FAST-TRACK COMMUNICATION
In May 2011, an enteroaggregative verocytotoxin-producing strain of Escherichia coli O104:H4 caused an outbreak of hemolytic uremic syndrome in Germany. Although traditional culture and phenotypic tests can identify the outbreak strain, rapid molecular testing is useful for timely diagnosis. While multitarget PCR assays have been reported by Bielaszewska et al. (1) and Cui et al. (5), such assays combined multiple pairs of primers, e.g., genes coding for O104 and H4 antigens, Shiga toxin (3), and the tellurite resistance gene terD (2). As none of the targets is unique to the outbreak strain, an isolate is identified only if PCR results for all loci are positive. Hence, such assays are limited to cultured isolates and have limited use in uncultured clinical, food, or environmental samples.
Recently, we described a proof-of-concept study on a pan-genomic analysis approach in single-target selection for PCR identification and detection of Burkholderia species (6). Using the specific targets, we designed a robust PCR assay capable of detecting and identifying the different Burkholderia species, not only from cultured isolates but also directly from soil and spiked sputum samples. We reckon that this approach may be valuable to the design of PCR assays for direct detection of outbreak bacterial strains from various uncultured samples. Here, we present an improved version of our algorithm, ssGeneFinder; we used the program to generate specific single targets for detecting the recent E. coli O104:H4 outbreak strain and subsequently validated the targets.
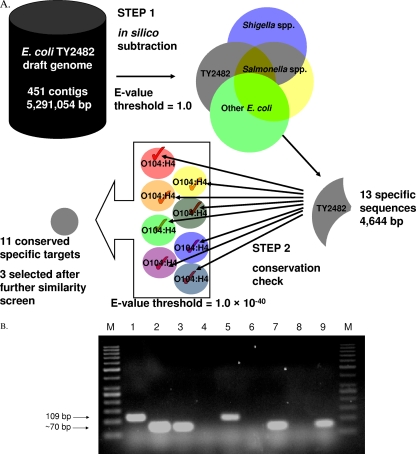
The algorithm is outlined in Fig. 1A. We downloaded unannotated genomic contigs (large DNA sequence fragments) of nine outbreak strain E. coli O104:H4 isolates to serve as target species (see Table S1 in the supplemental material). Gene annotation was not required, as the in silico subtraction by ssGeneFinder is applicable even to genome fragments. Among the nine isolates, the genome of TY2482 was automatically selected as the starting genome because of its small size, since a conserved target must be present even in the target genome with the smallest number of genes. Ninety-six, 39, and 10 strains of E. coli, Salmonella spp., and Shigella spp. for which complete or draft genome data were available from GenBank (see Table S2 in the supplemental material) were used in in silico subtraction: higher BLAST word size values were used initially to remove regions of obvious similarity, and the values gradually lowered to increase sensitivity, eliminating smaller regions which may cause nonspecific priming. After five rounds of in silico subtraction, the initial 5,291,054 bp of sequence data were reduced to 13 sequences (total length, 4,644 bp) specific to the starting genome and not found in any of the nontarget genomes. Those loci were further searched against the eight other outbreak isolates to determine whether they were present and conserved. Eleven of the 13 loci were of sufficient length and reported as potential targets (see Table S3 in the supplemental material). Little sequence variation is expected for a presumptively clonal outbreak: indeed, only one single-nucleotide deletion was observed in a poly(A) tract of isolate LB226692 upon multiple sequence alignments of the 11 loci.
Fig. 1.
(A) The ssGeneFinder process. Starting with the smallest target genome (Escherichia coli TY2482 by BGI) with 451 contigs, ssGeneFinder performed an in silico subtraction to remove sequences with significant similarity to those found in Shigella spp., Salmonella spp., and other E. coli genomes (step 1). Thirteen sequences, with a total length of 4,644 bp (approximately 0.09% of the starting genome size), remained after the subtraction and were searched against other draft genomes of the outbreak strain. Sequences that were deemed too short or nonconserved were removed (step 2). The 11 conserved and specific targets were subjected to similarity search using NCBI BLASTn. Targets with significant sequence similarity to other species in the GenBank database were also removed. Finally, three feasible targets were generated. (B) DNA products from PCR identification and detection of German outbreak strain E. coli RKI 11 2027. Lane M, molecular marker (GeneRuler 50-bp DNA ladder); lanes 1 to 3, PCR amplification products from E. coli RKI 11 2027 using primers specific to targets usid000007 (contig 69, bp 14714 to 14853), usid000006 (contig 69, bp 14491 to 14693), and usid000002 (contig 43, bp 1486 to 1633), respectively; lane 4, E. coli isolate AKLT01, using primers specific to target usid000007; lane 5, like lane 4 but spiked with an equal volume of genomic DNA extract from E. coli RKI 11 2027; lane 6, stool sample S01, using primers specific to target usid000006; lane 7, like lane 6, spiked as in lane 5; lane 8, soil sample SL01, using primers specific to target usid0000002; lane 9, like lane 8, spiked as in lane 5.
The ssGeneFinder run took 24 min on a Windows desktop computer (default settings, 2.93 GHz Intel CPU). The putative targets (see Table S3 in the supplemental material) were further analyzed by BLASTn search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to check for sequence similarity to other bacterial species or strains not included in the above-described analysis. Three of the 11 loci were considered feasible targets and subject to primer design using NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Table 1). Genomic DNA from the outbreak strain was obtained from the EU Reference Laboratory for E. coli. The specificity of the primers was assessed using a collection of 65 E. coli isolates, 25 culture-negative stool samples, and 7 environmental soil samples (see Table S4 and Methods in the supplemental material). All three targets were successfully amplified from the outbreak strain, and there was no false-positive result in the screened panel of samples (Fig. 1B).
Table 1.
Validated primers targeting the feasible molecular targets
| Target identification no. (location on reference assembly) | Product length (bp) | Primer | Position |
Length (bp) | Tm (°C)a | |
|---|---|---|---|---|---|---|
| Start | End | |||||
| usid000002 (contig 43, bp 1486–1633) | 73 | LPW18119 5′-TTCTGGCCCTGTGCACCGTAT-3′ | 21 | 41 | 21 | 66.1 |
| LPW18120 5′-CGGACTCGCGACACATTGCT-3′ | 93 | 74 | 20 | 65.4 | ||
| usid000006 (contig 69, bp 14491–14693) | 71 | LPW18117 5′-CCTAGTCTTGCTTGCTCTGTGGTCA-3′ | 110 | 134 | 25 | 66.2 |
| LPW18118 5′-GAATCAACAAAAATGCCTGGCG-3′ | 180 | 159 | 22 | 65.6 | ||
| usid000007 (contig 69, bp 14714–14853) | 109 | LPW18115 5′-CTTGCAGGCCTTTAAGATCGC-3′ | 23 | 43 | 21 | 63.1 |
| LPW18116 5′-TGGCAACAGTGAAAAATGAAACG-3′ | 131 | 109 | 23 | 64.0 | ||
Tm, melting temperature.
This enterohemorrhagic E. coli (EHEC) outbreak heralded the use of high-throughput sequencing in epidemiological investigation. Multiple isolates were sequenced, and draft genomes were publicly available within weeks (4). These genomes, however, were often at their fragmented, finishing stage, hindering the extraction of individual gene sequences for traditional probe design. ssGeneFinder circumvents such obstacles, as it analyses all regions of an input genome, extracting even conserved intergenic spacers as targets. It offers a user-friendly interface, employs a robust algorithm, and minimizes hands-on time. The upsurge in the availability of high-throughput bacterial sequencing data warrants the development of similar practical tools to exploit its full potential in clinical microbiology and infectious diseases.
The ssGeneFinder software and run data are available from http://147.8.74.24/tom/ssGeneFinder. The software is supported on Windows, Mac OS, and Linux.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 31 August 2011.
REFERENCES
- 1. Bielaszewska M., et al. 22 June 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. [Epub ahead of print.] doi:10.1016/S1473-3099(11)70165-7 [DOI] [PubMed] [Google Scholar]
- 2. Bielaszewska M., Tarr P. I., Karch H., Zhang W., Mathys W. 2005. Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates. J. Clin. Microbiol. 43:452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bielaszewska M., Zhang W., Tarr P. I., Sonntag A. K., Karch H. 2005. Molecular profiling and phenotype analysis of Escherichia coli O26:H11 and O26:NM: secular and geographic consistency of enterohemorrhagic and enteropathogenic isolates. J. Clin. Microbiol. 43:4225–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brzuszkiewicz E., et al. 29 June 2011. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: entero-aggregative-haemorrhagic Escherichia coli (EAHEC). Arch. Microbiol. [Epub ahead of print.] doi:10.1007/s00203-011-0725-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cui Y., et al. 13 July 2011. Identification of the hybrid strain responsible for Germany food-poisoning outbreak by polymerase chain reaction. J. Clin. Microbiol. [Epub ahead of print.] doi:10.1128/JCM.01312-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ho C.-C., et al. 2011. Novel pan-genomic analysis approach in target selection for multiplex PCR identification and detection of Burkholderia pseudomallei, Burkholderia thailandensis, and Burkholderia cepacia complex species: a proof-of-concept study. J. Clin. Microbiol. 49:814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.