2 min. All filter samples and blanks were stored frozen at −20°C until downstream analyses could be performed. All samples were analyzed individually (i.e., samples were not composited prior to analysis), yielding a total of 96 air samples analyzed for both bacterial concentrations and community composition.
Meteorological data (see Table S1 in the supplemental material) were downloaded from the NOAA National Weather Service website (http://www.crh.noaa.gov) for each of three metropolitan cities (Chicago, Detroit, and Cleveland), and the nearest weather station measurements for Mayville, WI, were taken from Milwaukee, WI, approximately 88 km southeast of Mayville.
Total bacterial abundance measurements via flow cytometry.
Filter punches, 5 cm2 in size, were taken from each sample filter (400.5-cm2 total filter size) and used for the determination of bacterial abundances using a Cyan ADP flow cytometer (Beckman-Coulter, Fullerton, CA). Particles (including bacteria) were removed from the filter punches by shaking the filters at 220 rpm in 10 ml high-performance liquid chromatography (HPLC)-grade water for 2 h at room temperature inside a sterile petri plate. Prior to analysis with the flow cytometer, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) at a final working concentration of 2 μg/ml DAPI in HPLC-grade water, and each sample tube was spiked with 25 μl fluorescent beads (stock concentration, 1,000 μl−1). Bacteria were counted using the side scatter (log) versus violet (log; 450 nm) scatterplot, and beads were counted using the forward scatter (log) versus fluorescein isothiocyanate (FITC; log; 530 nm) scatterplot. Both plots can clearly distinguish DAPI-labeled cells from fluorescent beads, as the multifluorophore-containing beads had an approximately 10-fold increase in fluorescence in the violet channel, an increase in both forward and side scatter, as the 10-μm beads were significantly larger than the majority of the aerosolized bacteria, and bead fluorescence at higher wavelengths such as in the FITC channel (530 nm), where the DAPI-stained cells will not fluoresce. Considering that environmental aerosol samples are a heterogeneous mixture of nonbiological and biological aerosol particles, the background fluorescence was set at 5 fluorescence units in the violet channel (where DAPI fluoresces) for all samples to allow consistent sample-to-sample comparisons. Particles with fluorescence above this threshold were counted and considered to be intact cells. The total number of cells per cubic meter of air was calculated by considering the bead count efficiency, the dilution factor, and the volume of air that had passed through each filter. HPLC-grade water blanks stained with DAPI were used as negative controls to assess reagent contamination, and field filter blanks were analyzed to assess potential human contamination during sampling procedures. Scatterplots were generated in Summit V4.3 software (Dako, Carpinteria, CA), where counts were then exported to a spreadsheet for further statistical analyses.
DNA extraction and PCR amplification.
Bacterial DNA was extracted directly from a second filter punch using the PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA). Individual filter punches were cut into small pieces by a sterile technique, loaded into the bead tube of the DNA extraction kit, and heated to 65°C for 10 min, followed by 2 min of vortexing. The remaining steps of the DNA extraction were performed according to the manufacturer's instructions. Bacterial community composition was determined using a barcoded pyrosequencing procedure, which facilitates multiplexed sequencing of partial 16S rRNA genes. The protocol used here is identical to that described previously (5, 6, 18), including both the PCR conditions and primer sequences. Negative controls (both no-template controls and template from blank filters) were included in all of the steps of the process, from DNA extraction to PCR amplification, to check for contamination. PCR amplicons from each sample were pooled in approximately equal amounts in a single tube and sent to the University of South Carolina Environmental Genomics Core Facility on a 454 Life Sciences Genome Sequence FLX (Roche, Florence, SC) for pyrosequencing. With this protocol, we obtained sequence read lengths that averaged 330 bp in length. Previous work has shown that reads of this length over the V2 region of the 16S rRNA gene region provide sufficient information for accurate phylogenetic clustering of bacterial communities (31) and accurate taxonomic assignment to at least the family level of resolution (31).
Sequence analysis.
Sequences were analyzed and processed using the QIIME package (9). Briefly, QIIME takes all of the sequences from a single pyrosequencing run and assigns sample identities using a mapping file and the barcode assigned to each sample. Sequences were removed from the analysis if they were <200 bp in length, had a quality score of <25, contained ambiguous characters or an uncorrectable barcode, or did not contain the primer sequence. The remaining sequences were clustered into phylotypes using UCLUST v1.2.21q (17), with a minimum coverage of 99% and a minimum identity of 97%. A representative sequence was chosen for each phylotype by selecting the longest sequence that had the highest number of hits to other sequences of that particular phylotype. Representative sequences were aligned using PyNAST (8) against the Greengenes core set (15). Taxonomic assignments were made using the Ribosomal Database Project classifier (43). A phylogenetic tree containing the aligned sequences was then produced using FastTree (39). To determine the phylogenetic similarity between sample types, the unweighted UniFrac metric was used (32). UniFrac distances are based on the fraction of branch length shared by two communities within a phylogenetic tree made from the 16S rRNA gene sequences of all of the bacterial communities being compared. The UniFrac algorithm provides an estimate of the overall phylogenetic similarity between each pair of communities and therefore avoids some of the pitfalls associated with taxon-based community analyses (32).
Analysis of communities from likely source environments.
To gain a better understanding of the relationship between the midwestern airborne bacterial communities to those communities from likely source environments, we compared the airborne communities to previously described 16S rRNA gene sequence data sets representing a diversity of soil types across North and South America (n = 88) (25), leaf surfaces spanning more than 60 unique tree species (n = 112) (40), and fecal communities from cows (n = 6) (16), humans (n = 45) (13), and dogs (n = 156). For this analysis, QIIME was used in a manner similar to that described above, with the following modifications. Because different fragments of the 16S rRNA gene were sequenced in different studies (Escherichia coli positions 799 to 1115 correspond to the leaf surface samples, whereas the E. coli 16S rRNA gene region encompassing positions 27 to 338 was sequenced for all of the other sample types), the sequences could not be clustered directly, so instead of using the default UCLUST algorithm for phylotype assignment, a UCLUST phylotype assignment based on the Greengenes core set (15) was performed. Due to unequal sampling efforts (differing numbers of sequences per sample), the data set was rarefied to 500 sequences per sample to remove sample heterogeneity. Alpha diversity was assessed by examining rarefaction curves, and a tree containing the same set of sequences from the Greengenes core set, available at http://qiime.wordpress.com/, was used to assess the phylogenetic relationships among the air, soil, leaf surface, and fecal data sets.
Statistical analyses.
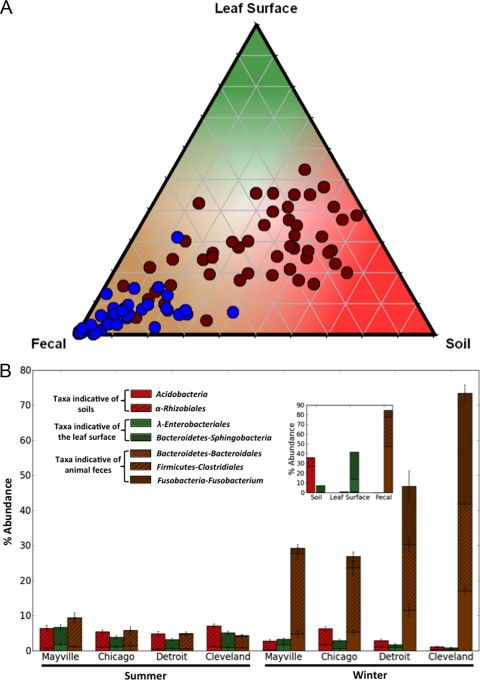
The analysis-of-similarities (ANOSIM) function in PRIMER V6 (12) was used on the unweighted UniFrac distance matrix to determine whether any categories of samples contained significantly different bacterial communities. Relationships among bacterial community similarity (determined using UniFrac), bacterial abundances, and meteorological parameters were investigated using Mantel tests as implemented in the PRIMER V6 software package (12). The SIMPER (similarity percentage) function in PRIMER V6 was used to identify bacterial taxa indicative of and unique to the soil, leaf surface, and fecal environments, with the relative abundances of these “indicator taxa” subsequently used to determine the relative contribution of each source environment to the bacterial communities in the air samples. Briefly, SIMPER determines the contribution of each taxonomic category to the average Bray-Curtis dissimilarity between pairs of grouped samples (e.g., the different environment types). A ternary plot (see Fig. 5A) was made in Sigma Plot 8.02, where the indicator taxa were fitted to an x-y-z coordinate system with each corner of the triangle representing a specific source environment. To create a heat map (see Fig. S3 in the supplemental material), the heat map function in R (V2.11.0) was used. The variation in bacterial abundance over the two seasons and across the four midwestern cities was assessed by analysis of variance, followed by pairwise comparisons.
Fig. 5.
Relative contributions of bacteria from likely source environments, i.e., soil, leaf surfaces, and animal feces. (A) Ternary plot representing the relative abundances of the indicator taxa in midwestern air samples color coded by season (maroon, summer; blue, winter), where the shading designates a given source environment (red, soil; green, leaf surface; brown, potential fecal origin). (B) Relative abundances of taxa indicative of the three source environments. (Inset) Relative abundances of these same indicator taxa in their native habitats, i.e., soil, leaf surfaces, and animal feces.
RESULTS AND DISCUSSION
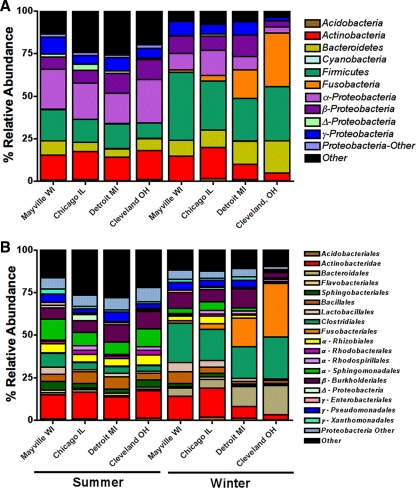
We found that the near-surface atmosphere harbors diverse bacterial communities (see Fig. S2 in the supplemental material) and individual samples contain ∼200 to 300 phylotypes, spanning seven bacterial phyla (Fig. 1A). These results contrast sharply with results obtained via culture-based analyses, which have long been the standard approach for studying airborne bacterial communities. Most culture-based analyses of atmospheric samples recover far fewer taxa, and the most commonly cultured airborne bacterial taxa are Gram-positive spore-forming bacteria (29), which we found to be relatively rare members of the airborne bacterial communities (Fig. 1; see Fig. S3 in the supplemental material). Although this “culturing bias” has been reported previously in many environments (37), studies that still rely on cultivation of airborne bacteria (including studies seeking potential allergens) will undoubtedly overlook dominant members of the airborne bacterial community.
Fig. 1.
Most abundant bacterial groups identified using barcoded pyrosequencing at the phylum level (A) and at the order level (B). Proteobacterial groups are designated by the letters α, β, γ, and Δ for the Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria, respectively.
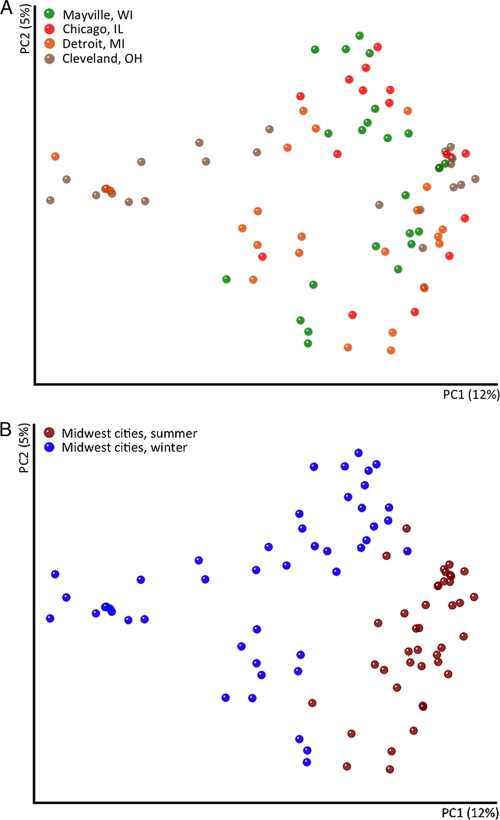
Samples collected at the four different locations did harbor distinct airborne bacterial communities; however, this association was relatively weak (Fig. 2A; ANOSIM Global R = 0.14, P < 0.001). When we examined summer and winter data separately, community composition was still weakly correlated with location during the summer sampling period (ANOSIM Global R = 0.23, P < 0.001). However, bacterial communities from the winter sampling period exhibited more predictable shifts among the four cities surveyed (ANOSIM Global R = 0.43, P < 0.001). Within a given season, there were no significant correlations between measured meteorological conditions (see Table S1 in the supplemental material) and bacterial community structure either within individual sites (r = <0.2 and P > 0.1 for each measured meteorological condition) or across all of the sites (r = <0.2 and P > 0.1in all cases). Other studies have also observed significant variability in bacterial community composition over time at individual locations (5, 7, 19), indicating that the dynamics of bacterial populations in the near-surface atmosphere are complex and likely influenced by factors (including rates of emission from source environments and rates of surface deposition of airborne bacteria) that are difficult to predict from simple meteorological parameters alone.
Fig. 2.
Relationships between bacterial communities in samples from the four locations (A) and across the summer and winter seasons (B). Bacterial communities were clustered using principal-coordinate analysis of the unweighted UniFrac distance matrix, with symbols that are closer together indicating air samples with more phylogenetically similar bacterial communities.
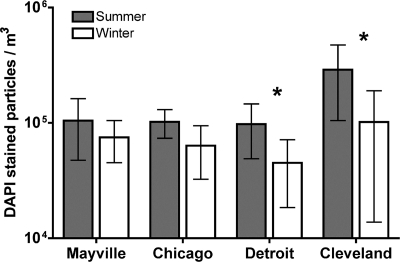
Strong seasonal patterns in bacterial community composition (Fig. 2B, ANOSIM Global R = 0.47, P < 0.001) and abundance were observed, with bacterial concentrations 52% lower, on average, in the winter than in the summer samples (Fig. 3). The summertime bacterial communities were dominated by taxa that prior culture-independent studies have also shown are common inhabitants of the atmosphere, including Pseudomonadales, Burkholderiales, Rhizobiales, and Sphingomonadales (5, 6). In most continental locations, soils (dust) and leaf surfaces are believed to be the dominant sources of bacteria in the near-surface atmosphere (7, 30), and this is likely true for the summer samples analyzed here as determined by directly comparing the air bacterial communities to previously analyzed communities from soils (25) and leaf surfaces (40) (Fig. 4A and 5). This finding differs from patterns observed in more arid regions (7), where reduced plant cover and lower soil moisture levels (which increase dust flux) lead to airborne bacterial communities almost entirely derived from soil. Also of note was the high relative abundance of members of the order Sphingomonadales in our summer samples (Fig. 1B). While this group did not make it into our source-tracking analysis (members of the order Sphingomonadales were not an abundant group in the phyllosphere study used in our current meta-analysis [40]), this particular group is known from other studies to be a common inhabitant of leaves (23, 36), supporting our hypothesis that leaves are an important source of bacteria in the atmosphere during the summer months. The nearby freshwater communities of the Great Lakes are another possible source of bacteria in the atmosphere in the summer months. However, freshwater systems contain few of the taxa found in our samples, so lake waters are not likely to be a dominant source of bacteria in either our summer or our winter air samples. In particular, freshwater bacterial communities are typically dominated by Actinobacteria (nonhuman associated), Cyanobacteria, Verrucomicrobia, and Bacteroidetes (non-gut-associated bacteria, e.g., members of the orders Sphingobacteriales and Flavobacteriales) (21, 47), which were rare in our samples, except for Actinobacteria, which appear to be more cosmopolitan across environments (Fig. 1A). Taken together, these results suggest that summer air bacterial communities are likely derived from both soil and leaf sources and that the harsh environmental conditions found in the atmosphere select for airborne communities that differ substantially from those found in other environments, a result consistent with previous studies (5, 6).
Fig. 3.
Total abundances of bacteria present in air samples collected from one small town (Mayville, WI) and three metropolitan cities (Chicago, IL; Detroit, MI; and Cleveland, OH) in the midwestern United States during the summer and winter. An asterisk denotes a statistically significant difference (P < 0.05) between summer and winter samples. Error bars indicate ±1 standard deviation.
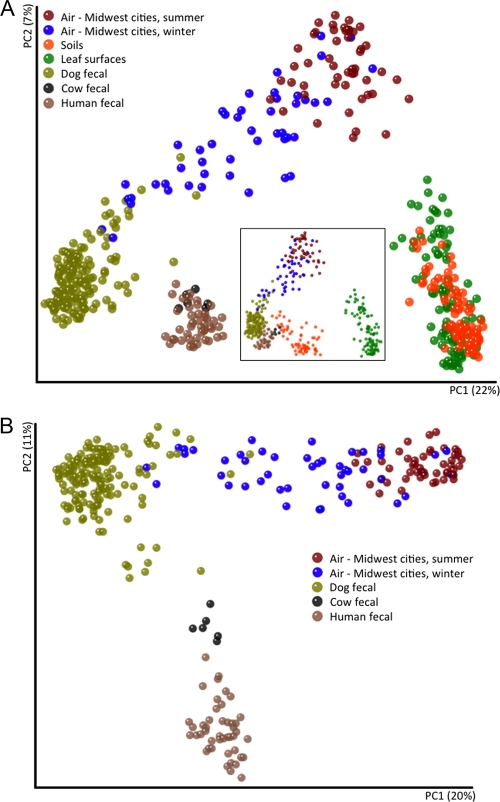
Fig. 4.
Relationships between summer and winter air samples (maroon and blue, respectively) and samples collected from a variety of possible source environments, including soils, leaf surfaces and various animal fecal samples. Communities were clustered using principal-coordinate analysis of the unweighted UniFrac distance matrix from a combined sequence data set of approximately 630,000 sequences. (A) Midwestern air samples alongside all three potential source environments (soil, leaf surfaces, and feces). (Inset) The same principal-coordinate analysis plot rotated along the first and third principal-coordinate axes. (B) Phylogenetic similarity between midwestern airborne bacterial communities and the communities of various fecal samples, including dog feces.
During the midwestern winter, the factors affecting airborne bacterial communities and concentrations are likely to be very different. Rates of bacterial aerosolization from plant, soil, and water sources are probably far lower because most plants are leafless, water bodies may be frozen, and the frozen or snow-covered ground likely reduces the flux of dust in the atmosphere. These factors may explain the reduced bacterial concentrations observed in air samples collected during the winter (Fig. 3). The composition of the bacterial communities found in winter air samples also differed dramatically from those found in summer samples (Fig. 2B). Many of the winter samples were dominated by bacterial taxa, including Bacteroidales, Clostridiales, and Fusobacteria, that were rare in previous molecular surveys of airborne bacteria (5, 6). Because these bacterial taxa are common inhabitants of the mammalian gut (27), we conducted a more detailed source-tracking analysis to directly compare the airborne bacterial communities to bacterial communities found in fecal matter collected from humans (n = 45), cows (n = 6), and dogs (n = 156) (Fig. 4). This analysis suggests that dog feces are likely the dominant source of aerosolized bacteria in the winter months in Cleveland, Detroit, and, to a lesser extent, Chicago (Fig. 4B; see Fig. S3 in the supplemental material). This is evident primarily from the high abundances of fusobacterial taxa, which are very common in the guts of dogs (33) (see Fig. S3) and, to a lesser extent, other carnivores, including house cats (27, 42, 46), but nearly absent from humans, cows, horses, chickens, and pigs (26, 27).
Our results point to feces, and dog feces in particular, as a dominant source of bacteria in the outdoor air in the metropolitan areas sampled here, particularly Cleveland and Detroit (see Fig. S3). Several additional lines of evidence support this conclusion. First, the dominance of common mammalian gut-associated bacteria in many of the winter air samples is striking, and these bacteria are very rare in other environments from which bacteria may be aerosolized, including soils, leaves, and lake waters (Fig. 4; see Fig. S3). Although there are sewage treatment facilities located within a 10-mile radius of the Chicago, Detroit, and Cleveland sampling locations that may emit fecal bacteria into the atmosphere, sewage treatment facilities typically harbor very low abundances of members of the phylum Fusobacteria (4). Second, most of the dominant taxa in the winter samples (including Bacteroidales, Clostridiales, and Fusobacteria; Fig. 5B) are obligate anaerobes, suggesting that they are unlikely to be derived from a wide range of source environments. Third, dog feces are likely abundant in these areas (dog populations are typically 37% of human populations in the United States [3]), and a large proportion of dog feces is likely to be deposited outside, where it could be aerosolized over time (particularly in Detroit, where recent reports suggest that there are >10,000 stray dogs within the city limits [11]). In the summer months, the feces-associated bacteria are still present in the air samples (Fig. 5) but they are far less abundant, as other bacterial sources (particularly leaves and soil) become relatively more important. Clearly, further investigation is required to understand the importance of dog fecal matter as a source of bacteria in the near-surface atmosphere of metropolitan areas. Further work is also required to test whether our finding that dog feces represent an important source of airborne bacteria in metropolitan areas is relevant to the health of individuals in these areas. We did not specifically test for human pathogens in our samples, and it is unknown whether these fecal bacteria are capable of triggering allergies or asthmatic reactions.
The results presented here have a number of important implications for the study of microbial life in the atmosphere. First, we demonstrated that comprehensive cross-environment analyses of bacterial communities can be useful for source tracking, as we can identify taxa unique to specific environments and use that information to determine the relative importance of various microbial habitats as sources of bacteria in the atmosphere. Second, our work suggests that the diversity of bacteria in outdoor air is high and that these bacteria sometimes come from unexpected sources, such as dog feces. Third, our work suggests that air quality standards should include a microbial component based largely on culture-independent methods and that perhaps apparently unrelated public health measures (such as enforcement of scoop laws) may also be important for meeting these standards. Finally, this work highlights the fact that the spatiotemporal variability of airborne bacteria needs to be assessed in integrative studies that bring microbiologists and atmospheric scientists together to understand the dynamics of bacterial cells in the near-surface atmosphere.
Supplementary Material
ACKNOWLEDGMENTS
Support for this work was provided by grants from the CIRES IRP program (to N.F.), the U.S. Environmental Protection Agency (to N.F.), the National Science Foundation (to N.F.), the Howard Hughes Medical Institute, and the National Institutes of Health, and aerosol sample collection for this project was supported by the Lake Michigan Air Directors Consortium (LADCO).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Albrecht A., Witzenberger R., Bernzen U., Jackel U. 2007. Detection of airborne microbes in a composting facility by cultivation based and cultivation-independent methods. Ann. Agric. Environ. Med. 14:81–85 [PubMed] [Google Scholar]
- 2. American Lung Association 2007. Trends in asthma morbidity and mortality. American Lung Association Epidemiology and Statistics Unit, Research and Program Services Division, Washington, DC: http://www.lungusa.org/finding-cures/our-research/trend-reports/asthma-trend-report.pdf [Google Scholar]
- 3. American Veterinary Medical Association 1997. U.S. pet ownership & demographics sourcebook. American Veterinary Medical Association, Schaumburg, IL [Google Scholar]
- 4. Bibby K., Viau E., Peccia J. 2010. Pyrosequencing of the 16S rRNA gene to reveal bacterial pathogen diversity in biosolids. Water Res. 44:4252–4260 [DOI] [PubMed] [Google Scholar]
- 5. Bowers R. M., et al. 2009. Characterization of airborne microbial communities at a high-elevation site and their potential to act as atmospheric ice nuclei. Appl. Environ. Microbiol. 75:5121–5130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bowers R. M., McLetchie S., Knight R., Fierer N. 2010. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 5:601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brodie E. L., et al. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 104:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caporaso J. G., et al. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caporaso J. G., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Christner B. C., Morris C. E., Foreman C. M., Cai R., Sands D. C. 2008. Ubiquity of biological ice nucleators in snowfall. Science 319:1214. [DOI] [PubMed] [Google Scholar]
- 11. Christoff C. 2011. Detroit won't bite on Discovery Channel show exploring the lives of stray dogs. Detroit Free Press, Detroit, MI [Google Scholar]
- 12. Clarke K. R., Warwick R. M. 2001. A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar. Ecol. Prog. Ser. 216:265–278 [Google Scholar]
- 13. Costello E. K., et al. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. D'Amato G. 2002. Environmental urban factors (air pollution and allergens) and the rising trends in allergic respiratory diseases. Allergy 57:30–33 [DOI] [PubMed] [Google Scholar]
- 15. DeSantis T. Z., et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Durso L. M., et al. 2010. Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl. Environ. Microbiol. 76:4858–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edgar R. C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461 [DOI] [PubMed] [Google Scholar]
- 18. Fierer N., Hamady M., Lauber C. L., Knight R. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:17994–17999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fierer N., et al. 2008. Short-term temporal variability in airborne bacterial and fungal populations. Appl. Environ. Microbiol. 74:200–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hirano S. S., Upper C. D. 1983. Ecology and epidemiology of foliar bacterial plant pathogens. Annu. Rev. Phytopathol. 21:243–269 [Google Scholar]
- 21. Humbert J.-F., et al. 2009. Comparison of the structure and composition of bacterial communities from temperate and tropical freshwater ecosystems. Environ. Microbiol. 11:2339–2350 [DOI] [PubMed] [Google Scholar]
- 22. Jaenicke R. 2005. Abundance of cellular material and proteins in the atmosphere. Science 308:73. [DOI] [PubMed] [Google Scholar]
- 23. Kim H., et al. 1998. High population of Sphingomonas species on plant surface. J. Appl. Microbiol. 85(4):731–736 [Google Scholar]
- 24. Lange J., Thorne P. S., Lynch N. 1997. Application of flow cytometry and fluorescent in situ hybridization for assessment of exposures to airborne bacteria. Appl. Environ. Microbiol. 63:1557–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lauber C. L., Hamady M., Knight R., Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75:5111–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee J. E., Lee S., Sung J., Ko G.-P. 2011. Analysis of human and animal fecal microbiota for microbial source tracking. ISME J. 5:362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ley R. E., et al. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lighthart B. 2000. Mini-review of the concentration variations found in the alfresco atmospheric bacterial populations. Aerobiologia 16:7–16 [Google Scholar]
- 29. Lighthart B. 1997. The ecology of bacteria in the alfresco atmosphere. FEMS Microbiol. Ecol. 23:263–274 [Google Scholar]
- 30. Lindemann J., Constantinidou H. A., Barchet W. R., Upper C. D. 1982. Plants as sources of airborne bacteria, including ice nucleation-active bacteria. Appl. Environ. Microbiol. 44:1059–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Z., DeSantis T. Z., Andersen G. L., Knight R. 2008. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 36(18):e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lozupone C. A., Hamady M., Kelley S. T., Knight R. 2007. Quantitative and qualitative (beta) diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Middelbos I. S., et al. 2010. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS One 5:e9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Möhler O., DeMott P. J., Vali G., Levin Z. 2007. Microbiology and atmospheric processes: the role of biological particles in cloud physics. Biogeosciences 4:1059–1071 [Google Scholar]
- 35. Moorman J. E., Zahran H., Truman B. I., Molla M. T. 2011. Current asthma prevalence—United States, 2006-2008. MMWR Surveill. Summ. 60(Suppl.):84–86 [PubMed] [Google Scholar]
- 36. Murakami Y., Otsuka S., Senoo K. 2010. Abundance and community structure of sphingomonads in leaf residues and nearby bulk soil. Microbes Environ. 25(3):183–189 [DOI] [PubMed] [Google Scholar]
- 37. Pace N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734–740 [DOI] [PubMed] [Google Scholar]
- 38. Pillai S. D., Ricke S. C. 2002. Bioaerosols from municipal and animal wastes: background and contemporary issues. Can. J. Microbiol. 48:681–696 [DOI] [PubMed] [Google Scholar]
- 39. Price M. N., Dehal P. S., Arkin A. P. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Redford A. J., Bowers R. M., Knight R., Linhart Y., Fierer N. 2010. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 12:2885–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rinsoz T., Duquenne P., Greff-Mirguet G., Oppliger A. 2008. Application of real-time PCR for total airborne bacterial assessment: comparison with epifluorescence microscopy and culture-dependent methods. Atmos. Environ. 42:6767–6774 [Google Scholar]
- 42. Ritchie L. E., Steiner J. M., Suchodolski J. S. 2008. Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis. FEMS Microbiol. Ecol. 66:590–598 [DOI] [PubMed] [Google Scholar]
- 43. Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wiedinmyer C., et al. 2009. The contribution of biological particles to observed particulate organic carbon at a remote high altitude site. Atmos. Environ. 43:4278–4282 [Google Scholar]
- 45. Womack A. M., Bohannan B. J. M., Green J. L. 2010. Biodiversity and biogeography of the atmosphere. Philos. Trans. R. Soc. B Biol. Sci. 365:3645–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang H., Chen L. 2010. Phylogenetic analysis of 16S rRNA gene sequences reveals distal gut bacterial diversity in wild wolves (Canis lupus). Mol. Biol. Rep. 37:4013–4022 [DOI] [PubMed] [Google Scholar]
- 47. Zwart G., Crump B. C., Kamst-van Agterveld M. P., Hagen F., Han S-K. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141–155 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.