Abstract
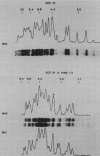
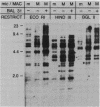
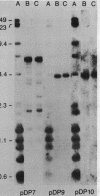
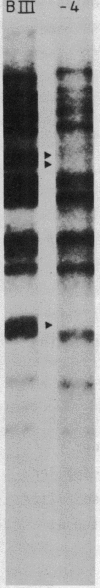
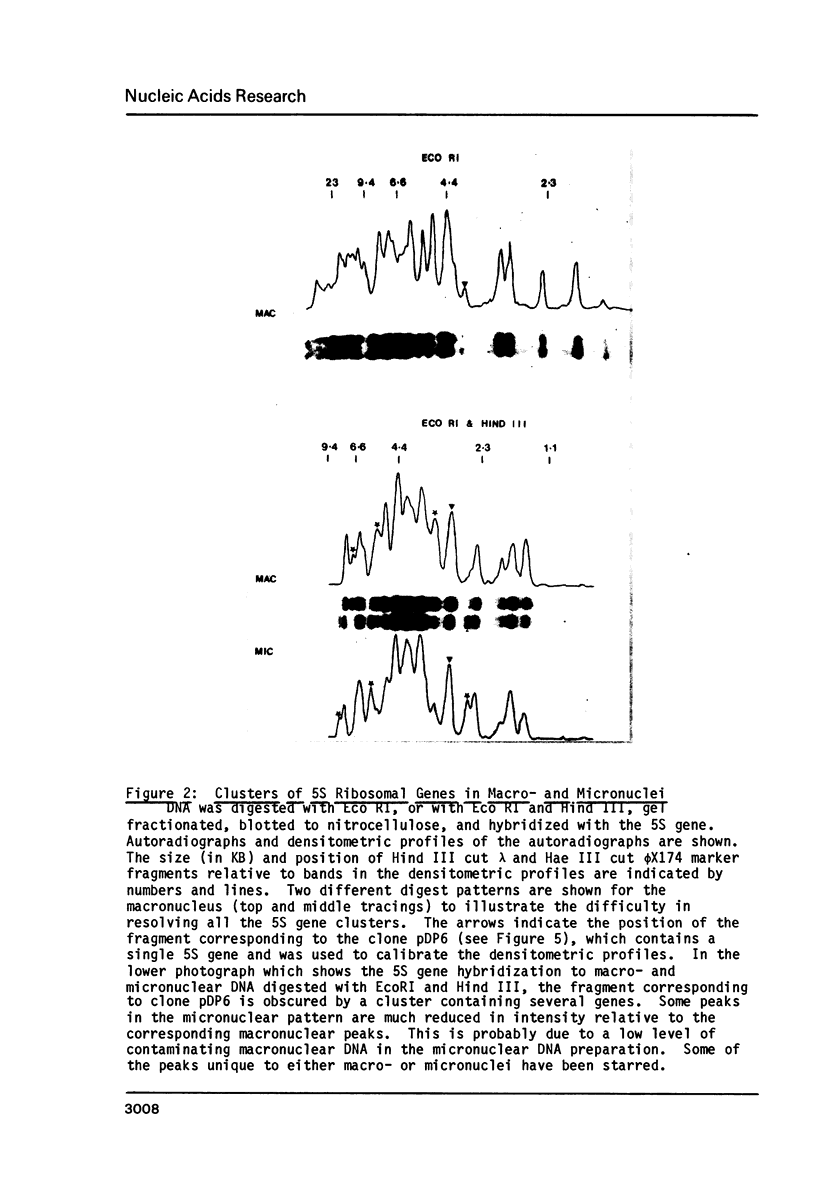
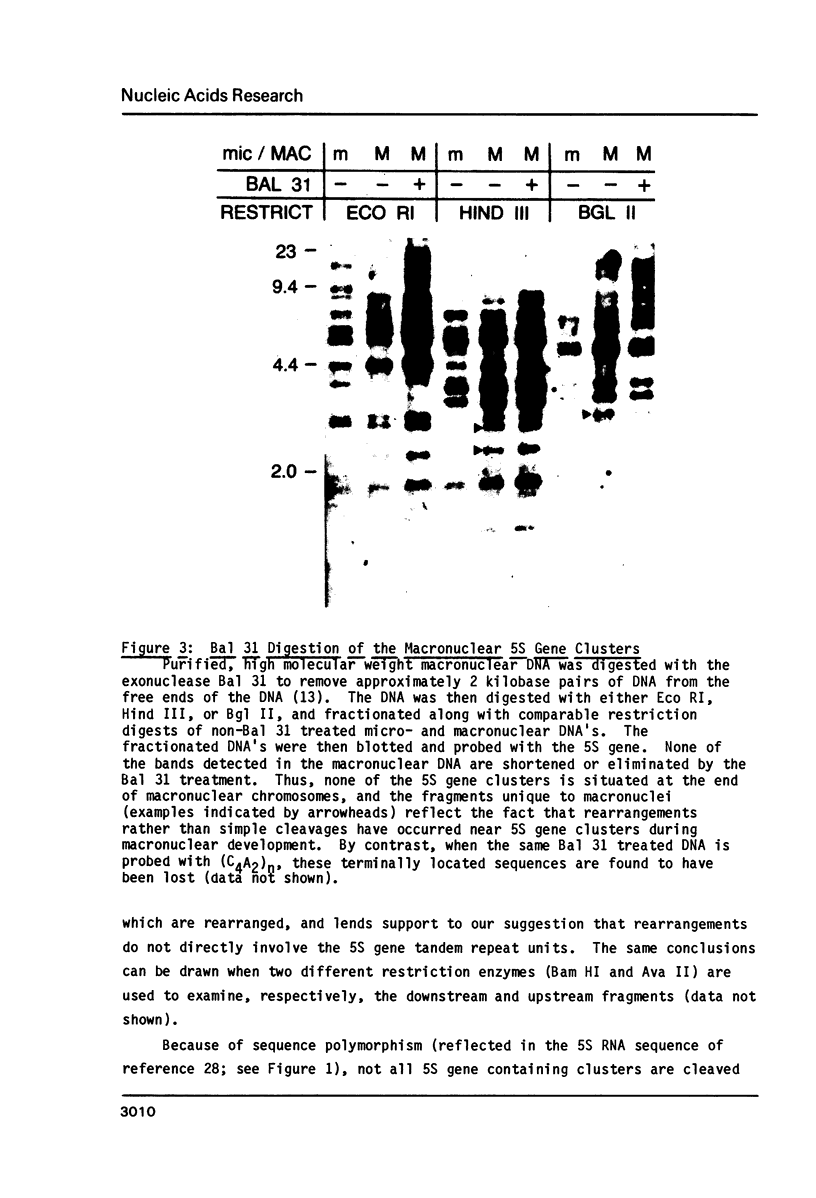
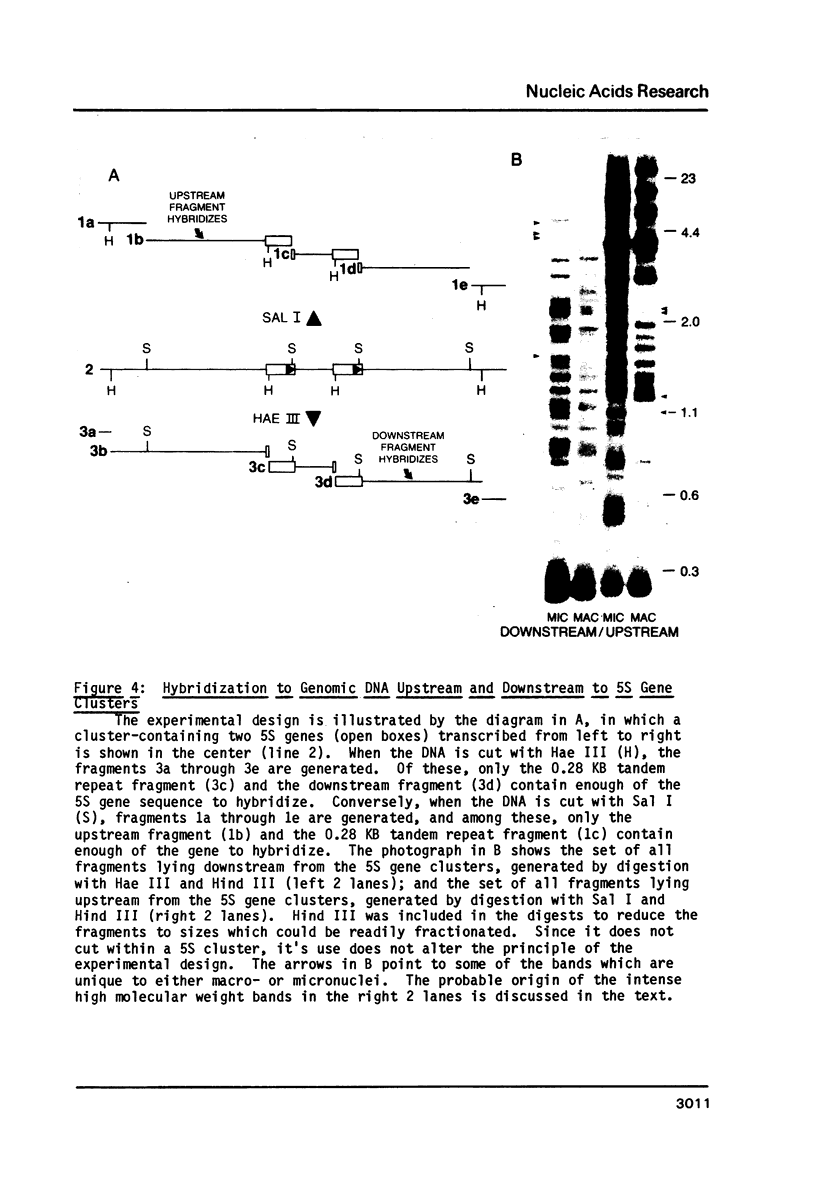
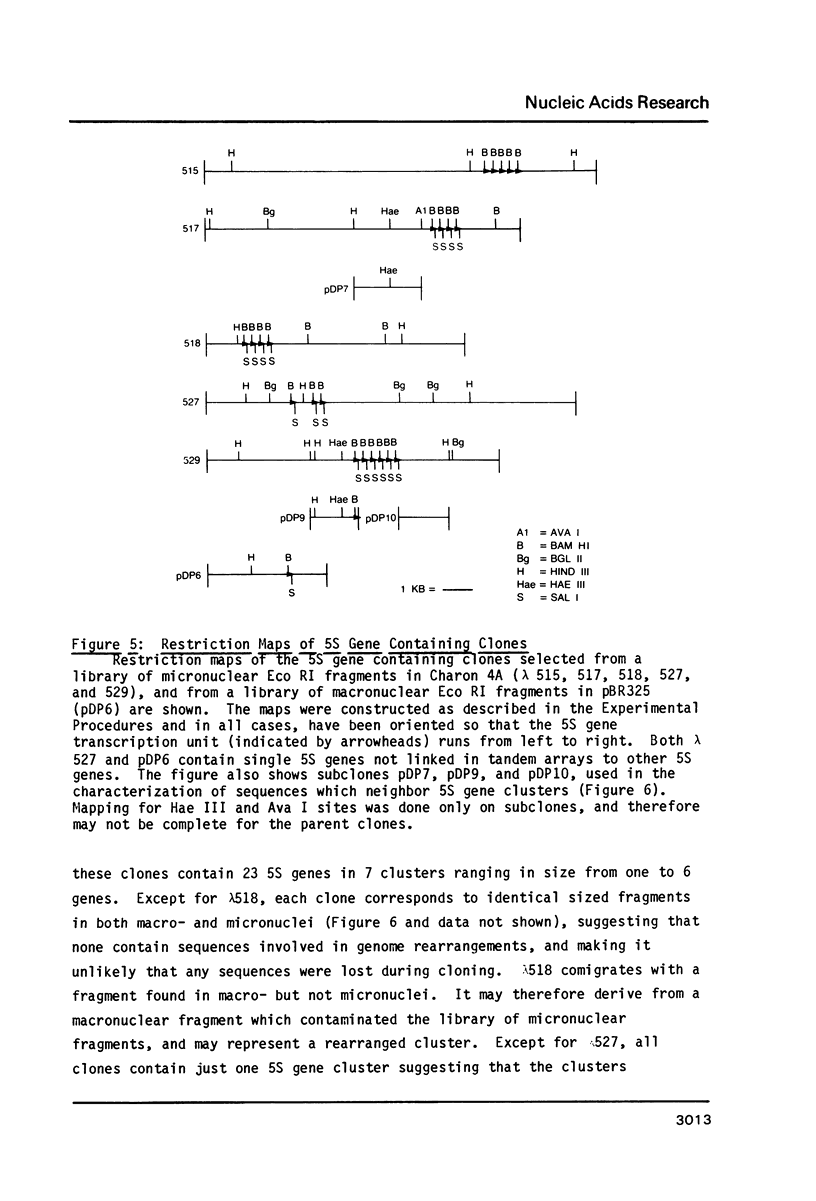
Macro- and micronuclei of Tetrahymena thermophila each contain approximately 30 clusters of 5S genes per haploid genome. Structural changes in DNA sequences associated with some of these clusters occur during the development of the transcriptionally active macronucleus from the transcriptionally inert micronucleus. Exonuclease digestion indicates that DNA fragmentation is not responsible for these changes, making it likely that sequence rearrangements occur near some 5S genes during macronuclear development. These rearrangements appear to be random in location with respect to the 5S genes and do not alter either the tandem repeat organization of the genes, the average number (five) or the range in number (one to about 16) of genes per cluster. The 5S gene clusters are not closely linked and are not flanked by common repeating elements large enough to cross-hybridize. Sequence analysis of one tandem repeat suggests that Tetrahymena 5S genes have intragenic promoters. These observations indicate that features other than DNA rearrangements or DNA sequence per se are responsible for the transcriptional activation of 5S genes during macronuclear development.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Bromberg S., Pratt K., Hattman S. Sequence specificity of DNA adenine methylase in the protozoan Tetrahymena thermophila. J Bacteriol. 1982 May;150(2):993–996. doi: 10.1128/jb.150.2.993-996.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns P. J., Brussard T. B. Positive selection for mating with functional heterokaryons in Tetrahymena pyriformis. Genetics. 1974 Nov;78(3):831–841. doi: 10.1093/genetics/78.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns P. J., Brussard T. E. Nullisomic tetrahymena: eliminating germinal chromosomes. Science. 1981 Jul 31;213(4507):549–551. doi: 10.1126/science.213.4507.549. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Jacobs H. T., Britten R. J. Very short repeats and coordinate induction of genes. Nature. 1983 Feb 10;301(5900):468–470. doi: 10.1038/301468a0. [DOI] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A. Genome organization and reorganization in Tetrahymena. Annu Rev Genet. 1980;14:203–239. doi: 10.1146/annurev.ge.14.120180.001223. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Hattman S., Pleger G. L. ( 6 N)methyl adenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J Cell Biol. 1973 Mar;56(3):697–701. doi: 10.1083/jcb.56.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky M. A. Macro- and micronuclei of Tetrahymena pyriformis: a model system for studying the structure and function of eukaryotic nuclei. J Protozool. 1973 Feb;20(1):19–25. doi: 10.1111/j.1550-7408.1973.tb05995.x. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J., Bloomer L. S. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982 Apr;28(4):781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- Gurdon J. B., Dingwall C., Laskey R. A., Korn L. J. Developmental inactivity of 5S RNA genes persists when chromosomes are cut between genes. Nature. 1982 Oct 14;299(5884):652–653. doi: 10.1038/299652a0. [DOI] [PubMed] [Google Scholar]
- Hart R. P., Folk W. R. Structure and organization of a mammalian 5 S gene cluster. J Biol Chem. 1982 Oct 10;257(19):11706–11711. [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Kaney A. R., Speare V. J. An amicronucleate mutant of Tetrahymena thermophila. Exp Cell Res. 1983 Feb;143(2):461–467. doi: 10.1016/0014-4827(83)90074-5. [DOI] [PubMed] [Google Scholar]
- Katzen A. L., Cann G. M., Blackburn E. H. Sequence-specific fragmentation of macronuclear DNA in a holotrichous ciliate. Cell. 1981 May;24(2):313–320. doi: 10.1016/0092-8674(81)90321-4. [DOI] [PubMed] [Google Scholar]
- Kimmel A. R., Gorovsky M. A. Numbers of 5S and tRNA genes in macro- and micronuclei of Tetrahymena pyriformis. Chromosoma. 1976 Mar 10;54(4):327–337. doi: 10.1007/BF00292813. [DOI] [PubMed] [Google Scholar]
- Kimmel A. R., Gorovsky M. A. Organization of the 5S RNA genes in macro- and micronuclei of Tetrahymena pyriformis. Chromosoma. 1978 Jun 23;67(1):1–20. doi: 10.1007/BF00285644. [DOI] [PubMed] [Google Scholar]
- King B. O., Yao M. C. Tandemly repeated hexanucleotide at Tetrahymena rDNA free end is generated from a single copy during development. Cell. 1982 Nov;31(1):177–182. doi: 10.1016/0092-8674(82)90417-2. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Gurdon J. B. The reactivation of developmentally inert 5S genes in somatic nuclei injected into Xenopus oocytes. Nature. 1981 Feb 5;289(5797):461–465. doi: 10.1038/289461a0. [DOI] [PubMed] [Google Scholar]
- Korn L. J. Transcription of Xenopus 5S ribosomal RNA genes. Nature. 1982 Jan 14;295(5845):101–105. doi: 10.1038/295101a0. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Maizels N. Dictyostelium 17S, 25S, and 5S rDNAs lie within a 38,000 base pair repeated unit. Cell. 1976 Nov;9(3):431–438. doi: 10.1016/0092-8674(76)90088-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Moss T. Transcription of cloned Xenopus laevis ribosomal DNA microinjected into Xenopus oocytes, and the identification of an RNA polymerase I promoter. Cell. 1982 Oct;30(3):835–842. doi: 10.1016/0092-8674(82)90288-4. [DOI] [PubMed] [Google Scholar]
- Pan W. C., Blackburn E. H. Single extrachromosomal ribosomal RNA gene copies are synthesized during amplification of the rDNA in Tetrahymena. Cell. 1981 Feb;23(2):459–466. doi: 10.1016/0092-8674(81)90141-0. [DOI] [PubMed] [Google Scholar]
- Pan W. C., Orias E., Flacks M., Blackburn E. H. Allele-specific, selective amplification of a ribosomal RNA gene in Tetrahymena thermophila. Cell. 1982 Mar;28(3):595–604. doi: 10.1016/0092-8674(82)90214-8. [DOI] [PubMed] [Google Scholar]
- Peterson R. C., Doering J. L., Brown D. D. Characterization of two xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980 May;20(1):131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Thomas M., Kramer R. A., Davis R. W. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978 Aug 15;123(3):387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Pratt K., Hattman S. Deoxyribonucleic acid methylation and chromatin organization in Tetrahymena thermophila. Mol Cell Biol. 1981 Jul;1(7):600–608. doi: 10.1128/mcb.1.7.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Yanofsky C., Driftmier K., Metzenberg R. L., Alzner-DeWeerd B., RajBhandary U. L. Dispersed 5S RNA genes in N. crassa: structure, expression and evolution. Cell. 1981 Jun;24(3):819–828. doi: 10.1016/0092-8674(81)90107-0. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N., Sussman J. L. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M. C., Gorovsky M. A. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma. 1974;48(1):1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- Yao M. C. Ribosomal RNA gene amplification in Tetrahymena may be associated with chromosome breakage and DNA elimination. Cell. 1981 Jun;24(3):765–774. doi: 10.1016/0092-8674(81)90102-1. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Yao C. H. Repeated hexanucleotide C-C-C-C-A-A is present near free ends of macronuclear DNA of Tetrahymena. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7436–7439. doi: 10.1073/pnas.78.12.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R. W., Yao M. C. Elimination of DNA sequences during macronuclear differentiation in Tetrahymena thermophila, as detected by in situ hybridization. Chromosoma. 1982;85(1):11–22. doi: 10.1007/BF00344591. [DOI] [PubMed] [Google Scholar]
- Zasloff M., Ginder G. D., Felsenfeld G. A new method for the purification and identification of covalently closed circular DNA molcules. Nucleic Acids Res. 1978 Apr;5(4):1139–1152. doi: 10.1093/nar/5.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M., Santos T. Reiteration frequency mapping: analysis of repetitive sequence organization within cloned DNA fragments containing the human initiator methionine tRNA gene. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5668–5672. doi: 10.1073/pnas.77.10.5668. [DOI] [PMC free article] [PubMed] [Google Scholar]