Abstract
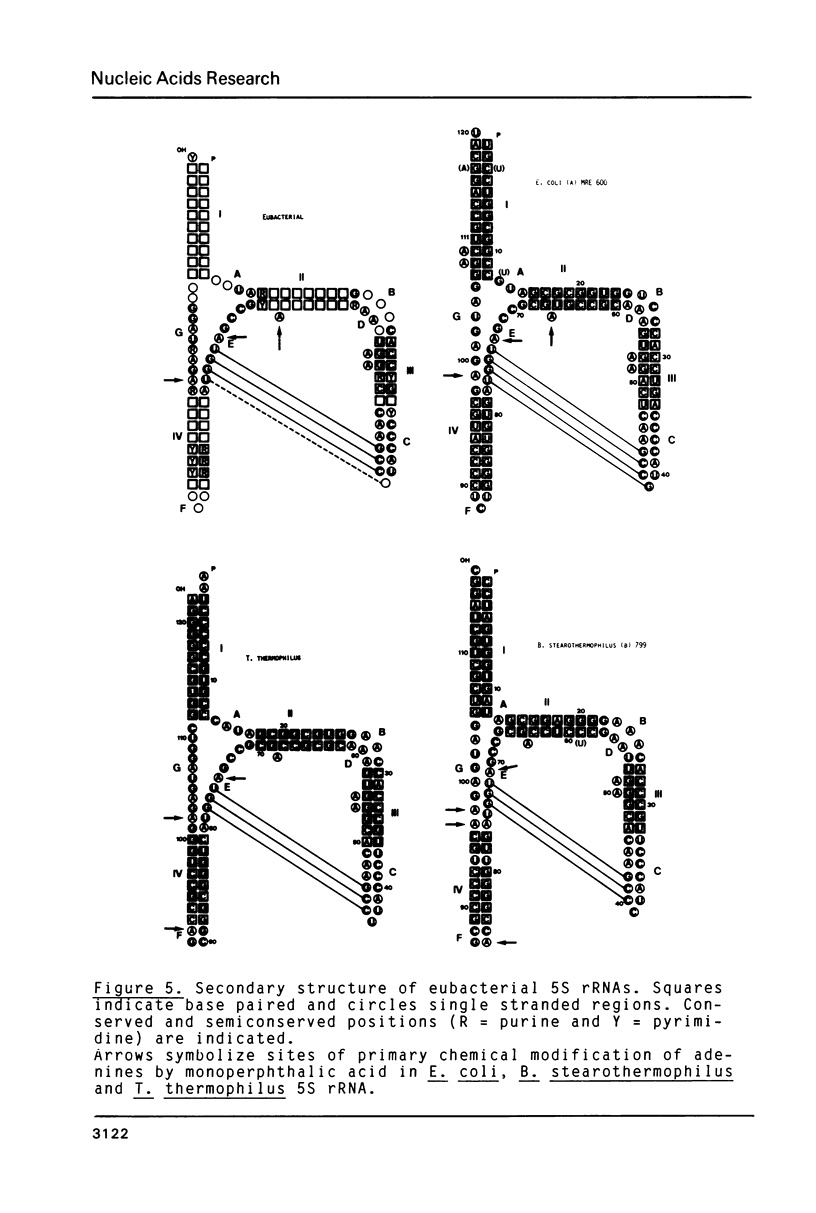
Adenines in free 5S rRNA from Escherichia coli, Bacillus stearothermophilus and Thermus thermophilus have been oxidized at their N-1 position using monoperphthalic acid. The determination of the number of adenine 1-N-oxides was on the basis of UV spectroscopic data of the intact molecule. Identification of the most readily accessible nucleotides by sequencing gel analysis reveals that they are located in conserved positions within loops, exposed hairpin loops and single-base bulge loops. Implications for the structure and function of 5S rRNA will be discussed on the basis of this comparative analysis.
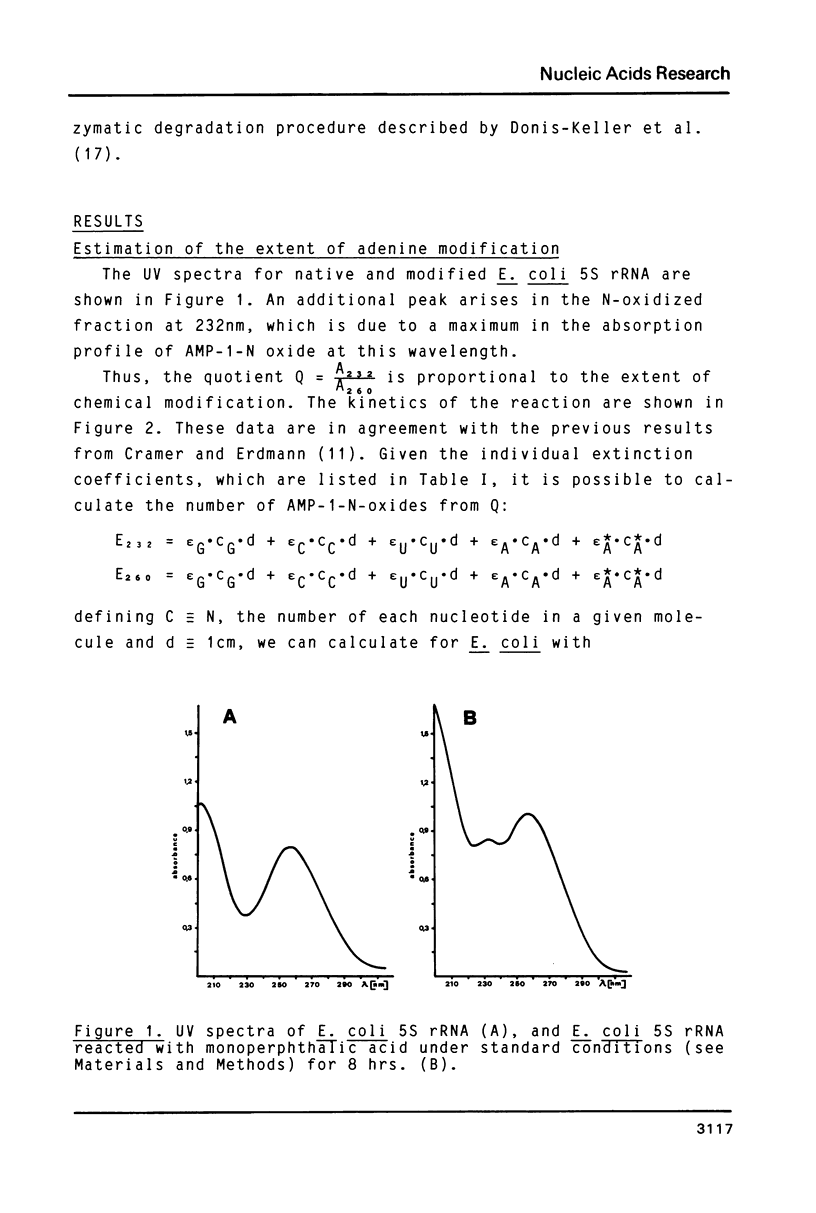
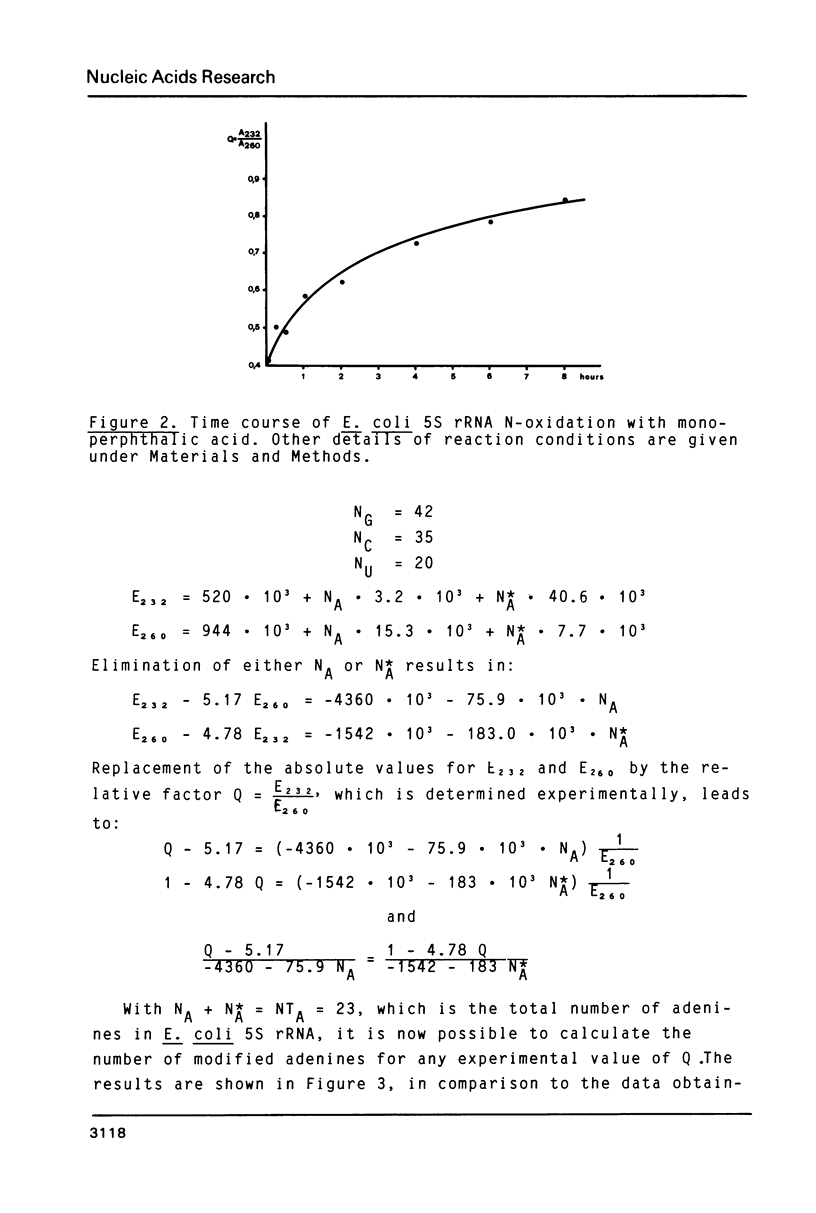
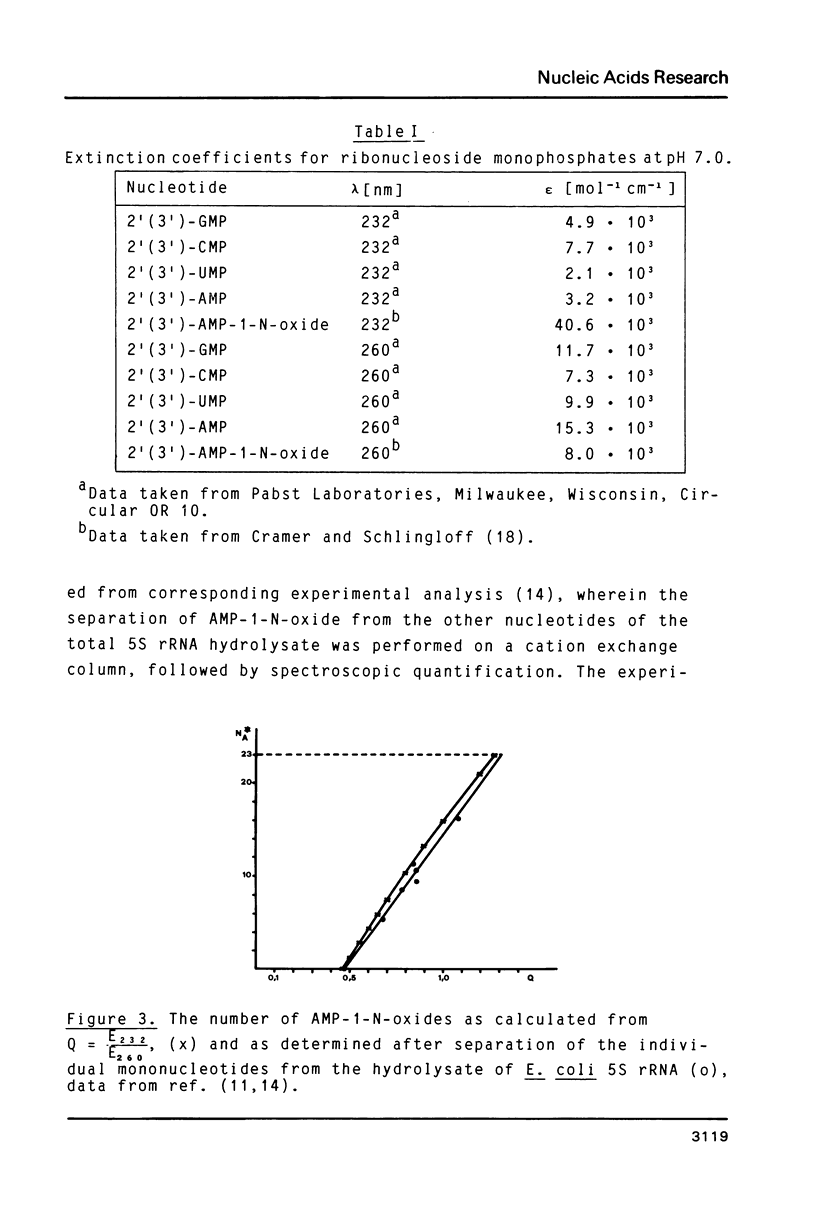
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aubert M., Scott J. F., Reynier M., Monier R. Rearrangement of the conformation of Escherichia coli 5S RNA. Proc Natl Acad Sci U S A. 1968 Sep;61(1):292–299. doi: 10.1073/pnas.61.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm S., Fabian H., Venyaminov SYu, Matveev S. V., Lucius H., Welfle H., Filimonov V. V. Structural analysis of the A and B conformers of Escherichia coli 5 S ribosomal RNA by infrared spectroscopy. FEBS Lett. 1981 Sep 28;132(2):357–361. doi: 10.1016/0014-5793(81)81197-0. [DOI] [PubMed] [Google Scholar]
- Cramer F., Erdmann V. A. Amount of adenine and uracil base pairs in E. coli 23S, 16S and 5S ribosomal RNA. Nature. 1968 Apr 6;218(5136):92–93. doi: 10.1038/218092a0. [DOI] [PubMed] [Google Scholar]
- De Wachter R., Chen M. W., Vandenberghe A. Conservation of secondary structure in 5 S ribosomal RNA: a uniform model for eukaryotic, eubacterial, archaebacterial and organelle sequences is energetically favourable. Biochimie. 1982 May;64(5):311–329. doi: 10.1016/s0300-9084(82)80436-7. [DOI] [PubMed] [Google Scholar]
- Delihas N., Andersen J. Generalized structures of the 5S ribosomal RNAs. Nucleic Acids Res. 1982 Nov 25;10(22):7323–7344. doi: 10.1093/nar/10.22.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A., Doberer H. G. Structure and function of 5S RNA: the role of the 3' terminus in 5S RNA function. Mol Gen Genet. 1972;114(2):89–94. doi: 10.1007/BF00332779. [DOI] [PubMed] [Google Scholar]
- Farber N. M., Cantor C. R. A slow tritium exchange study of the solution structure of Escherichia coli 5 S ribosomal RNA. J Mol Biol. 1981 Feb 25;146(2):223–239. doi: 10.1016/0022-2836(81)90433-2. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Hancock J., Wagner R. A structural model of 5S RNA from E. coli based on intramolecular crosslinking evidence. Nucleic Acids Res. 1982 Feb 25;10(4):1257–1269. doi: 10.1093/nar/10.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao T. H., Crothers D. M. A proton-coupled conformational switch of Escherichia coli 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3360–3364. doi: 10.1073/pnas.77.6.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kime M. J., Moore P. B. NMR evidence for the existence of two native conformations of 5S RNA. Nucleic Acids Res. 1982 Aug 25;10(16):4973–4983. doi: 10.1093/nar/10.16.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K. R., Fox G. E. Secondary structure of eukaryotic cytoplasmic 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2150–2154. doi: 10.1073/pnas.78.4.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Douthwaite S., Garrett R. A., Noller H. F. A "bulged" double helix in a RNA-protein contact site. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7331–7335. doi: 10.1073/pnas.78.12.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieler T., Erdmann V. A. Three-dimensional structural model of eubacterial 5S RNA that has functional implications. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4599–4603. doi: 10.1073/pnas.79.15.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin D., Kao T., Crothers D. M. A characterization of the low temperature structural transition of Escherichia coli 5 S RNA by partial enzymatic digestion. J Biol Chem. 1983 Sep 25;258(18):10813–10816. [PubMed] [Google Scholar]
- Silberklang M., RajBhandary U. L., Lück A., Erdmann V. A. Chemical reactivity of E. coli 5S RNA in situ in the 50S ribosomal subunit. Nucleic Acids Res. 1983 Feb 11;11(3):605–617. doi: 10.1093/nar/11.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Luehrsen K. R., Woese C. R., Pace N. R. An unusual 5S rRNA, from Sulfolobus acidocaldarius, and its implications for a general 5S rRNA structure. Nucleic Acids Res. 1981 Nov 25;9(22):6129–6137. doi: 10.1093/nar/9.22.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studnicka G. M., Eiserling F. A., Lake J. A. A unique secondary folding pattern for 5S RNA corresponds to the lowest energy homologous secondary structure in 17 different prokaryotes. Nucleic Acids Res. 1981 Apr 24;9(8):1885–1904. doi: 10.1093/nar/9.8.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]



