Abstract
A key element in iron-dependent regulation of iron metabolism and virulence-related functions for Pseudomonas aeruginosa is the sigma factor PvdS. PvdS expression itself is also influenced by iron-independent stimuli. We show that pyoverdine production and pvdS expression depend on one of the two lipases of P. aeruginosa.
TEXT
Due to its fundamental functions in cell metabolism, iron is an essential element for most living cells. However, although this element is present in high abundance, but with very low bioavailability in aerobic environments at neutral pH, both prokaryotes and eukaryotes must mitigate this paradox in order to acquire this metal for key metabolic processes.
The Gram-negative bacterial pathogen Pseudomonas aeruginosa has become one of the best-understood bacterial models to study regulatory processes related to iron acquisition and metabolism (26). P. aeruginosa is a ubiquitously distributed bacterium that has an enormous metabolic versatility which enables it to survive in a range of diverse environmental niches. It has evolved a variety of strategies for both iron acquisition and for the regulation of its metabolism (3, 4, 11, 18).
In Gram-negative bacteria, the repressor Fur (ferric uptake regulator) plays the master role in regulation of iron metabo-lism (7, 27). Fur and its cofactor Fe(II) bind to so-called Fur boxes within promoters of iron-regulated genes (encoding, e.g., proteins for siderophore biosynthesis and uptake), thereby repressing their expression. In P. aeruginosa more than 200 genes are regulated in response to changes in iron availability (17). However, only a subpopulation of these genes are directly repressed by Fur (17), and several Fur-dependent mechanisms have been described to act downstream of Fur to regulate a subset of iron-regulated genes (reviewed in references 26 and 30). Among them is the extracytoplasmic function sigma factor PvdS, which plays a central role in regulation of at least 26 iron-repressible genes or operons in P. aeruginosa involved in iron uptake and metabolism (17, 24). PvdS controls the expression of major P. aeruginosa virulence factors, including pyoverdine, the extracellular protease V (PrpL), and exotoxin A, suggesting a prominent role of PvdS in regulation of virulence (5, 21, 29, 31). PvdS is known to be directly regulated by Fur and is expressed under iron limitation (13, 16). However, additional mechanisms control PvdS sigma factor activity at posttranslational levels (12, 28), and there are indications that expression of PvdS itself and PvdS-regulated genes are directly or indirectly affected by additional environmental conditions, like carbon sources, oxygen tension, copper starvation, and population density (8, 15, 22), suggesting a functional integration of different signals into an appropriate physiological and regulatory answer to a given sum of different stimuli. Recently, CysB, the first non-iron-stimulated regulator with an effect on PvdS expression, was described (10).
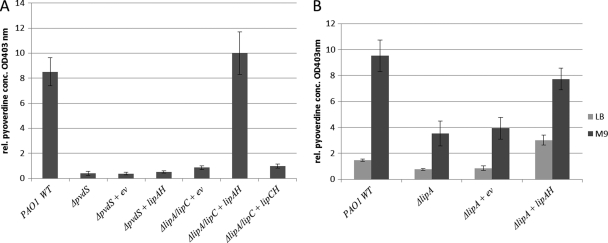
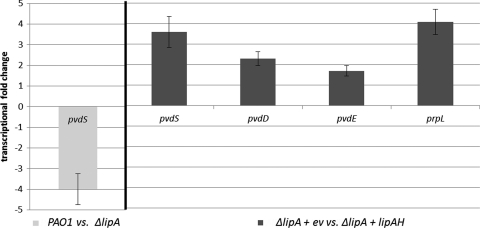
Lipolytic enzymes have been suggested to be involved in lipid signaling pathways, since they influence virulence-relevant phenotypes of P. aeruginosa (1, 14, 25, 33). Among those enzymes is the lipase LipC (PA4813), which was found to affect motility, biofilm formation, and rhamnolipid production (19), whereas the second lipase, LipA (PA2862), was unobtrusive with respect to virulence-related phenotypes. However, mutant strains (ΔlipA [32] and ΔlipA ΔlipC [ΔlipA] with an additional GM cassette in lipC [19]) with deletions of lipase genes lack the characteristic green color of P. aeruginosa, thereby suggesting reduced pyoverdine production. Comparison of the pyoverdine concentration in cultures of the wild type, a pvdS mutant (16), and a lipC lipA double mutant grown under iron-limiting conditions in M9 medium (20) with 3 g/liter succinate as the carbon source (20 ml in 250-ml flasks; 8 h, 37°C, 150 rpm) proved this assumption (Fig. 1A). Pyoverdine production in this mutant was reduced to levels comparable to those observed with the pvdS mutant and could be restored by complementation with LipA, but not with LipC (Fig. 1A). Complementation was performed using plasmids pVLT33-lipAH and pVLT33-lipCH, with the target genes transcriptionally regulated from the plasmid-carried tac promoter (6), and lipase expression was verified by spectrophotometric activity measurement with p-nitrophenyl-palmitate as the substrate (23, 34) (data not shown). As expected, in LB as a rich medium, pyoverdine production was decreased compared to cultivation in M9 medium without extra iron (Fig. 1B). Interestingly, expression of lipA in the LipA-deficient mutant led to increased pyoverdine levels, even in the non-iron-restricted rich LB medium (Fig. 1B). As the wild type and the empty vector control appeared not to produce pyoverdine under these non-iron limitation conditions, this finding suggests a decoupling of pyoverdine production from iron limitation. Moreover, the results show that reaching full pyoverdine production capacity under iron limitation conditions requires the presence of LipA. To distinguish whether LipA was needed up or downstream of PvdS, this lipase was also expressed in the pvdS mutant strain. In fact, LipA expression in this iron regulation mutant could not restore pyoverdine production, thus strongly suggesting that LipA acts upstream of PvdS. Expression of the pvdS gene was measured in the lipA mutant by real-time PCR with SYBR green assays. Total RNA was isolated using the RNA minikit (Qiagen, Hilden, Germany) and reverse transcribed into cDNA with the High Cap RNA-cDNA kit (Life Technology, Darmstadt, Germany), which was then used as template for quantitative PCR on an HT7900 cycler (Applied Biosystems, Darmstadt, Germany). Changes were determined using the comparative threshold cycle (CT) method by normalizing to the levels of mRNA transcribed from the reference gene rpoD (2). The transcript level of pvdS was found to be 4-fold reduced in the lipA mutant compared to the wild type. Consequently, complementation with functional LipA (as determined by the lipase activity assay [data not shown]) led to an increase not only of the transcript levels for pvdS but also of the pvdD (pyoverdine synthetase D), pvdE (pyoverdine biosynthesis protein PvdE), and prpL genes (Fig. 2), which are known as PvdS-regulated target genes and play important roles in pyoverdine biosynthesis (10, 17) or, in the case of prpL, function in virulence (24). These results demonstrate that the lipase LipA, or a yet-uncharacterized lipase-dependent signaling pathway, is involved in regulation of pvdS gene expression at the transcriptional level, with all consequences for PvdS-dependent regulation of downstream genes. Consequently, with an influence on iron acquisition, LipA affects one of the key processes required for full virulence of P. aeruginosa. This effect appears to be specific for the lipase LipA, and not for the second lipase, LipC, which has been shown to influence other virulence-related functions. Accordingly, these data show that both lipases are likely important for P. aeruginosa pathogenicity while they appear to have very distinct functions.
Fig. 1.
Pyoverdine concentrations in P. aeruginosa culture supernatants. (A) Wild-type (WT), ΔpvdS mutant, ΔlipA ΔlipC double mutant, and the double mutant with a lipA or lipC expression plasmid, which also harbored the gene for the lipase-specific chaperone lipH for proper folding of the lipases, were grown for 8 h at 37°C in M9 minimal medium without iron supplementation, and pyoverdine concentrations were determined in cell-free supernatants (9). (B) The wild type and a ΔlipA mutant harboring the empty vector (ev) or a lipA expression plasmid were grown in rich LB or minimal M9 medium, and the relative pyoverdine concentrations were determined after 8 h of growth.
Fig. 2.
Change in gene expression of pvdS in a P. aeruginosa ΔlipA mutant relative to the wild type (light gray). The right side of the graph compares (based on the fold change) the regulation of pvdS and PvdS-regulated genes (pvdD, pvdE, and prpL) in a ΔlipA mutant with empty vector (ev) versus a ΔlipA mutant which harbored the lipA gene and the gene for the lipase-specific chaperone lipH on plasmid pVLT33-LipAH (lipAH; dark gray). In all cases transcription was normalized to rpoD. Cultures were grown for 8 h in LB medium at 37°C before RNA was isolated. Error bars represent standard deviations from three independent experiments.
Acknowledgments
Horst Funken was supported by grant no. 0313980B from the German Federal Ministry of Education and Research within the program “Systems Biology of Microorganisms—SysMO” and the grant AZ13235-1 from the Deutsche Bundesstiftung Umwelt. Portions of this study were also supported by a grant (R37 AI15940) from the U.S. National Institutes of Health to Michael Vasil.
Footnotes
Published ahead of print on 12 August 2011.
REFERENCES
- 1. Barker A. P., et al. 2004. A novel extracellular phospholipase C of Pseudomonas aeruginosa is required for phospholipid chemotaxis. Mol. Microbiol. 53:1089–1098 [DOI] [PubMed] [Google Scholar]
- 2. Bartels K. M., et al. 2011. Glycosylation is required for outer membrane localization of the lectin LecB in Pseudomonas aeruginosa. J. Bacteriol. 193:1107–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cornelis P. 2010. Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 86:1637–1645 [DOI] [PubMed] [Google Scholar]
- 4. Cornelis P., Matthijs S., Van Oeffelen L. 2009. Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22:15–22 [DOI] [PubMed] [Google Scholar]
- 5. Cunliffe H. E., Merriman T. R., Lamont I. L. 1995. Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. J. Bacteriol. 177:2744–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Lorenzo V., Eltis L., Kessler B., Timmis K. N. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17–24 [DOI] [PubMed] [Google Scholar]
- 7. Escolar L., Perez-Martin J., de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frangipani E., Slaveykova V. I., Reimmann C., Haas D. 2008. Adaptation of aerobically growing Pseudomonas aeruginosa to copper starvation. J. Bacteriol. 190:6706–6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hofte M., Buysens S., Koedam N., Cornelis P. 1993. Zinc affects siderophore-mediated high affinity iron uptake systems in the rhizosphere Pseudomonas aeruginosa 7NSK2. Biometals 6:85–91 [DOI] [PubMed] [Google Scholar]
- 10. Imperi F., Tiburzi F., Fimia G. M., Visca P. 2010. Transcriptional control of the pvdS iron starvation sigma factor gene by the master regulator of sulfur metabolism CysB in Pseudomonas aeruginosa. Environ. Microbiol. 12:1630–1642 [DOI] [PubMed] [Google Scholar]
- 11. Imperi F., Tiburzi F., Visca P. 2009. Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 106:20440–20445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lamont I. L., Beare P. A., Ochsner U., Vasil A. I., Vasil M. L. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 99:7072–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leoni L., Ciervo A., Orsi N., Visca P. 1996. Iron-regulated transcription of the pvdA gene in Pseudomonas aeruginosa: effect of Fur and PvdS on promoter activity. J. Bacteriol. 178:2299–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miller R. M., et al. 2008. Pseudomonas aeruginosa twitching motility-mediated chemotaxis towards phospholipids and fatty acids: specificity and metabolic requirements. J. Bacteriol. 190:4038–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ochsner U. A., Vasil M. L. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. U. S. A. 93:4409–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ochsner U. A., Johnson Z., Lamont I. L., Cuncliffe H. E., Vasil M. L. 1996. Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol. Microbiol. 21:1019–1028 [DOI] [PubMed] [Google Scholar]
- 17. Ochsner U. A., Wilderman P. J., Vasil A. I., Vasil M. L. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277–1287 [DOI] [PubMed] [Google Scholar]
- 18. Poole K., McKay G. A. 2003. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 8:D661–D686 [DOI] [PubMed] [Google Scholar]
- 19. Rosenau F., et al. 2010. Lipase LipC affects motility, biofilm formation and rhamnolipid production in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 309:25–34 [DOI] [PubMed] [Google Scholar]
- 20. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21. Shigematsu T., et al. 2001. Iron-mediated regulation of alkaline proteinase production in Pseudomonas aeruginosa. Microbiol. Immunol. 45:579–590 [DOI] [PubMed] [Google Scholar]
- 22. Stintzi A., Evans K., Meyer J. M., Poole K. 1998. Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol. Lett. 166:341–345 [DOI] [PubMed] [Google Scholar]
- 23. Stuer W., Jaeger K. E., Winkler U. K. 1986. Purification of extracellular lipase from Pseudomonas aeruginosa. J. Bacteriol. 168:1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tiburzi F., Imperi F., Visca P. 2008. Intracellular levels and activity of PvdS, the major iron starvation sigma factor of Pseudomonas aeruginosa. Mol. Microbiol. 67:213–227 [DOI] [PubMed] [Google Scholar]
- 25. Tielen P., et al. 2010. Extracellular enzymes affect biofilm formation of mucoid Pseudomonas aeruginosa. Microbiology 156:2239–2252 [DOI] [PubMed] [Google Scholar]
- 26. Vasil M. L. 2007. How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals 20:587–601 [DOI] [PubMed] [Google Scholar]
- 27. Vasil M. L., Ochsner U. A. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34:399–413 [DOI] [PubMed] [Google Scholar]
- 28. Visca P. 2004. Iron regulation and siderophore signalling in virulence by Pseudomonas aeruginosa, p. 69–123 In Ramos J.-L. (ed.), Pseudomonas. Kluwer Academic Publishers, Boston, MA [Google Scholar]
- 29. Visca P., Imperi F., Lamont I. L. 2007. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 15:22–30 [DOI] [PubMed] [Google Scholar]
- 30. Visca P., Leoni L., Wilson M. J., Lamont I. L. 2002. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45:1177–1190 [DOI] [PubMed] [Google Scholar]
- 31. Wilderman P. J., et al. 2001. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 69:5385–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilhelm S., Tommassen J., Jaeger K. E. 1999. A novel lipolytic enzyme located in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 29:6977–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilhelm S., Gdynia A., Tielen P., Rosenau F., Jaeger K. E. 2007. The autotransporter esterase EstA of Pseudomonas aeruginosa is required for rhamnolipid production, cell motility, and biofilm formation. J. Bacteriol. 189:6695–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winkler U. K., Stuckmann M. 1979. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J. Bacteriol. 138:663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]