Abstract
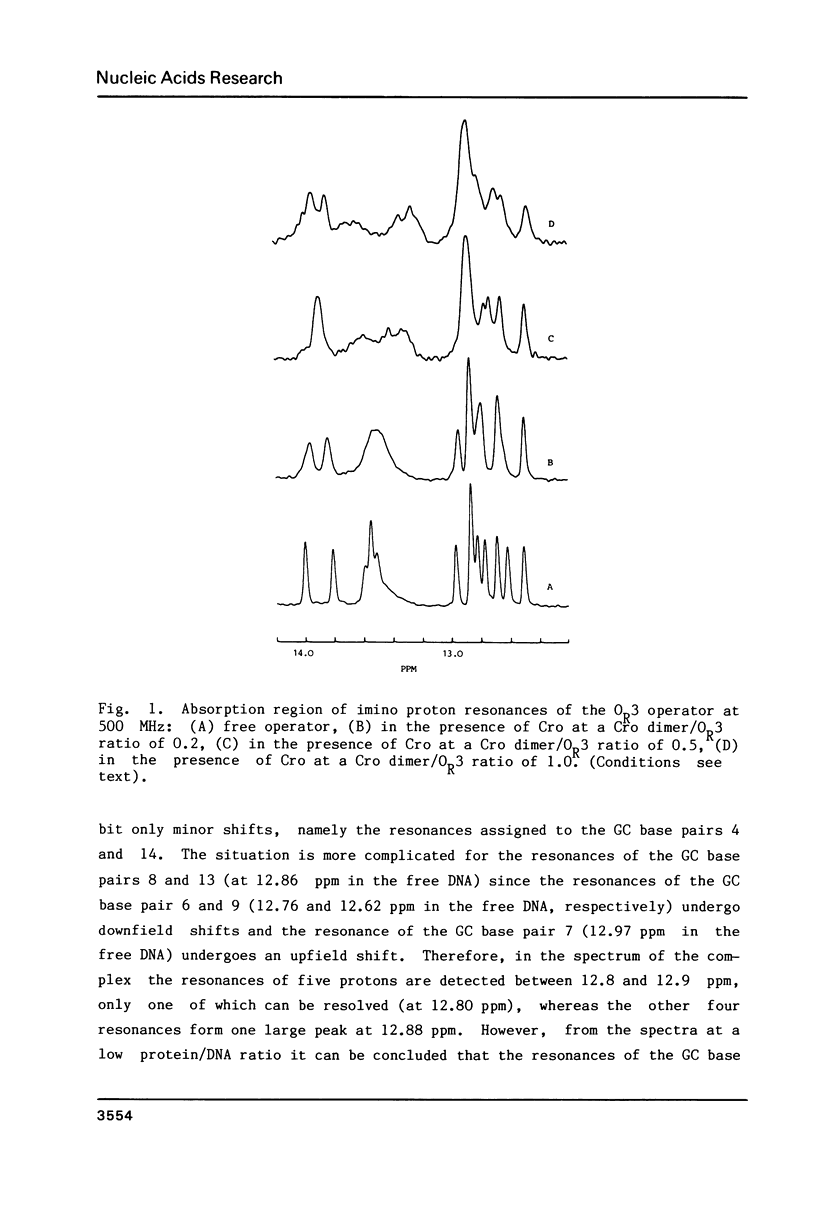
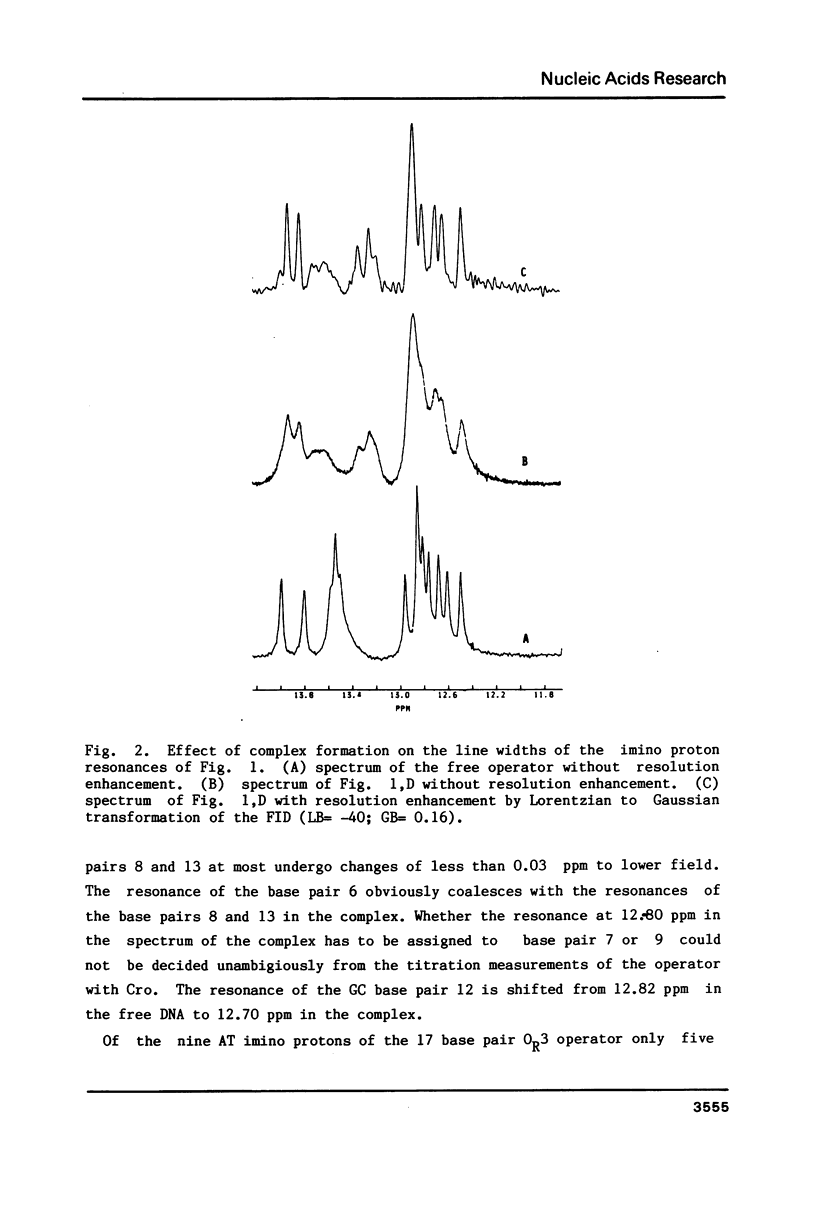
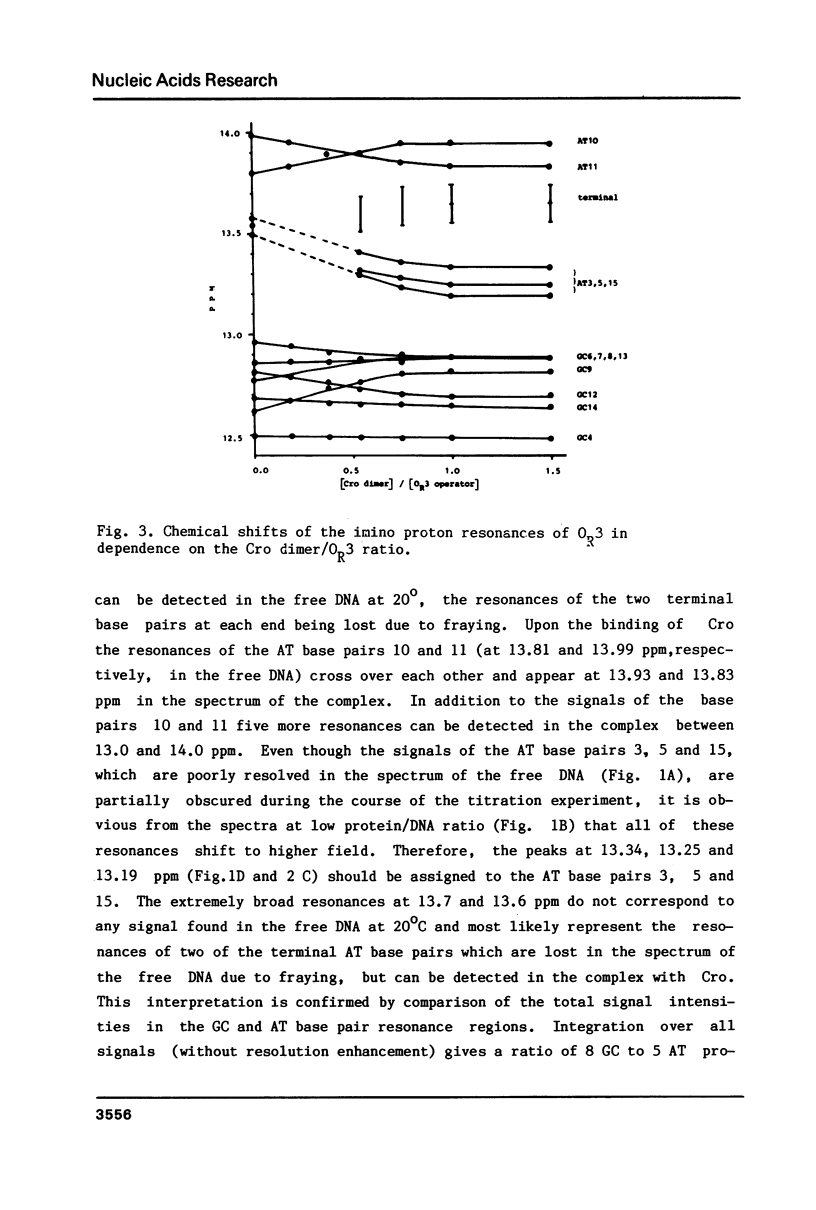
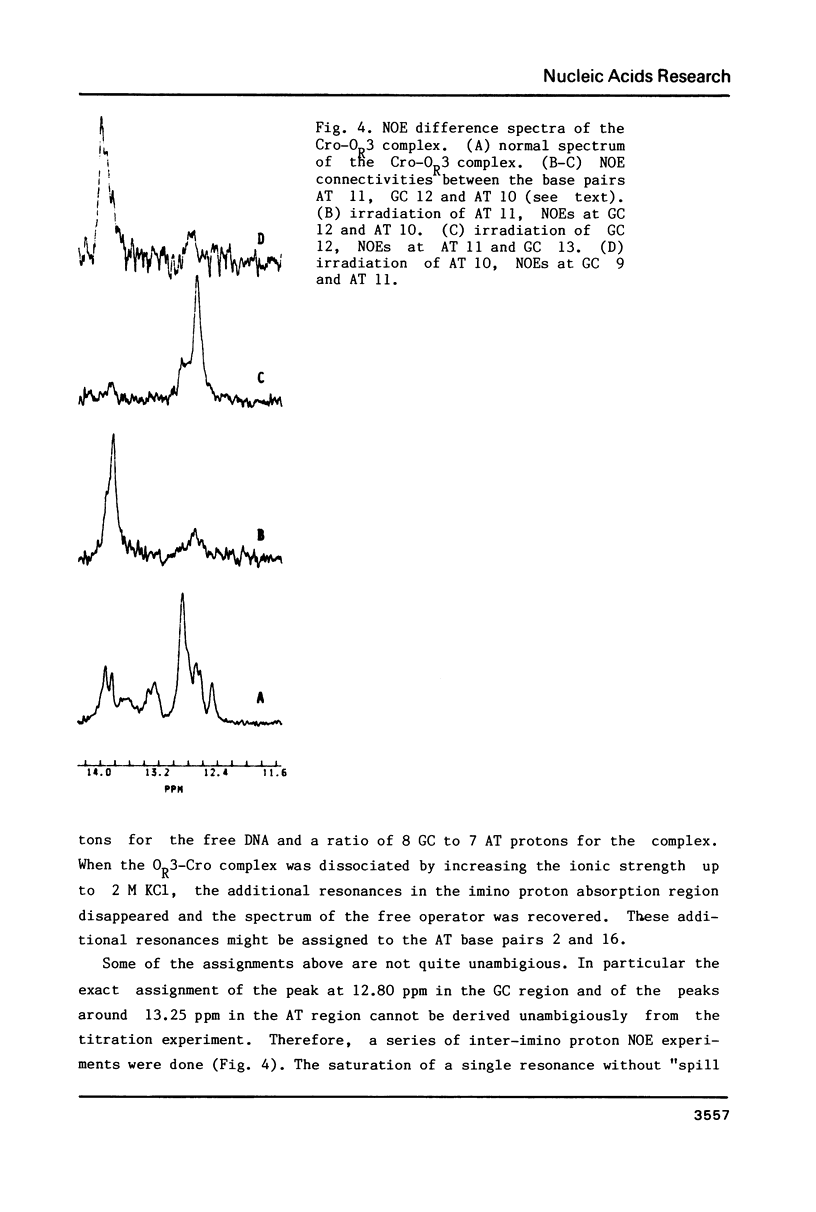
The specific complex between the lambda phage OR3 operator and the Cro protein has been studied by proton NMR spectroscopy at 500 MHz. The DNA imino proton resonances of this complex have been assigned to specific base pairs using the known assignments of these resonances for the free operator. Increase of the protein/DNA ratio to complete saturation of the OR3 operator with the Cro protein made it possible to follow the shift changes of the resonances. Ambiguities were resolved by nuclear Overhauser effect measurements on the complex. The shifts of the imino proton resonance positions provide information on the changes induced in the conformation of the operator upon complex formation with a dimer of the Cro protein. The most striking shift occurs for the central (GC 9) base pair, which is known to have no direct contacts with the Cro protein. This shift may be induced by a bend in the OR3 operator DNA at the GC 9 base pair to accommodate the operator for the binding of the Cro protein dimer. The imino proton resonances of two additional base pairs can be observed in the complex, demonstrating an overall stabilization of the DNA structure by the binding of the Cro protein.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Arndt K. T., Boschelli F., Cook J., Takeda Y., Tecza E., Lu P. lambda Phage cro repressor interaction with DNA. J Biol Chem. 1983 Apr 10;258(7):4177–4183. [PubMed] [Google Scholar]
- Bolotina I. A., Kurochkin A. V., Kirpichnikov M. P. Studies of the structure of bacteriophage lambda cro protein in solution. Analysis of the circular dichroism data. FEBS Lett. 1983 May 8;155(2):291–294. doi: 10.1016/0014-5793(82)80623-6. [DOI] [PubMed] [Google Scholar]
- Buck F., Hahn K. D., Zemann W., Rüterjans H., Sadler J. R., Beyreuther K., Kaptein R., Scheek R., Hull W. E. NMR study of the interaction between the lac repressor and the lac operator. Eur J Biochem. 1983 May 2;132(2):321–327. doi: 10.1111/j.1432-1033.1983.tb07365.x. [DOI] [PubMed] [Google Scholar]
- Chou S. H., Hare D. R., Wemmer D. E., Reid B. R. Sequence-specific recognition of deoxyribonucleic acid. Chemical synthesis and nuclear magnetic resonance assignment of the imino protons of lambda OR3 operator deoxyribonucleic acid. Biochemistry. 1983 Jun 21;22(13):3037–3041. doi: 10.1021/bi00282a002. [DOI] [PubMed] [Google Scholar]
- Culard F., Maurizot J. C. Lac repressor - lac operator interaction. Circular dichroism study. Nucleic Acids Res. 1981 Oct 10;9(19):5175–5184. doi: 10.1093/nar/9.19.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E., Kopka M. L., Pjura P. A random-walk model for helix bending in B-DNA. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7099–7103. doi: 10.1073/pnas.80.23.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giessner-Prettre C., Pullman B. Intermolecular nuclear shielding due to the aromatic amino acids of proteins and to porphyrins. J Theor Biol. 1971 May;31(2):287–294. doi: 10.1016/0022-5193(71)90188-3. [DOI] [PubMed] [Google Scholar]
- Heerschap A., Haasnoot C. A., Hilbers C. W. Nuclear magnetic resonance studies on yeast tRNAPhe I. Assignment of the iminoproton resonances of the acceptor and D stem by means of Nuclear Overhauser Effect experiments at 500 MHz. Nucleic Acids Res. 1982 Nov 11;10(21):6981–7000. doi: 10.1093/nar/10.21.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helene C., Maurizot J. C. Interactions of oligopeptides with nucleic acids. CRC Crit Rev Biochem. 1981;10(3):213–258. doi: 10.3109/10409238109113600. [DOI] [PubMed] [Google Scholar]
- Herskowitz I., Hagen D. The lysis-lysogeny decision of phage lambda: explicit programming and responsiveness. Annu Rev Genet. 1980;14:399–445. doi: 10.1146/annurev.ge.14.120180.002151. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Meyer B. J., Ptashne M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Poteete A. R., Lauer G., Sauer R. T., Ackers G. K., Ptashne M. lambda Repressor and cro--components of an efficient molecular switch. Nature. 1981 Nov 19;294(5838):217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- Johnson A., Meyer B. J., Ptashne M. Mechanism of action of the cro protein of bacteriophage lambda. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1783–1787. doi: 10.1073/pnas.75.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirpichnikov M. P., Kurochkin A. V., Skryabin K. G. Studies of the structure of bacteriophage lambda cro protein in solution. Analysis of the aromatic region of the 1H NMR spectrum. FEBS Lett. 1982 Dec 27;150(2):407–410. doi: 10.1016/0014-5793(82)80778-3. [DOI] [PubMed] [Google Scholar]
- Kurochkin A. V., Kirpichnikov M. P. Studies of the structure of bacteriophage lambda cro protein in solution. Globular structure of cro protein. FEBS Lett. 1982 Dec 27;150(2):411–415. doi: 10.1016/0014-5793(82)80779-5. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Ohlendorf D. H., Anderson W. F., Takeda Y. Structure of the DNA-binding region of lac repressor inferred from its homology with cro repressor. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1428–1432. doi: 10.1073/pnas.79.5.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer R., Meyer B., Ptashne M. Gene regulation at the right operator (OR) bacteriophage lambda. I. OR3 and autogenous negative control by repressor. J Mol Biol. 1980 May 15;139(2):147–161. doi: 10.1016/0022-2836(80)90302-2. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Meyer B. J., Maurer R., Ptashne M. Gene regulation at the right operator (OR) of bacteriophage lambda. II. OR1, OR2, and OR3: their roles in mediating the effects of repressor and cro. J Mol Biol. 1980 May 15;139(2):163–194. doi: 10.1016/0022-2836(80)90303-4. [DOI] [PubMed] [Google Scholar]
- Meyer B. J., Ptashne M. Gene regulation at the right operator (OR) of bacteriophage lambda. III. lambda repressor directly activates gene transcription. J Mol Biol. 1980 May 15;139(2):195–205. doi: 10.1016/0022-2836(80)90304-6. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Fisher R. G., Takeda Y., Matthews B. W. The molecular basis of DNA-protein recognition inferred from the structure of cro repressor. Nature. 1982 Aug 19;298(5876):718–723. doi: 10.1038/298718a0. [DOI] [PubMed] [Google Scholar]
- Ohtsuka E., Ikehara M., Söll D. Recent developments in the chemical synthesis of polynucleotides. Nucleic Acids Res. 1982 Nov 11;10(21):6553–6570. doi: 10.1093/nar/10.21.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. L. Ethidium bromide-(dC-dG-dC-dG)2 complex in solution: intercalation and sequence specificity of drug binding at the tetranucleotide duplex level. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3343–3347. doi: 10.1073/pnas.73.10.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M., Backman K., Humayun M. Z., Jeffrey A., Maurer R., Meyer B., Sauer R. T. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976 Oct 8;194(4261):156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Jeffrey A., Johnson A. D., Maurer R., Meyer B. J., Pabo C. O., Roberts T. M., Sauer R. T. How the lambda repressor and cro work. Cell. 1980 Jan;19(1):1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- Ptitsyn O. B., Finkelstein A. V., Kirpichnikov M. P., Skryabin K. G. cI and lexA repressors consist of three cro-like domains. FEBS Lett. 1982 Oct 4;147(1):11–15. doi: 10.1016/0014-5793(82)81001-6. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Scheek R. M., Zuiderweg E. R., Klappe K. J., van Boom J. H., Kaptein R., Rüterjans H., Beyreuther K. lac Repressor headpiece binds specifically to half of the lac operator: a proton nuclear magnetic resonance study. Biochemistry. 1983 Jan 4;22(1):228–235. doi: 10.1021/bi00270a033. [DOI] [PubMed] [Google Scholar]
- Tropp J., Redfield A. G. Environment of ribothymidine in transfer ribonucleic acid studied by means of nuclear Overhauser effect. Biochemistry. 1981 Apr 14;20(8):2133–2140. doi: 10.1021/bi00511a010. [DOI] [PubMed] [Google Scholar]
- Ulrich E. L., John E. M., Gough G. R., Brunden M. J., Gilham P. T., Westler W. M., Markley J. L. Imino proton assignments in the proton nuclear magnetic resonance spectrum of the lambda phage OR3 deoxyribonucleic acid fragment. Biochemistry. 1983 Sep 13;22(19):4362–4365. doi: 10.1021/bi00288a003. [DOI] [PubMed] [Google Scholar]


