Abstract
Cells of the gliding bacterium Flavobacterium johnsoniae move rapidly over surfaces. Transposon mutagenesis was used to identify sprE, which is involved in gliding. Mutations in sprE resulted in the formation of nonspreading colonies on agar. sprE mutant cells in wet mounts were almost completely deficient in attachment to and movement on glass, but a small percentage of cells exhibited slight movements, indicating that the motility machinery was not completely disrupted. SprE is a predicted lipoprotein with a tetratricopeptide repeat domain. SprE is similar in sequence to Porphyromonas gingivalis PorW, which is required for secretion of gingipain protease virulence factors. Disruption of F. johnsoniae sprE resulted in decreased extracellular chitinase activity and decreased secretion of the cell surface motility protein SprB. Reduced secretion of cell surface components of the gliding machinery, such as SprB, may account for the defects in gliding. Orthologs of sprE are found in many gliding and nongliding members of the phylum Bacteroidetes, suggesting that similar protein secretion systems are common among members of this large and diverse group of bacteria.
INTRODUCTION
Cells of Flavobacterium johnsoniae cells crawl over surfaces in a process called gliding motility. This type of motility is common among members of the phylum Bacteroidetes, of which F. johnsoniae is a member. Cells of F. johnsoniae move at speeds of approximately 2 μm/s over wet glass surfaces. As a result of these cell movements, colonies of F. johnsoniae have thin spreading edges.
Gliding of F. johnsoniae appears to be mediated by the rapid movement of adhesins, such as SprB, along the cell surface (15, 32). GldK, GldL, GldM, GldN, SprA, and SprT are required for secretion of SprB to the cell surface (38, 41). They also appear to be involved in secretion of extracellular chitinase (38, 41). GldK, GldL, GldM, GldN, SprA, and SprT are similar in sequence to proteins of Porphyromonas gingivalis that are involved in secretion of gingipain proteases, which are virulence factors of this periodontal pathogen (40–42). The P. gingivalis proteins appear to form a complex, referred to as the Por secretion system (PorSS). Known proteins secreted by the P. gingivalis and F. johnsoniae PorSSs have N-terminal signal peptides that are typical of proteins targeted to the Sec system. It is thought that these proteins are transported across the cytoplasmic membrane by the Sec system and utilize the PorSS for transit of the outer membrane (38, 41). The PorSS is not closely related to other bacterial protein secretion systems, such as the type I to VI secretion systems (7, 10). Other proteins required for F. johnsoniae gliding that are not thought to be part of the PorSS include GldA, GldB, GldD, GldF, GldG, GldH, GldI, and GldJ (1, 5, 11–13, 26, 27). Some of these proteins may comprise the “motor” that propels SprB along the cell surface. The physical relationship of this motor to the PorSS is not known. They may be intimately associated, as is the case for swimming bacteria, where a type III protein secretion system is an integral part of the bacterial flagellum and is involved in its assembly (16). Gliding motility is not unique to members of the phylum Bacteroidetes and is observed in many bacteria outside this phylum (24). Gliding of Myxococcus xanthus and of Mycoplasma species has been studied at the molecular level (20, 23, 29, 43–45). These bacteria lack orthologs to most of the proteins that are required for F. johnsoniae gliding, and the mechanism of cell movement is thought to be different for each of these organisms (4, 15).
Previously, 294 Tn4351-, HimarEm1-, and HimarEm2-induced mutants of F. johnsoniae that formed nonspreading colonies were isolated (4, 11–13). The mutants were screened for motility defects in wet mounts and were divided into 3 groups. Nineteen of the mutants exhibited cell division defects resulting in the production of filamentous cells that were completely nonmotile. Several of these had insertions in ftsX (18), and the rest had insertions in other genes predicted to be involved in cell division (unpublished results). Another 51 mutants were completely nonmotile and had normal cell morphology. These mutants each had mutations in gld genes required for motility (1, 4, 5, 11–13, 26, 27). The remaining 224 mutants formed nonspreading colonies but retained some ability to move in wet mounts. The sites of transposon insertions were determined for 32 of these motile nonspreading mutants that were randomly selected for analysis. Six of the mutants had insertions in sprA (33), 2 had insertions in secDF (31), 16 had insertions in sprB (32), 2 had insertions in sprC (36), and 1 had an insertion in sprD (36), as previously described. Two of the remaining mutants (CJ101-285 and FJ140) had insertions in fjoh_2111, which is predicted to encode the lipoprotein localization protein LolA (M. McBride, unpublished results). GldB, GldD, GldH, GldI, GldJ, and GldK are lipoproteins (4, 5, 26, 27), which may account for the motility defects exhibited by lolA mutants. In this study, the remaining three mutants were analyzed, resulting in the identification of sprE. Cells with mutations in sprE formed nonspreading colonies and were severely defective in gliding. SprE is similar in sequence to P. gingivalis PorSS protein PorW. F. johnsoniae sprE mutants were defective in extracellular chitinase and in localization of SprB to the cell surface, suggesting that SprE is a component of the F. johnsoniae PorSS.
MATERIALS AND METHODS
Bacterial and bacteriophage strains, plasmids, and growth conditions.
F. johnsoniae strains MM101 and FJ1, which are direct descendants of the F. johnsoniae type strain ATCC 17061, were the wild-type strains used in this study (26). F. johnsoniae strains MM101 and FJ1 are essentially identical, except that strain MM101 has a partial defect in chitin utilization. F. johnsoniae FJ114 and FJ156 (32, 36) were the sprB mutant strains used. F. johnsoniae strains were grown in Casitone-yeast extract (CYE) medium at 30°C, as previously described (25). To observe colony spreading, F. johnsoniae was grown on PY2 agar medium (1) at 25°C. MM medium was used to observe movement of individual cells in wet mounts (21). The F. johnsoniae bacteriophages used in this study were φCj1, φCj13, φCj23, φCj28, φCj29, φCj42, φCj48, and φCj54 (6, 35, 46). Sensitivity to F. johnsoniae bacteriophages was determined as previously described by spotting 5 μl of phage lysates (109 PFU/ml) onto lawns of cells in CYE overlay agar (38). Escherichia coli strains were grown in Luria-Bertani medium at 37°C. The following antibiotics were used at the indicated concentrations when needed: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; erythromycin, 100 μg/ml; kanamycin, 35 μg/ml; and tetracycline, 20 μg/ml.
Transposon mutagenesis and identification of sites of insertion.
Tn4351 and HimarEm2 were introduced into wild-type F. johnsoniae by conjugation from E. coli as previously described (4, 11, 13). Mutants were selected by plating cells on PY2 agar containing erythromycin, and nonspreading colonies were isolated. Chromosomal DNA was isolated from mutants, and sites of transposon insertion were determined as previously described (4, 11, 18).
Sequence analysis.
Sequences were analyzed with MacVector software (Cary, NC), and comparisons to database sequences were made using the BLAST algorithm (2). Predictions regarding cellular localization were made using the PSORTb (8) TMpredict (9), and CELLO (47) tools, and tetratricopeptide repeat (TPR) domains were predicted using the TPRpred tool (17). The phylogenetic distribution of sprE orthologs was determined using the U.S. Department of Energy Joint Genome Institute Integrated Microbial Genomes (IMG; version 3.3) gene profile tools (22) and by BLASTP analyses of completely sequenced microbial genomes available via NCBI (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi).
Cloning of sprE.
A 2.92-kb region of F. johnsoniae DNA which spans sprE was amplified using Fideli Taq DNA polymerase (USB Corp.) and primers 712 (5′ TTTGCCGGATCCGACCCAACAACAGTAAGCCG 3′; the BamHI site is underlined) and 713 (5′ CACTTTGTCGACCATTATATTATGGGTTTTTTGGGG 3′; the SalI site is underlined). This fragment was digested with SalI and BamHI and ligated into pBCSK+ (Stratagene) that had been cut with the same enzymes to generate pNap1. pNap1 was digested with XbaI and KpnI, and the fragment containing sprE was inserted into pCP23 (1), which had been digested with the same enzymes to generate pNap2.
Protein expression and antibody production.
A 2,568-bp fragment encoding the C-terminal 825 amino acids of SprE was amplified using ExTaq (Takara Bio Inc., Otsu, Japan) and primers 921 (5′ GCTAGGGATCCATGGAGGTCTTGGACTTGAG 3′; the BamHI site is underlined) and 923 (5′ GCTAGGTCGACGTCTCCTACAATAGAGGTTCC 3′; the SalI site is underlined). The PCR product was digested with BamHI and SalI and cloned into pET30-C that had been digested with the same enzymes, generating pEVG11D. pEVG11D was introduced into E. coli Rosetta 2(DE3) cells (Novagen, Madison, WI), which expressed seven rare tRNAs required for the efficient expression of SprE. To isolate recombinant SprE, cells were grown to mid-log phase at 37°C in rich medium plus glucose (10 g tryptone, 5 g yeast extract, 5 g NaCl, 2 g glucose/liter), induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and incubated for 4 h at 37°C. Cells were disrupted using a French press, and inclusion bodies containing recombinant SprE were collected by centrifugation at 6,000 × g for 15 min at 4°C. The inclusions were boiled in SDS-PAGE loading buffer, and SprE was visualized by CuCl2 staining, after separation on a 10% acrylamide gel by SDS-PAGE (19). The SprE band was cut from the gel and destained in 0.25 M Tris (pH 9.0), 0.25 M EDTA, and the protein was electroeluted at 60 mA for 5 h into 25 mM Tris, 192 mM glycine, 0.1% SDS using a model 422 Electro-Eluter (Bio-Rad). Polyclonal antibodies against recombinant SprE were produced and affinity purified using the recombinant protein by Proteintech Group, Inc. (Chicago, IL).
Cell fractionation and Western blot analysis.
F. johnsoniae cells were grown to mid-log phase in MM medium at 25°C without shaking. Cells were disrupted using a French press and fractionated into soluble and membrane fractions as previously described by centrifugation at 352,900 × g for 30 min (13). When whole cells were analyzed, they were pelleted at 4,000 × g, resuspended in SDS-PAGE loading buffer, and boiled for 5 min. Proteins (50 μg) were separated by SDS-PAGE, and Western blot analyses were performed as previously described (38) using crude antisera against SprB and SprE at 1:1,000 dilutions.
Microscopic observations of cell attachment to glass and of gliding motility.
Attachment of wild-type and mutant cells to glass was measured as previously described (33). In brief, cells were grown in MM medium without agitation overnight at 25°C to a density of 5 × 108 cells/ml. Cells (2.5 μl) were added to a Petroff-Hausser counting chamber, covered with a glass coverslip, and allowed to incubate for 2 min at 25°C. The number of cells attached to 12 randomly selected 0.03-mm2 regions of the glass coverslip was determined. Wild-type and mutant cells were examined for movement on glass by phase-contrast microscopy at 25°C essentially as previously described (32), except that standard glass slides and coverslips were used instead of Palmer cells.
Binding and movement of anti-SprB-coated polystyrene spheres.
Purified antibody against SprB (1 μl of a 1:10 dilution), 0.5-μm-diameter protein G-coated polystyrene spheres (1 μl of a 0.1% stock; Spherotech Inc., Libertyville, IL), and bovine serum albumin (BSA; 1 μl of a 1% solution) were added to 7 μl of cells (approximately 5 × 108 cells per ml) in MM medium. The cell mixture was spotted on a glass slide and covered with a glass coverslip, and images were recorded and analyzed using MetaMorph software as previously described (32).
Immunofluorescent localization of SprB.
Immunofluorescence was used to visualize SprB on the surface of formaldehyde-fixed cells as previously described (38), except that cells were collected on white 0.8-μm-pore-size Isopore membrane filters (Millipore, Billerica, MA). In brief, fixed cells were exposed to purified anti-SprB, followed by the F(ab′) fragment of goat anti-rabbit IgG conjugated to Alexa-488 (Invitrogen). InSpeck relative intensity fluorescent beads (Invitrogen-Molecular Probes, Eugene, OR) were added as controls. The filters were mounted on a glass slide with VectaShield with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Inc., Burlingame, CA), and samples were observed using a Nikon Eclipse 50i microscope. Images were captured with a Photometrics CoolSNAPES camera with exposure times of 100 to 200 ms (DAPI) and 300 ms (Alexa-488).
Chitin utilization.
Chitin utilization on agar was observed as previously described (38). Chitinase activities in culture medium and in cell extracts were measured as previously described (38) using the synthetic substrate 4-methylumbelliferyl β-d-N,N′-diacetyl-chitobioside [4-MU-(GlcNAc)2; obtained from Sigma-Aldrich (St. Louis, MO)]. Activities in the cell-free supernatants (secreted chitinase) and in cell extracts are indicated as pmol 4-methylumbelliferone released per μg total protein in the original cell suspension. Protein concentrations were determined by the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Waltham, MA).
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in the GenBank database (accession no. EF111025).
RESULTS
Identification and analysis of sprE.
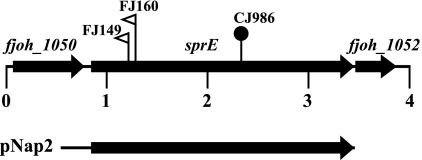
Analysis of Tn4351- and HimarEm2-induced mutants that formed nonspreading colonies composed of cells that retained some ability to move resulted in the identification of three mutants (FJ149, FJ160, and CJ986) that had insertions in fjoh_1051, which we named sprE (Fig. 1). sprE encodes a predicted primary product of 870 amino acids. The amino-terminal sequence contains a hydrophobic region terminated by a cysteine, which is characteristic of bacterial lipoproteins. The region between amino acids 119 and 379 displays six tetratricopeptide repeats (TPRs) as determined by TPRpred (17). TPR domains are degenerate 34-amino-acid repeated sequences found in proteins from many organisms and are often involved in protein-protein interactions (3). SprE is similar in sequence to P. gingivalis PorW, which is required for secretion of gingipain proteases (41). F. johnsoniae SprE and P. gingivalis PorW exhibit 22% amino acid identity and 42% similarity over the entire length of SprE (BLASTP E value of 4 × 10−39). P. gingivalis PorW is 290 amino acids longer than SprE, as a result of a C-terminal extension that is not found on the F. johnsoniae protein. fjoh_1050, which lies upstream of sprE, encodes a predicted ATP-binding-cassette transporter, and fjoh_1052, which is downstream of sprE, encodes a protein of unknown function. There is no evidence linking fjoh_1050 and fjoh_1052 to motility, protein secretion, or SprE function.
Fig. 1.
Map of the sprE region. Numbers below the map refer to kilobase pairs of sequence. The sites of HimarEm2 and Tn4351 insertions are indicated by open triangles and closed circles, respectively. The orientations of HimarEm2 insertions are indicated by the direction in which the triangles are pointing. As drawn, FJ149 and FJ160 both have inverted repeat IR1 on the right side of the transposons. The sprE mutants FJ149 and FJ160 were derived from wild-type strain FJ1, and the sprE mutant CJ986 was derived from wild-type strain MM101. The region of DNA carried by the complementing plasmid pNap2 is indicated beneath the map.
Mutations in sprE result in defects in attachment and motility.
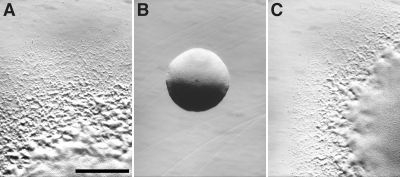
Cells of sprE mutants formed completely nonspreading colonies on agar, whereas wild-type cells formed colonies with thin spreading edges (Fig. 2). Introduction of sprE on pNap2 restored the ability to form spreading colonies. Wild-type and sprE mutant cells of F. johnsoniae were examined for attachment to and movement on glass, as previously described (33). Wild-type cells of F. johnsoniae readily attached to glass (Table 1) and glided at speeds of 2 μm/s (see Movie S1 in the supplemental material). Under these conditions, greater than 95% of the cells that attached to the glass exhibited gliding movements over a 2-min period. Cells of sprE mutants attached poorly to glass (Table 1; see Movie S1 in the supplemental material). The few cells that did attach occasionally exhibited brief gliding movements that were similar to those exhibited by wild-type cells (see Movie S2 in the supplemental material), indicating that unlike completely nonmotile gld mutants, cells of sprE mutants retain some limited ability to move. Complementation with pNap2 restored the ability to attach to and move efficiently on glass surfaces. The attachment and motility defects exhibited by cells of sprE mutants were similar to those observed for cells of sprA mutants (33). In contrast, cells of sprB mutants attached well to glass (Table 1), and most cells exhibited limited movements (32).
Fig. 2.
sprE is required for the formation of spreading colonies. Colonies were grown for 48 h at 25°C on PY2 agar medium. Photomicrographs were taken with a Photometrics CoolSNAPcf2 camera mounted on an Olympus IMT-2 phase-contrast microscope. (A) Wild-type F. johnsoniae FJ1; (B) sprE mutant FJ149; (C) FJ149 complemented with pNap2. Cells in panels A and B carried control vector pCP23. Bar in panel A, 1 mm (the bar applies to all panels).
Table 1.
Effect of mutations in sprE on attachment of cells to glass coverslips
| Strain | Description | Avg no. of cells attached to 0.03-mm2 region of glass coverslipa |
|---|---|---|
| FJ1 with control plasmid pCP23 | Wild type | 52.9 (11.6) |
| FJ114 with pCP23 | sprB mutant | 44.4 (14.8) |
| FJ149 with pCP23 | sprE mutant | 2.0 (1.2) |
| FJ149 with pNap2 | Complemented sprE mutant | 58.3 (12.7) |
Approximately 106 cells in 2.5 μl of MM medium were introduced into a Petroff-Hausser counting chamber and incubated for 2 min at 25°C. Samples were observed using an Olympus BH-2 phase-contrast microscope, and cells attached to a 0.03-mm2 region of the cover glass were counted. Numbers in parentheses are standard deviations calculated from 12 measurements.
SprB is improperly localized in cells of sprE mutants.
Components of the F. johnsoniae PorSS such as GldN and SprT are required for surface localization of SprB (38, 41). Since SprE is similar in sequence to the P. gingivalis PorSS protein PorW, we examined the effect of mutations in sprE on surface localization of SprB. Antibodies were used to detect SprB on wild-type cells and on cells of sprE mutants. Each of these strains produced SprB protein (see Fig. S1 in the supplemental material). Protein G-coated polystyrene latex spheres and antibodies against SprB were used to detect surface-exposed SprB on live cells and to observe movement of SprB protein along the cell surface (Table 2; see Movie S3 in the supplemental material). As previously reported (32), antibody-coated spheres attached specifically to wild-type cells expressing SprB and were rapidly propelled along the surfaces of the cells. Also as previously reported (32), such spheres failed to attach to sprB mutant cells, and protein G-coated spheres without antibodies failed to bind to wild-type cells. Antibody-coated spheres failed to bind to cells of the sprE mutants (Table 2; see Movie S3 in the supplemental material), indicating that SprB was not exposed on the surface of these cells. Complementation of the mutants with pNap2, which carries sprE, restored surface localization of SprB.
Table 2.
Effect of mutations in sprE on binding of protein G-coated polystyrene spheres carrying antibodies against SprB
| Strain | Description | Antibody added | Avg % of cells with spheres attacheda |
|---|---|---|---|
| FJ1 + control plasmid pCP23 | Wild type | No antibody | 0.3 (0.6) |
| FJ1 + control plasmid pCP23 | Wild type | Anti-SprB | 72.3 (7.6) |
| FJ114 + pCP23 | sprB mutant | Anti-SprB | 2.0 (1.0) |
| FJ149 + pCP23 | sprE mutant | Anti-SprB | 0.7 (0.6) |
| FJ149 + pNap2 | sprE mutant complemented with pNap2 | Anti-SprB | 67.7 (3.2) |
Affinity-purified anti-SprB- and 0.5-μm-diameter protein G-coated polystyrene spheres were added to cells as described in Materials and Methods. Samples were spotted on a glass slide, covered with a glass coverslip, incubated for 1 min at 25°C, and examined using a phase-contrast microscope. Images were recorded for 30 s, and 100 randomly selected cells were examined for the presence of attached spheres during this period. Numbers in parentheses are standard deviations calculated from three measurements.
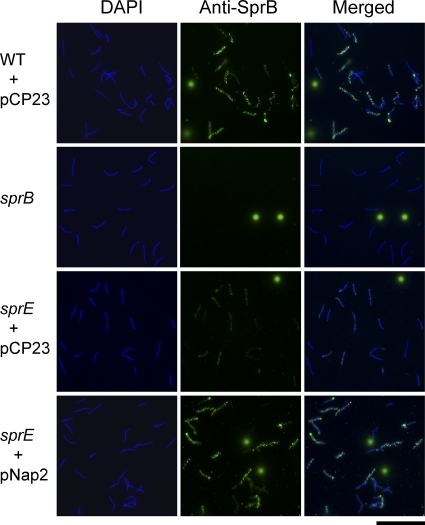
Analysis of surface exposure of SprB by immunofluorescence microscopy of fixed cells confirmed that sprE mutant cells are defective in surface localization of SprB (Fig. 3). However, small amounts of SprB were detected on cells of the sprE mutants by immunofluorescence microscopy. This may be the result of damage to cells during fixation and staining or could indicate that the immunofluorescence assay is more sensitive than the antibody-coated sphere assay and that the sprE mutant cells are only partially defective in secretion of SprB. In any case, the decreased levels of SprB and perhaps of other motility proteins on the cell surface may explain some of the motility defects exhibited by cells of sprE mutants.
Fig. 3.
Detection of surface-localized SprB protein by immunofluorescence microscopy. Cells of wild-type and mutant F. johnsoniae were exposed to DAPI and to anti-SprB antibodies, followed by secondary antibodies conjugated to Alexa-488. The fluorescent spheres are InSpeck relative intensity control fluorescence beads. WT, wild-type F. johnsoniae FJ1; sprB, sprB mutant FJ156; sprE, sprE mutant FJ149. pCP23 is a control vector; pNap2 carries sprE. Bar, 20 μm.
Cells of sprE mutants are defective in chitin utilization.
F. johnsoniae produces a suite of enzymes for chitin utilization, including at least one extracellular chitinase, encoded by fjoh_4555, and several cell-bound enzymes (28, 41). Mutations in F. johnsoniae porSS genes, such as gldN and sprT, result in defects in chitin utilization and defects in secretion of extracellular chitinase (38, 41). Cells of the sprE mutant FJ149 were deficient in chitin utilization, and complementation with pNap2 restored the ability to digest chitin to wild-type levels (see Fig. S2 in the supplemental material). The defect in chitin utilization appears to be a result of a deficiency in extracellular chitinase activity (Table 3). These results suggest that SprE is involved in secretion not only of SprB but also of extracellular chitinase by the F. johnsoniae PorSS.
Table 3.
Effect of mutations in sprE on chitinase activity
| Strain | Chitinase activitya |
|
|---|---|---|
| Extracellular | In cell extracts | |
| FJ1 (wild type)b | 223 (54) | 241 (32) |
| FJ149 (sprE mutant)b | 0 (0) | 272 (51) |
| FJ149 complemented with pNap2 | 234 (86) | 279 (50) |
Chitinase activities were determined using the synthetic substrate 4-MU-(GlcNAc)2. Equal amounts of each sample, based on the protein content of the original cell suspension, were incubated with 10 nmol of 4-MU-(GlcNAc)2 for 4 h at 37°C, and the amount of 4-MU released was determined by measuring fluorescence emission at 460 nm following excitation at 360 nm. Activities in the cell-free supernatants (extracellular chitinase) and in cell extracts are indicated as pmol 4-methylumbelliferone released per μg total protein in the original cell suspension. Numbers in parentheses are standard errors calculated from three measurements.
Strains without pNap2 carried control vector pCP23 so that all cultures could be grown under identical conditions in the presence of tetracycline.
Bacteriophage resistance of sprE mutants.
Sensitivity to F. johnsoniae bacteriophages was determined as previously described (38) by spotting phage lysates onto lawns of cells. Wild-type cells were lysed by all of the bacteriophages, whereas cells of sprE mutants exhibited complete resistance to four of the bacteriophages (φCj1, φCj13, φCj23, and φCj29) and partial resistance to three others (φCj42, φCj48, and φCj54) (see Fig. S3 in the supplemental material). Introduction of sprE on pNap2 restored phage sensitivity. The cell surface motility protein SprB is thought to function as a receptor for φCj1, φCj13, φCj23, and φCj29 (32), and additional SprB-like motility proteins may function as receptors for other phages (38). Mutations in sprE result in defects in secretion of SprB and perhaps of other cell surface proteins, which may explain the phage resistance exhibited by sprE mutants.
Identification and localization of SprE.
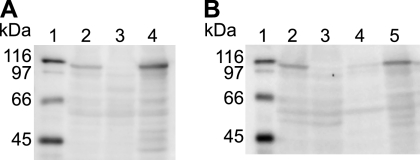
Recombinant SprE was produced in E. coli, and polyclonal antiserum was raised against it. The antiserum was used to detect SprE in cell extracts. SprE migrated with an apparent molecular mass of approximately 105 kDa, which was slightly larger than the predicted size of 98.7 kDa for the processed protein after removal of the signal peptide (Fig. 4A). SprE is a predicted lipoprotein, and the covalently attached fatty acids may account for the apparent increase in molecular mass. SprE was detected in extracts of wild-type cells but was absent from extracts of sprE mutants. Introduction of sprE on pNap2 restored production of SprE. SprE was found in the particulate (membrane) fraction of cell extracts (Fig. 4B), as expected for a lipoprotein.
Fig. 4.
Immunodetection of SprE. (A) Whole-cell extracts were examined for SprE by Western blot analysis. Lane 1, molecular mass markers; lane 2, wild-type F. johnsoniae FJ1 with control vector pCP23; lane 3, sprE mutant FJ149 with pCP23; lane 4, FJ149 complemented with pNap2. Fifty micrograms protein was loaded in each lane. (B) Cell fractions were examined for SprE by Western blot analysis. Lane 1, molecular mass markers; lane 2, whole cells of wild-type F. johnsoniae FJ1; lane 3, whole cells of sprE mutant FJ149; lane 4, wild-type F. johnsoniae FJ1, soluble fraction; lane 5, wild-type F. johnsoniae FJ1, membrane fraction. Equal amounts of each fraction based on the starting material were loaded in each lane. All strains in panel B carried empty control vector pCP23, which had no effect on expression of SprE.
DISCUSSION
Gram-negative bacteria have evolved many different machines to transport proteins across biological membranes. The sec and tat systems transport proteins into or across the cytoplasmic membrane (30), and a wide variety of protein secretion systems facilitate movement of proteins across the outer membrane. These include the type I to type VI protein secretion systems, as well as systems that have not been codified with a numeric type, such as the chaperone-usher pathway (7, 10). Some of these rely on the sec or tat systems for delivery of proteins across the cytoplasmic membrane, whereas others secrete proteins across both membranes. Recently, an apparently novel protein secretion system, the PorSS, was identified in two distantly related members of the phylum Bacteroidetes, F. johnsoniae and P. gingivalis (41). GldK, GldL, GldM, GldN, SprA, and SprT are components of the F. johnsoniae PorSS. Disruption of the genes encoding any of these proteins results in defects in secretion of the cell surface motility protein SprB and of extracellular chitinase. SprF is also required for secretion of SprB, but not of chitinase (36). SprF may function as an adapter to the PorSS to specifically allow secretion of SprB. The results presented in this paper suggest that F. johnsoniae SprE is involved in secretion of SprB and chitinase and thus may be another component of its PorSS. P. gingivalis has orthologs to each of the F. johnsoniae PorSS proteins mentioned above, and these proteins are required for gingipain secretion (40, 41). Additional P. gingivalis proteins that appear to be required for gingipain secretion and that may be part of its PorSS include PorU, PG0027, and PG0534 (14, 39, 41). F. johnsoniae has orthologs to the genes encoding each of these proteins, but their roles in protein secretion have not been examined.
Disruption of F. johnsoniae sprE results in decreased attachment to glass, severe defects in gliding, an inability to digest chitin, and resistance to some bacteriophages. Each of these defects can be explained by disruption of a protein secretion system that transports the motility protein SprB, related SprB-like adhesins, and chitinase. The decreased attachment to glass suggests that cell surface adhesins in addition to SprB may be mislocalized in sprE mutants, since sprB mutants do not exhibit an attachment defect to glass (Table 1) (32). Numerous sprB-like genes are present in the genome, and it has recently been suggested that these may encode semiredundant cell surface adhesins involved in motility that may be secreted by the PorSS (36, 37). sprE mutants also display greater phage resistance than sprB mutants. SprB is thought to be a receptor for some phages, such as φCj1, φCj13, φCj23, and φCj29 (32). Other SprB-like proteins secreted by the PorSS may serve as receptors for additional phages. The phenotypes of sprE mutants are similar to those of cells with mutations in genes encoding other components of the PorSS, such as gldK, gldL, gldM, gldN, sprA, and sprT. gldK, gldL, gldM, and gldN are colocalized on the genomes of each of the bacteroidetes that have these genes. This suggests that the proteins encoded by these genes function together. Biochemical evidence for a complex formed by the P. gingivalis orthologs of GldK, GldL, GldM, and GldN has been presented (41). F. johnsoniae SprA, SprE, and SprT may interact with the predicted GldK, GldL, GldM, and GldN complex, but this has not been examined. Disruption of gldK, gldL, and gldM and deletion of the region spanning gldN and its paralog, gldO, each results in complete loss of motility and complete resistance to bacteriophages. In contrast, cells with mutations in sprA, sprE, and sprT each exhibit limited motility of individual cells in wet mounts and exhibit sensitivity to some bacteriophages. GldK, GldL, GldM, and GldN may be core components of the PorSS, whereas SprA, SprE, and SprT may be less crucial, with some limited function occurring in their absence. None of the proteins are similar to proteins of known function, so their exact roles in protein secretion are difficult to predict. SprE contains a TPR domain, and other TPR domain proteins are involved in protein-protein interactions (3). SprE may be involved in protein-protein interactions among PorSS proteins and could be necessary for formation of a stable secretion system complex.
Each of the proteins of the PorSS is predicted to reside in the cell envelope, as expected for components of a protein secretion system. GldL is predicted to be a cytoplasmic membrane protein (4), and its P. gingivalis paralog, PorL, has been demonstrated to localize to the cytoplasmic membrane (41). SprA and SprT are predicted integral outer membrane proteins, and biochemical evidence confirms that F. johnsoniae SprA is partially exposed on the surface of the outer membrane (33) and that the P. gingivalis SprT paralog PorT localizes to the outer membrane (34). SprE and GldK are predicted outer membrane lipoproteins. SprE localizes to the membrane fraction, as expected of a lipoprotein, as does the P. gingivalis GldK paralog PorK (41). A model of the PorSS complex is depicted in Fig. S4 in the supplemental material. Additional experimentation is required to determine the stoichiometry of the secretion system proteins and to elucidate the dynamic interactions between the proteins that result in secretion of SprB, chitinase, and other proteins.
Complete genome sequences are available for 40 species belonging to 29 genera of the phylum Bacteroidetes. sprE orthologs are present in 35 of the 40 species (see Table S1 in the supplemental material). Thirty-two of the 35 sprE ortholog-containing strains also had orthologs for gldK, gldL, gldM, gldN, sprA, and sprT, which encode components of the PorSS. SprE and its orthologs may function with the products of these genes in protein secretion. The five species that lacked sprE orthologs were all closely related members of the genus Bacteroides. These Bacteroides species also lacked orthologs of sprA and sprT, and four of the five lacked orthologs of gldL, gldM, and gldN, suggesting that absence of functional PorSSs may be characteristic of members of the genus. sprE orthologs were not found in organisms outside the phylum Bacteroidetes, although not surprisingly, predicted proteins of unknown function with more limited similarity restricted to the TPR domain of SprE were identified. Orthologs of the other porSS genes were also not found in organisms outside the phylum Bacteroidetes. Each of the 35 bacteroidetes SprE orthologs mentioned above had a cysteine at or near the end of its hydrophobic signal peptide, which is a requirement for bacterial lipoproteins. This suggests that SprE and its orthologs are all lipoproteins and that membrane anchoring of the proteins by their lipid tails is important for function.
The PorSS of P. gingivalis is involved in secretion of gingipain virulence factors, and the PorSS of F. johnsoniae is involved in secretion of chitinase and of the cell surface gliding motility protein SprB. Further study will likely identify additional functions for PorSSs. The ease with which F. johnsoniae can be genetically manipulated makes it uniquely well suited to serve as a model system for studies of the mechanism of PorSS-mediated protein secretion. Given the novelty of the proteins involved, the PorSS is likely to exhibit significant differences in structure and function from other well-studied bacterial protein secretion systems.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants MCB-0641366 and MCB-1021721 from the National Science Foundation.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 22 July 2011.
REFERENCES
- 1. Agarwal S., Hunnicutt D. W., McBride M. J. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl. Acad. Sci. U. S. A. 94:12139–12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Blatch G. L., Lassle M. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932–939 [DOI] [PubMed] [Google Scholar]
- 4. Braun T. F., Khubbar M. K., Saffarini D. A., McBride M. J. 2005. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J. Bacteriol. 187:6943–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braun T. F., McBride M. J. 2005. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J. Bacteriol. 187:2628–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang L. Y. E., Pate J. L., Betzig R. J. 1984. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 159:26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Economou A., et al. 2006. Secretion by numbers: protein traffic in prokaryotes Mol. Microbiol. 62:308–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gardy J. L., et al. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617–623 [DOI] [PubMed] [Google Scholar]
- 9. Hofmann K., Stoffel W. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166 [Google Scholar]
- 10. Holland I. B. 2010. The extraordinary diversity of bacterial protein secretion mechanisms. Methods Mol. Biol. 619:1–20 [DOI] [PubMed] [Google Scholar]
- 11. Hunnicutt D. W., Kempf M. J., McBride M. J. 2002. Mutations in Flavobacterium johnsoniae gldF and gldG disrupt gliding motility and interfere with membrane localization of GldA. J. Bacteriol. 184:2370–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hunnicutt D. W., McBride M. J. 2001. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes gldD and gldE. J. Bacteriol. 183:4167–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hunnicutt D. W., McBride M. J. 2000. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes, gldB and gldC. J. Bacteriol. 182:911–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ishiguro I., Saiki K., Konishi K. 2009. PG27 is a novel membrane protein essential for a Porphyromonas gingivalis protease secretion system. FEMS Microbiol. Lett. 292:261–267 [DOI] [PubMed] [Google Scholar]
- 15. Jarrell K. F., McBride M. J. 2008. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6:466–476 [DOI] [PubMed] [Google Scholar]
- 16. Journet L., Hughes K. T., Cornelis G. R. 2005. Type III secretion: a secretory pathway serving both motility and virulence (review). Mol. Membr. Biol. 22:41–50 [DOI] [PubMed] [Google Scholar]
- 17. Karpenahall M. R., Lupas A. N., Söding J. 2007. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kempf M. J., McBride M. J. 2000. Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J. Bacteriol. 182:1671–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee C., Levin A., Branton D. 1987. Copper staining: a five minute protein stain for sodium dodecyl sulfate-polyacrylamide gels. Anal. Biochem. 166:308–312 [DOI] [PubMed] [Google Scholar]
- 20. Leonardy S., Bulyha I., Søgaard-Andersen L. 2008. Reversing cells and oscillating motility proteins. Mol. Biosyst. 4:1009–1014 [DOI] [PubMed] [Google Scholar]
- 21. Liu J., McBride M. J., Subramaniam S. 2007. Cell surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J. Bacteriol. 189:7503–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Markowitz V. M., et al. 2010. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38:D382–D390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mauriello E. M., Mignot T., Yang Z., Zusman D. R. 2010. Gliding motility revisited: how do the myxobacteria move without flagella? Microbiol. Mol. Biol. Rev. 74:229–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McBride M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49–75 [DOI] [PubMed] [Google Scholar]
- 25. McBride M. J., Kempf M. J. 1996. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 178:583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McBride M. J., Braun T. F. 2004. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J. Bacteriol. 186:2295–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McBride M. J., Braun T. F., Brust J. L. 2003. Flavobacterium johnsoniae GldH is a lipoprotein that is required for gliding motility and chitin utilization. J. Bacteriol. 185:6648–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McBride M. J., et al. 2009. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl. Environ. Microbiol. 75:6864–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miyata M. 2010. Unique centipede mechanism of Mycoplasma gliding. Annu. Rev. Microbiol. 64:519–537 [DOI] [PubMed] [Google Scholar]
- 30. Natale P., Brüser T., Driessen A. J. 2008. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—distinct translocases and mechanisms. Biochim. Biophys. Acta 1778:1735–1756 [DOI] [PubMed] [Google Scholar]
- 31. Nelson S. S., McBride M. J. 2006. Mutations in Flavobacterium johnsoniae secDF result in defects in gliding motility and chitin utilization. J. Bacteriol. 188:348–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nelson S. S., Bollampalli S., McBride M. J. 2008. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J. Bacteriol. 190:2851–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nelson S. S., Glocka P. P., Agarwal S., Grimm D. P., McBride M. J. 2007. Flavobacterium johnsoniae SprA is a cell surface protein involved in gliding motility. J. Bacteriol. 189:7145–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nguyen K. A., et al. 2009. Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiology 155:328–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pate J. L., Petzold S. J., Chang L.-Y. E. 1979. Phages for the gliding bacterium Cytophaga johnsonae that infect only motile cells. Curr. Microbiol. 2:257–262 [Google Scholar]
- 36. Rhodes R. G., Nelson S. S., Pochiraju S., McBride M. J. 2011. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J. Bacteriol. 193:599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rhodes R. G., Pucker H. G., McBride M. J. 2011. Development and use of a gene deletion strategy for Flavobacterium johnsoniae to identify the redundant motility genes remF, remG, remH, and remI. J. Bacteriol. 193:2418–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rhodes R. G., et al. 2010. Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB. J. Bacteriol. 192:1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saiki K., Konishi K. 2010. Identification of a novel Porphyromonas gingivalis outer membrane protein, PG534, required for the production of active gingipains. FEMS Microbiol. Lett. 310:168–174 [DOI] [PubMed] [Google Scholar]
- 40. Saiki K., Konishi K. 2007. Identification of a Porphyromonas gingivalis novel protein Sov required for the secretion of gingipains. Microbiol. Immunol. 51:483–491 [DOI] [PubMed] [Google Scholar]
- 41. Sato K., et al. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 107:276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sato K., et al. 2005. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis J. Biol. Chem. 280:8668–8677 [DOI] [PubMed] [Google Scholar]
- 43. Sun H., Zusman D. R., Shi W. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143–1146 [DOI] [PubMed] [Google Scholar]
- 44. Sun M., Wartel M., Cascales E., Shaevitz J. W., Mignot T. 2011. Motor-driven intracellular transport powers bacterial gliding motility. Proc. Natl. Acad. Sci. U. S. A. 108:7559–7564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wall D., Kaiser D. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1–10 [DOI] [PubMed] [Google Scholar]
- 46. Wolkin R. H., Pate J. L. 1985. Selection for nonadherent or nonhydrophobic mutants co-selects for nonspreading mutants of Cytophaga johnsonae and other gliding bacteria. J. Gen. Microbiol. 131:737–750 [Google Scholar]
- 47. Yu C. S., Chen Y. C., Lu C. H., Hwang J. K. 2006. Prediction of protein subcellular localization. Proteins 64:643–651 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.