Abstract
In higher eukaryotes, mitosis proceeds with nuclear envelope breakdown (NEBD) and disassembly of the nuclear pore complex (NPC); this is designated “open” mitosis. On the other hand, in many fungi, mitosis and chromosome segregation takes place without NEBD; this is designated “closed” mitosis. In a recent study on Schizosaccharomyces pombe, a closed mitosis organism, we reported a novel phenomenon that is equivalent to NEBD: a mixing of nuclear proteins and cytoplasmic proteins occurred transiently for a few minutes in meiosis without physical breakdown of the nuclear envelope. We designated this event virtual nuclear envelope breakdown (V-NEBD). In S. pombe, nuclear translocation of Rna1, a RanGAP1 homolog in S. pombe, occurs during meiosis, and this translocation of Rna1 leads to collapse of the Ran-GTP gradient across the nuclear envelope and occurs coincidently with V-NEBD. Here, we describe possible roles of RanGAP1 in V-NEBD in S. pombe and provide insights into the roles V-NEBD may play in meiosis.
Key words: RanGAP1, RanGTP gradient, nuclear envelope breakdown, nuclear pore complex, open-mitosis, closed-mitosis
In eukaryotic cells, nuclear and cytoplasmic compartmentalization is maintained by the activity of Ran, a small Ras-like GTPase, regulated by asymmetric localization of Ran guanine nucleotide exchange factor (RanGEF/RCC1) in the nucleus and Ran activating protein1 (RanGAP1) in the cytoplasm. The balance between the opposing activities of RanGEF/RCC1 and RanGAP1 generates a gradient of Ran-GTP across the NE, and this gradient controls the directionality of nucleocytoplasmic transport.1–4 In open mitosis, NEBD leads to collapse of the Ran-GTP gradient and diffusion of nuclear and cytoplasmic macromolecules (Fig. 1A) and allows microtubules to attach to the centromeres for chromosome segregation. On the other hand, in closed mitosis, the nuclear envelope remains intact throughout the mitotic cell cycle. The fission yeast Schizosaccharomyces pombe is one of the closed mitosis organisms: the nuclear envelope and NPC remain intact throughout the meiotic and mitotic cell cycles and the mitotic spindle forms within the nucleus during mitosis (Fig. 1D); in closed mitosis, tubulins are transported into the nucleus by importin.5
Figure 1.
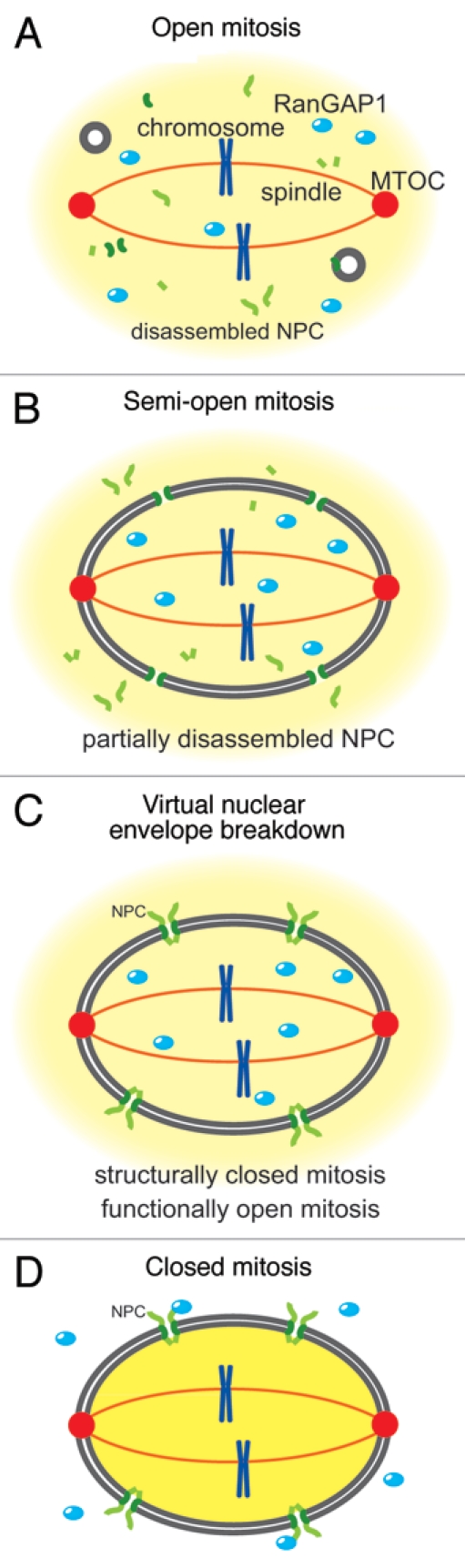
Four modes of nuclear envelope morphology during mitosis in eukaryotes. (A) In higher eukaryotes, both the nuclear membrane and the NPCs disassemble at the beginning of mitosis, and subsequently chromosomes are segregated by the mitotic spindle during mitosis. (B) In a filamentous fungus Aspergillus nidulans, the nuclear membrane does not disassemble, but the NPCs are partially disassembled and RanGAP1 enters into the nucleus. (C) In the fission yeast Schizosaccharomyces pombe, both the nuclear membrane and the NPCs remain intact but RanGAP1 enters into the nucleus, resulting in V-NE BD during meiosis II. In (B and C), the collapse of the Ran-GTP gradient results in scrambling of nuclear and cytoplasmic materials. (D) Many lower eukaryotes, like fungi, undergo closed mitosis. During closed mitosis both the nuclear membrane and the NPCs remain intact, RanGAP1 remains localized in the cytoplasm, and the Ran-GTP gradient across the NE remains intact.
Recently, we reported that in S. pombe nuclear proteins diffuse into the cytoplasm transiently in anaphase of the second meiotic division (anaphase II) as if NEBD had occurred in this closed mitosis organism: The nuclear membrane and NPCs remain intact throughout meiosis,6,7 but the permeability of the nuclear envelope during anaphase II indicates that the nuclear envelope is structurally closed but functionally open. Thus, we named this phenomenon virtual nuclear envelope breakdown (V-NEBD) (Fig. 1C). Strikingly, our study also revealed that RanGAP1 changes its localization from the cytoplasm to the nucleus at the onset of anaphase II in exact correlation with the timing of nuclear protein diffusion into the cytoplasm (Fig. 1C). Because the timing of the nuclear localization of RanGAP1 coincides with the diffusion of nuclear proteins out of the nucleus and because ectopic expression of NLS-conjugated RanGAP1 can induce diffusion of nuclear proteins to the cytoplasm,6 we speculate that the nuclear localization of RanGAP1 triggers diffusion of nuclear proteins out of the nucleus during anaphase II, although we currently have no direct evidence for this hypothesis.
Both closed mitosis and V-NEBD proceed with structurally intact nuclear envelopes and NPCs. Except for the nuclear localization of RanGAP1, the molecular basis generating the difference between closed mitosis and V-NEBD remains unclear, however, there are a number of other possible factors. For example, a meiosis-specific modification of NPC, such as phosphorylation, may be involved in changing the permeability barrier function of the NPC. Modification and/or alteration of transport machinery, such as importin β-s, may also be a factor in the permeability barrier change of the NPC. A third possible factor in V-NEBD is the formation of forespore membranes. Since the forespore membranes form during this period, the nuclear envelope may be weakened by sequestration of membrane components into forespore membranes during their assembly, as suggested by Arai et al.7 In this addendum, we will concentrate on possible roles of the nuclear translocation of RanGAP1.
The molecular structure of RanGAP1 suggests a possible regulatory mechanism specific to closed mitosis organisms. In metazoans, the RanGAP1 protein is modified by SUMO1 and interacts with RanBP2/Nup358 at the cytoplasmic surface of NPCs.8–10 In the plant Arabidopsis thaliana, RanGAP1 lacks the SUMO1 attachment domain and instead contains a plant-specific WPP domain which interacts with NE-associated WIP proteins.11,12 These domains result in concentrated localization of RanGAP1 at the NE. The majority of closed mitosis organisms also lack a SUMO1 attachment domain in their RanGAP1 proteins, but unlike plant RanGAP1 their RanGAP1 proteins have no alternative domains for NE attachment and do not show significant localization at the NE. These features may allow for translocation of RanGAP1 across the nuclear envelope. This idea is supported by the fact that the RanGAP1 homolog in Saccharomyces cerevisiae contains both NLS and NES signal motifs by which nuclear localization can be switched.13 Another type of fungi, Aspergillus nidulans, undergoes “semi-open” mitosis (Fig. 1B). During this type of mitosis, some of the nucleoporins disassemble from the NPC,14–16 and partial disassembly of the NPCs changes RanGAP1 localization from the cytoplasm into the nucleus (Fig. 1B).14 Considering these facts, the translocation of RanGAP1 seems a common strategy in closed mitosis and semi-open mitosis organisms; and in closed mitosis organisms, this translocation abates the Ran-GTP gradient and leads to virtual breakdown of the nuclear envelope without physical breakdown of the nuclear envelope. Thus, localization of RanGAP1 may act as a molecular switch to control the barrier function of the nuclear envelope.
Another interesting genetic phenomenon associated with nuclear localization of RanGAP, called segregation distortion, occurs in Drosophila. In the segregation distorter (SD) system of Drosophila, heterozygous SD/SD+ males produce only SD-bearing sperm during spermatogenesis.17 It has been shown that the SD-causing locus corresponds to a partial duplication of the RanGAP gene and encodes a C-terminal truncation of RanGAP (Sd-RanGAP).18 The truncated Sd-RanGAP is enzymatically active but is mis-localized into the nucleus, and this nuclear localization of enzymaticallyactive RanGAP is necessary for segregation distortion.19,20 Although it is unknown how this event results in selective elimination of SD+ sperm, it has been inferred that nuclear localization of RanGAP affects nuclear events through changes in concentrations of nuclear Ran-GTP.21,22 While the underlying mechanisms of segregation distortion and V-NEBD may be different, the segregation distortion phenomenon in Drosophila is reminiscent of V-NEBD in S. pombe in the sense that both events are associated with nuclear localization of enzymatically-active RanGAP1.
In addition to its role in V-NEBD, nuclear translocation of RanGAP1 may have a direct role in nuclear function independent of its RanGAP activity. In S. pombe, it has been reported that a fraction of RanGAP1 is localized on chromatin where RanGAP1 interacts with histone H3 and histone H3 methyltransferase Clr4 and enhances histone methyltransferase activity, suggesting a role of RanGAP1 in heterochromatin assembly.23 Interestingly, RanGAP activity is not required for this function.23 In addition to these findings, we reported that the nuclear translocation of RanGAP1 might be correlated to spore formation.6 Spores are small dormant state cells and chromosomes must be packed into a small volume in the nuclei of spores. Although it is unknown how spore chromatin is formed, one fascinating idea is that anaphase II-specific nuclear translocation of RanGAP1 in S. pombe correlates with a process which produces dormant chromatin.
The exact roles of V-NEBD in meiosis are unknown. Possibilities include (1) allowing nuclear proteins to exert their activities in the cytoplasm, for example in forespore membrane assembly and (2) allowing cytoplasmic proteins to exert their activities in the nucleus, for example in dormant chromatin formation. Another possible role of V-NEBD may be to decrease the volume of the spore nucleus by allowing nuclear proteins to move into the cytoplasm. A final possible role of V-NEBD may be to decrease specific proteins in the nucleus. For example, although cyclins are targeted for degradation at anaphase in both mitosis and meiosis via ubiquitination by the anaphase promoting complex, V-NEBD could further ensure braking of cell cycle progression by rapidly decreasing the number of cyclin molecules in the nucleus.
Acknowledgements
We thank D.B. Alexander for critical reading of the manuscript. This work was supported by grants from the Japan Science and Technology Agency (to T.H.) and the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.H. and Y.H.).
Abbreviations
- NE
nuclear envelope
- NEBD
nuclear envelope breakdown
- NES
nuclear export signal
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- RanGAP1
RanGTPase activating protein1
- RanGEF
Ran guanine nucleotide-exchange factor
- V-NEBD
virtual nuclear envelope breakdown
- MTOC
microtubule organizing center
References
- 1.Joseph J. Ran at a glance. J Cell Sci. 2006;119:3481–3484. doi: 10.1242/jcs.03071. [DOI] [PubMed] [Google Scholar]
- 2.Ciciarello M, Mangiacasale R, Lavia P. Spatial control of mitosis by the GTPase Ran. Cell Mol Life Sci. 2007;64:1891–1914. doi: 10.1007/s00018-007-6568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- 4.Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 5.Sato M, Toda T. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature. 2007;447:334–337. doi: 10.1038/nature05773. [DOI] [PubMed] [Google Scholar]
- 6.Asakawa H, Kojidani T, Mori C, Osakada H, Sato M, Ding DQ, et al. Virtual breakdown of the nuclear envelope in fission yeast meiosis. Curr Biol. 2010;20:1919–1925. doi: 10.1016/j.cub.2010.09.070. [DOI] [PubMed] [Google Scholar]
- 7.Arai K, Sato M, Tanaka K, Yamamoto M. Nuclear compartmentalization is abolished during fission yeast meiosis. Curr Biol. 2010;20:1913–1918. doi: 10.1016/j.cub.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 10.Saitoh H, Pu R, Cavenagh M, Dasso M. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc Natl Acad Sci USA. 1997;94:3736–3741. doi: 10.1073/pnas.94.8.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose A, Meier I. A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci USA. 2001;98:15377–15382. doi: 10.1073/pnas.261459698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu XM, Meulia T, Meier I. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol. 2007;17:1157–1163. doi: 10.1016/j.cub.2007.05.076. [DOI] [PubMed] [Google Scholar]
- 13.Feng W, Benko AL, Lee JH, Stanford DR, Hopper AK. Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J Cell Sci. 1999;112:339–347. doi: 10.1242/jcs.112.3.339. [DOI] [PubMed] [Google Scholar]
- 14.De Souza CP, Osmani AH, Hashmi SB, Osmani SA. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr Biol. 2004;14:1973–1984. doi: 10.1016/j.cub.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 15.Osmani AH, Davies J, Liu HL, Nile A, Osmani SA. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol Biol Cell. 2006;17:4946–4961. doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Souza CP, Osmani SA. Double duty for nuclear proteins—the price of more open forms of mitosis. Trends Genet. 2009;25:545–554. doi: 10.1016/j.tig.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyttle TW. Segregation distorters. Annu Rev Genet. 1991;25:511–557. doi: 10.1146/annurev.ge.25.120191.002455. [DOI] [PubMed] [Google Scholar]
- 18.Merrill C, Bayraktaroglu L, Kusano A, Ganetzky B. Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila. Science. 1999;283:1742–1745. doi: 10.1126/science.283.5408.1742. [DOI] [PubMed] [Google Scholar]
- 19.Kusano A, Staber C, Ganetzky B. Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Dev Cell. 2001;1:351–361. doi: 10.1016/s1534-5807(01)00042-9. [DOI] [PubMed] [Google Scholar]
- 20.Kusano A, Staber C, Ganetzky B. Segregation distortion induced by wild-type RanGAP in Drosophila. Proc Natl Acad Sci USA. 2002;99:6866–6870. doi: 10.1073/pnas.102165099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusano A, Staber C, Chan HY, Ganetzky B. Closing the (Ran)GAP on segregation distortion in Drosophila. Bioessays. 2003;25:108–115. doi: 10.1002/bies.10222. [DOI] [PubMed] [Google Scholar]
- 22.Presgraves DC. Does genetic conflict drive rapid molecular evolution of nuclear transport genes in Drosophila? Bioessays. 2007;29:386–391. doi: 10.1002/bies.20555. [DOI] [PubMed] [Google Scholar]
- 23.Nishijima H, Nakayama J, Yoshioka T, Kusano A, Nishitani H, Shibahara K, et al. Nuclear RanGAP is required for the heterochromatin assembly and is reciprocally regulated by histone H3 and Clr4 histone methyltransferase in Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:2524–2536. doi: 10.1091/mbc.E05-09-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]


