ABSTRACT
The Sec translocase pathway is the major route for protein transport across and into the cytoplasmic membrane of bacteria. Previous studies reported that the SecA translocase ATP-binding subunit and the cell surface HtrA protease/chaperone formed a single microdomain, termed “ExPortal,” in some species of ellipsoidal (ovococcus) Gram-positive bacteria, including Streptococcus pyogenes. To investigate the generality of microdomain formation, we determined the distribution of SecA and SecY by immunofluorescent microscopy in Streptococcus pneumoniae (pneumococcus), which is an ovococcus species evolutionarily distant from S. pyogenes. In the majority (≥75%) of exponentially growing cells, S. pneumoniae SecA (SecASpn) and SecYSpn located dynamically in cells at different stages of division. In early divisional cells, both Sec subunits concentrated at equators, which are future sites of constriction. Further along in division, SecASpn and SecYSpn remained localized at mid-cell septa. In late divisional cells, both Sec subunits were hemispherically distributed in the regions between septa and the future equators of dividing cells. In contrast, the HtrASpn homologue localized to the equators and septa of most (>90%) dividing cells, whereas the SrtASpn sortase located over the surface of cells in no discernable pattern. This dynamic pattern of Sec distribution was not perturbed by the absence of flotillin family proteins, but was largely absent in most cells in early stationary phase and in ∆cls mutants lacking cardiolipin synthase. These results do not support the existence of an ExPortal microdomain in S. pneumoniae. Instead, the localization of the pneumococcal Sec translocase depends on the stage of cell division and anionic phospholipid content.
IMPORTANCE
Two patterns of Sec translocase distribution, an ExPortal microdomain in certain ovococcus-shaped species like Streptococcus pyogenes and a spiral pattern in rod-shaped species like Bacillus subtilis, have been reported for Gram-positive bacteria. This study provides evidence for a third pattern of Sec localization in the ovococcus human pathogen Streptococcus pneumoniae. The SecA motor and SecY channel subunits of the Sec translocase localize dynamically to different places in the mid-cell region during the division cycle of exponentially growing, but not stationary-phase, S. pneumoniae. Unexpectedly, the S. pneumoniae HtrA (HtrASpn) protease/chaperone principally localizes to cell equators and division septa. The coincident localization of SecASpn, SecYSpn, and HtrASpn to regions of peptidoglycan (PG) biosynthesis in unstressed, growing cells suggests that the pneumococcal Sec translocase directs assembly of the PG biosynthesis apparatus to regions where it is needed during division and that HtrASpn may play a general role in quality control of proteins exported by the Sec translocase.
Introduction
The Sec translocase is the most important molecular machine for the secretion of proteins and insertion of membrane proteins in bacteria (1, 2). The Sec translocase apparatus consists of the highly conserved SecYEG conducting channel imbedded in the membrane, the peripherally bound SecA ATP-dependent motor protein that mediates posttranslational export, the SRP-FtsY system that mediates cotranslational export, and a variety of ancillary protein factors (reviewed in references 1 and 2). To understand whether protein delivery is coupled to cellular functions, the subcellular location of the Sec translocase has been examined in several bacteria. In the Gram-negative bacterium Escherichia coli, the SecY and SecE subunits showed spiral-like, polar, or no localization, possibly depending on experimental expression levels (3). In the rod-shaped Gram-positive bacterium Bacillus subtilis, subcellular localization studies of SecA and SecY indicated that the Sec translocase is present in a spiral-like structure along the cell length (4).
In the ovococcus (American football-shaped) Gram-positive bacterium Streptococcus pyogenes, immunogold electron microscopy (IG-EM) was used to locate S. pyogenes SecA (SecASpy) and HtrASpy, which is thought to function as a dual-function protease and protein chaperone (5, 6). SecASpy and HtrASpy were found to colocalize at only one specific site in each cell, termed the “ExPortal,” which was defined as a microdomain specialized for secretion. It was proposed that concentrated secretion at a specific site might enable some bacteria to coordinate protein translocation and subsequent folding, especially in bacterial pathogens like S. pyogenes that secrete many adhesins, toxins, and virulence factors to their cell surfaces (6). More recent IG-EM studies of the ovococcus species Enterococcus faecalis (7) and Streptococcus mutans (8) also indicate that their homologues of SecA and the general “housekeeping” sortase SrtA localize to a single locus, similar to the ExPortal reported for SecA and HtrA in S. pyogenes (6). In contrast, the SrtA sortase of S. pyogenes is not located in a single microdomain and seems to distribute around cell peripheries with some localization at the septa of dividing cells (9).
However, not all studies support the existence of a Sec translocase ExPortal. In one IG-EM study, SecA of S. pyogenes was distributed throughout the cell periphery and was not confined to a microdomain (10). In other studies of S. pyogenes (10) and the spherical coccus bacterium Staphylococcus aureus (11), sortase-attached surface proteins were directed to mid-cell or polar locations by a mechanism that involves specific motifs in signal peptides. Secretion at two disparate locations in cells is not consistent with a single ExPortal per cell. In addition, the localization of the Sec apparatus seems to be influenced by the growth phase of some bacteria. The Sec apparatus was localized in spirals in B. subtilis cells growing exponentially, but this pattern disappeared in stationary-phase cells (4). In other bacteria, like E. faecalis, SecA location in an ExPortal is independent of growth stage (7). To date, no clear picture has emerged about the localization of the Sec translocase during different stages of cell division and culture growth.
Like S. pyogenes, Streptococcus pneumoniae (pneumococcus) is an important opportunistic, ovococcus-shaped, Gram-positive human pathogen (12, 13). However, S. pneumoniae (mitis group) is evolutionarily distant from other species of Streptococcus in which SecA and HtrA homologues have been localized, including S. pyogenes (pyogenic group) and S. mutans (mutans group) (reviewed in reference 14). This evolutionary distance is reflected by the remarkably different sets of virulence factors used and different diseases caused by these different commensal species of Streptococcus (14–17). S. pneumoniae is an aerotolerant anaerobe that colonizes the nasopharyngeal cavities of children and adults (12, 13, 16). Besides acting as a commensal, S. pneumoniae is a human opportunistic pathogen that causes several serious invasive diseases, including pneumonia, otitis media (earache), meningitis, and bacteremia, that result in at least 1.6 million deaths annually worldwide (13). The majority of pneumococcal virulence factors, including choline binding proteins, pneumococcal surface proteins A and C (PspA and PspC, respectively), proteinase maturation protein A (PpmA), autolysin B (LytB), and metal receptor binding proteins (e.g., PiaA, PiuA, and PsaA), contain signal peptides and are transported to the cell surface by the S. pneumoniae Sec (SecSpn) translocase (16, 18). Essential proteins involved in pneumococcal cell division, such as PcsB, are also delivered to the cell surface by the Sec system (19).
We report here the first determination of the distribution of two Sec translocase subunits, SecA and SecY, as well as HtrA and SrtA in S. pneumoniae during different stages of cell division and phases of growth. In this study, we constructed strains that express the respective FLAG-tagged proteins from their native chromosomal loci in an unencapsulated derivative of the prototypic virulent serotype 2 strain D39 (20). Lack of capsule causes these strains to form mainly diplococci (19) that can be binned according to stage of division before immunofluorescent images are analyzed (21, 22). We show that during exponential growth, SecASpn and SecYSpn distribute dynamically to mid-cell and division regions active in peripheral and septal peptidoglycan (PG) synthesis in ovococcus-shaped cells (see references 21, 23, and 24). Likewise, the HtrASpn protease/chaperone localizes to regions of PG biosynthesis and cell division, whereas the SrtASpn general sortase is present over the entire cell periphery. We also provide evidence that entry into stationary phase and ∆cls mutations that block synthesis of the anionic lipid cardiolipin lead to loss of localization of SecASpn and HtrASpn. Besides not supporting the existence of an ExPortal microdomain in S. pneumoniae, these results suggest that Sec-mediated secretion and membrane protein insertion and HtrA-catalyzed protein quality control localize to regions of active PG biosynthesis and cell division.
RESULTS
SecASpn-L-FLAG3-, SecYSpn-L-FLAG3-, HtrASpn-L-FLAG3-, and SrtASpn-L-FLAG3-expressing strains exhibit wild-type phenotypes.
We constructed strains expressing FLAG3-tagged proteins by fusing a 10-amino-acid linker followed by three tandem copies of the FLAG epitope (FLAG3) to the carboxyl terminus of SecASpn, SecYSpn, HtrASpn, or SrtASpn. Each resulting markerless construct was expressed at its native chromosomal locus in place of the native gene (see Tables S1 and S2 and Fig. S1 in the supplemental material). The resulting strains, IU4468 (D39 ∆cps rpsL1 htrA-L-FLAG3), IU4470 (D39 ∆cps rpsL1 srtA-L-FLAG3), IU4472 (D39 ∆cps rpsL1 secA-L-FLAG3), and IU5471 (D39 ∆cps rpsL1 secY-L-FLAG3) showed no aberrant growth phenotypes compared to the parent strain IU1824 (D39 ∆cps not expressing a FLAG3 fusion protein) at either 37°C or 41.2°C in brain heart infusion (BHI) broth (data not shown). The four strains expressing FLAG3-tagged proteins showed no differences in colony morphology on Trypticase soy agar II (TSAII) blood agar plates or cell morphology as observed by phase-contrast microscopy compared to the IU1824 parent strain (column 1 in Fig. 1 and 4; see Fig. S2 and S5 in the supplemental material). Since S. pneumoniae ∆htrA mutants exhibit a temperature-sensitive growth phenotype (25), the normal growth of IU4468 (D39 ∆cps htrA-L-FLAG3) at 41.2°C indicated that the HtrA-L-FLAG3 protein was functional. Likewise, the normal growth of IU4472 (D39 ∆cps secA-L-FLAG3) and IU5471 (D39 ∆cps secY-L-FLAG3) indicated that the FLAG3 tag did not significantly impede function of the essential SecA and SecY proteins. We confirmed that the SrtA-L-FLAG3 fusion sortase was functional in strain IU4470 (D39 ∆cps srtA-L-FLAG3) based on its wild-type level of cell-associated β-galactosidase activity compared to a ∆srtA mutant, which secretes this activity into the growth medium (see Text S2 in the supplemental material). Finally, Western blotting (see Text S2) showed that the FLAG3-tagged SecA, HtrA, and SrtA proteins were present as single bands with the expected molecular masses and that the relative cellular amount of each protein did not change in cells in exponential or transition phase (see Text S2) sampled for immunofluorescent microscopy (IFM) studies (Fig. 1–4).
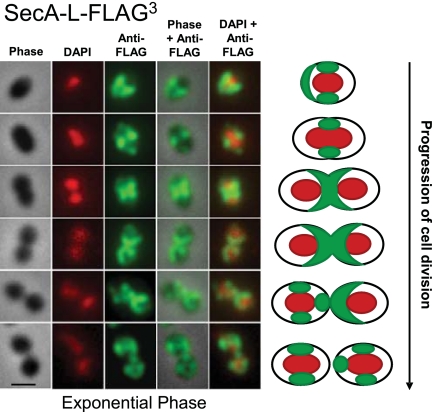
FIG 1 .
SecA-L-FLAG3 localizes primarily to equators, septa, and hemispheres of exponentially growing S. pneumoniae cells. Strain IU4472 (D39 ∆cps rpsL1 secA-L-FLAG3) was grown to mid-exponential phase in BHI broth, and DAPI staining and IFM of cells were performed as described in Materials and Methods. Cells at different stages of division were binned based on images from phase-contrast microscopy (column 1), and localization patterns of DAPI (pseudocolored red; column 2) and SecA-L-FLAG3 (pseudocolored green; column 3) were determined from images obtained by epifluorescent microscopy. Columns 4 and 5 show overlays of columns 1 and 3 or columns 1 and 2, respectively. Images of representative cells are shown at different stages of division. A schematic summary at right shows the SecASpn localization pattern for >75% of the cell population at each stage (Fig. 2). The experiment was performed more than 5 times independently. Multiple fields were observed in each experiment, with similar results. Scale bar = 1 µm.
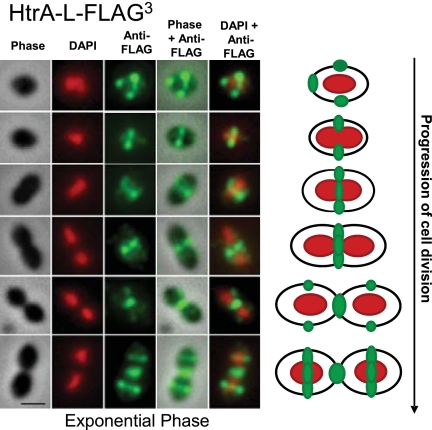
FIG 4 .
HtrA-L-FLAG3 localizes to equators and septa of exponentially growing S. pneumoniae cells. Strain IU4468 (D39 ∆cps rpsL1 htrA-L-FLAG3) was grown to mid-exponential phase in BHI broth, and DAPI staining and IFM of cells were performed as described in Materials and Methods. Representative images are shown with the same row and column arrangements as in Fig. 1. A schematic summary at right shows the HtrASpn distribution pattern in >90% of the cell population (Fig. 2). The experiment was performed more than 3 times independently. Multiple fields were observed in each experiment, with similar results. Scale bar = 1 µm.
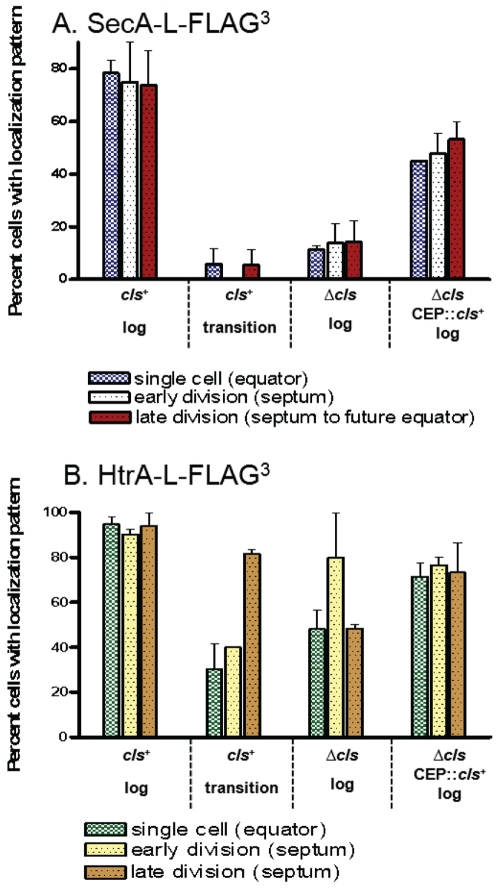
FIG 2 .
Distribution of predivisional, early divisional, and late-divisional cells with SecA-L-FLAG3 (A) and HtrA-L-FLAG3 (B) localized at equators, division septa, or hemispheres between septa and future equators, as indicated. Cells were grown exponentially (log) or to the transition to stationary phase, and IFM was performed as described in Materials and Methods. (A) cls+ parent strain IU4472 (D39 ∆cps rpsL1 secA-L-FLAG3), ∆cls::Pc-erm mutant IU5093, and complemented ∆cls mutant IU5138 (Δcls::Pc-erm//CEP::cls+, where “//” indicates a separate ectopic location in the chromosome). (B) cls+ parent strain IU4468 (D39 ∆cps rpsL1 htrA-L-FLAG3), ∆cls::Pc-erm mutant IU5089, and complemented ∆cls mutant IU5136 (∆cls::Pc-erm/CEP::cls+). See the text for additional details.
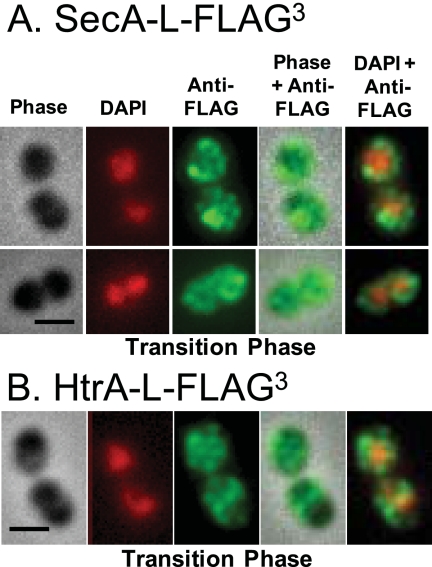
FIG 3 .
SecA-L-FLAG3 (A) and HtrA-L-FLAG3 (B) do not localize in cells during the transition from exponential to stationary phase in BHI broth. Strains IU4472 (Fig. 1) and IU4468 (Fig. 4) were grown to transition phase, and DAPI staining and IFM of cells were performed as described in Materials and Methods. Representative cells are shown with the same column arrangement as in Fig. 1. The experiment was performed more than 2 times independently. Multiple fields were observed in each experiment, with similar results. Scale bar = 1 µm.
SecASpn and SecYSpn localize dynamically during cell division.
The localization patterns of SecA-L-FLAG3 were determined by IFM in cells at the following three stages of division (Materials and Methods): predivisional single cells (oval cells) (Fig. 1, rows 1 and 2), early divisional cells (slight indentation at mid-cell) (Fig. 1, row 3), and late divisional cells (septum width of less than half the cell width) (Fig. 1, row 4). Visual patterns of localization were confirmed by determining pixel intensities in different regions of cells (Materials and Methods; data not shown). Among predivisional single cells, 78% showed localization of SecA-L-FLAG3 primarily at equators in the region between separating nucleoids (Fig. 1, row 2; Fig. 2A). An additional 15% of predivisional cells had SecA-L-FLAG3 spreading from equators to over half of the cell (Fig. 1; row 1). In cells with this hemispheric distribution, nucleoids appeared condensed and spherical, suggesting that these cells might have been newly divided. Besides this prominent equatorial localization of SecA-L-FLAG3 in 93% of predivisional cells, weaker labeling of SecA-L-FLAG3 was sometimes present at one pole (36%; e.g., see Fig. 1, row 2), both poles (16%), or at other areas (46%), consistent with a dynamic redistribution of SecASpn and the translocase apparatus during division.
This conclusion was supported by patterns seen in early and late divisional cells. About 75% of cells at these stages (Fig. 2A) had a “χ” (Chi)-like distribution of SecA-L-FLAG3 along the division septum between the divided nucleoids, eventually forming two hemispheres surrounding the middle halves of the daughter cells (Fig. 1, rows 3 and 4). Finally, in postdivisional diplococci (Fig. 1, rows 5 and 6), SecA-L-FLAG3 was distributed in each cell similar to the patterns observed in separated single cells (Fig. 1, rows 1 and 2). Daughter cells that contained spherical nucleoids showed a half-cell labeling pattern of SecA-L-FLAG3 (Fig. 1, row 5, right cell), similar to that of late divisional cells (Fig. 1, row 4), while daughter cells with duplicating nucleoids showed the strongest SecA-L-FLAG3 labeling at mid-cell (Fig. 1, row 4, left cell; row 6, both cells). Taken together, these images indicate that SecASpn distributes dynamically during the cell cycle and goes through stages where it is distributed over the hemispherical halves between the separated nucleoids of dividing pneumococcal cells (Fig. 1, right).
The localization patterns of SecY-L-FLAG3 determined by IFM were remarkably similar to those of SecA-L-FLAG3 at all three stages of division of exponentially growing cells (see Fig. S2 in the supplemental material). Similar to SecA-L-FLAG3, SecY-L-FLAG3 localized chiefly at equators of single predivisional cells, at septa of early divisional cells, and as hemispheres in late-divisional cells in the majority (>75%) of cells at each stage of division (data not shown). This result indicates that SecASpn tracks with SecYSpn, which is a member of the translocase channel complex, in exponentially growing pneumococcal cells.
SecASpn and SecYSpn are not localized in transition-phase cells.
We performed similar IFM and DAPI (4′,6-diamidino-2-phenylindole) labeling of cells grown to transition phase before reaching stationary phase (optical density at 620 nm [OD620] of 0.78, 15 min before the maximal OD620 of 0.82). The localization patterns of SecA-L-FLAG3 and SecY-L-FLAG3 observed in exponentially growing cells (Fig. 1 and 2A; see Fig. S2 in the supplemental material) were present in fewer than 6% of transition-phase cells (Fig. 2A and 3A) (data not shown). Instead, SecA-L-FLAG3 and SecY-L-FLAG3 appeared as puncta in no discernable pattern in a large majority of transition-phase cells, including single cells and diplococci at different stages of division (Fig. 3A) (data not shown).
SecASpn is not localized in ∆cls (cardiolipin synthase) mutants.
The anionic lipids phosphatidylglycerol (PGly) and cardiolipin (CL) have been implicated in protein translocation across the bacterial membrane (3, 4, 26), and depletion of anionic lipids results in rapid loss of the spiral localization of the Sec components in B. subtilis (4). In addition, the ExPortal microdomain of S. pyogenes was postulated to be enriched in anionic phospholipids (27). Consequently, we determined the distribution of SecA-L-FLAG3 in cls (spd_0185) deletion mutants lacking cardiolipin synthase, which converts PGly to CL. A ∆cls::Pc-erm deletion/insertion mutation and a nonpolar markerless ∆cls mutation were constructed (see Tables S1 and S2 in the supplemental material), and both types of mutations caused similar phenotypes. Growth of unencapsulated D39 ∆cps ∆cls::Pc-erm, D39 ∆cps ∆cls::Pc-erm secA-L-FLAG3, and D39 ∆cps ∆cls secA-L-FLAG3 mutants was impaired in BHI broth compared to that of their cls+ parents, with longer lags and lower (63 to 86%) growth yields (see Fig. S3 in the supplemental material) (data not shown). At low (OD620 of <0.1) densities, ∆cls mutant cells often formed long chains (>30 cells), which occasionally contained lysed cells (block arrows, column 1 in Fig. S4B in the supplemental material) (data not shown) and typically were wider and more spherical than the cls+ parent cells (column 1 in Fig. S4A and S4B in the supplemental material) (data not shown). SecASpn-L-FLAG3 was mislocalized in most cells containing either type of ∆cls mutation (see Fig. S4B) (data not shown), and only 11 to 27% of ∆cls mutant cells showed the patterns of SecASpn-L-FLAG3 localization observed in the cls+ parent (Fig. 1 and 2A; see Fig. S4A and S4B) (data not shown). At higher culture densities, the number of lysed ∆cls cells within chains increased, leading to shorter chains among many lysed cells (data not shown).
These defects in growth yield, chain length, cell lysis, and SecASpn-L-FLAG3 localization caused by both ∆cls mutations were largely complemented by expression of Cls+ protein from the PfcsK promoter in the ectopic CEP site (Fig. 2A; see Fig. S3 and S4C in the supplemental material) (data not shown) (28, 29). Complementation levels for both ∆cls mutations were similar in the presence or absence of 0.8% (wt/vol) fucose, which is an inducer of the PfcsK promoter (19, 21, 30). This result suggests that sufficient Cls+ enzymatic activity is expressed by leakiness of the PfcsK promoter in the absence of inducer to complement the phenotypes. Since the complementation levels were similar for the ∆cls::Pc-erm and the nonpolar ∆cls mutations, the lack of full complementation is not likely due to polarity of the ∆cls mutations on expression of the downstream spd_0186 and spd_0187 genes of unknown function. The nearly complete complementation of SecA-L-FLAG3 mislocalization (Fig. 2A; see Fig. S4A and S4C) (data not shown) supports the notion that CL (or less likely, the Cls+ protein itself) plays a role in proper localization of the protein translocase in S. pneumoniae.
HtrA localizes at equatorial rings and division septa in exponentially growing pneumococcus.
Despite its importance to S. pneumoniae physiology and virulence (see Discussion), the cellular localization of HtrASpn on cell surfaces has never been reported. We determined the localization of HtrA-L-FLAG3 at different stages of the cell cycle in exponentially growing cells (Fig. 4). Strikingly, HtrA-L-FLAG3 localized to the equators and division septa of >90% of cells (Fig. 2B and 4). Among single cells (Fig. 4, rows 1 and 2), HtrA-L-FLAG3 was located at equators, and 20% of these cells showed additional strong labeling at one pole, possibly from the septum of the previous division (e.g., Fig. 4, row 1). As equators became division septa, HtrA-L-FLAG3 retained its mid-cell localization (Fig. 4, rows 3 and 4). HtrA-L-FLAG3 remained at closing septa, after daughter cells appeared to be separated, and then started to appear at the equators of newly divided cells (Fig. 4, rows 5 and 6).
Like SecA-L-FLAG3, HtrA-L-FLAG3 delocalized in cultures transitioning to stationary phase (Fig. 2B and 3B). Although some transition-phase cells still contained HtrA-L-FLAG3 at equators and septa, HtrA-L-FLAG3 was also located randomly over the surfaces of many cells (Fig. 3B). The localization of HtrA-L-FLAG3 to equators and septa observed in the exponentially growing parent strains (Fig. 4) was diminished in the ∆cls::Pc-erm mutant, but to a lesser extent than for SecA-L-FLAG3 (Fig. 2A and B). The mislocalization of HtrA-L-FLAG3 in the ∆cls::Pc-erm mutant was again complemented by ectopic expression of Cls+ (Fig. 2B). Because HtrASpn localizes to the sites of peptidoglycan (PG) biosynthesis and cell division (see references 21 and 24), we examined the cell morphologies of isogenic ∆htrA and htrA+ strains during growth at 37°C and 41.2°C in BHI broth at culture densities with OD620s of 0.1 (mid-log) and 0.4 (late-log phase). We did not observe any obvious differences in cell shape, cell size, or chaining under these conditions between the two strains (data not shown).
The SrtASpn sortase locates randomly on cell surfaces.
Sortase was reported to be in the ExPortal structure of some bacteria (7, 8). Consequently, we determined the distribution of SrtA-L-FLAG3 during the cell cycle. In contrast to SecA-L-FLAG3, SecY-L-FLAG3, and HtrA-L-FLAG3, SrtA-L-FLAG3 did not localize and was distributed in an apparently random punctate pattern over the surface of cells at all stages of division (see Fig. S5 in the supplemental material). This conclusion was supported by preliminary three-dimensional (3D)-reconstructed images showing SrtA-L-FLAG3 located randomly on the pneumococcal cell surface (data not shown).
SecASpn, HtrASpn, and SrtASpn locations are not affected by the rpsL1 allele or absence of flotillin family proteins or each other.
Markerless unencapsulated strains expressing SecA-L-FLAG3, HtrA-L-FLAG3, and SrtA-L-FLAG3 from their native loci (IU4468, IU4470, and IU4472; see Table S1 in the supplemental material) contained the rpsL1 allele imparting streptomycin (Sm) resistance used in allele exchange. We complemented the rpsL1 mutation by a dominant Pc-[Kanr-rpsL+] insertion into the ectopic CEP site (strains IU4648, IU4650 and IU4652) (see Table S1), restoring Sm sensitivity. The cellular locations of SecA-L-FLAG3, HtrA-L-FLAG3, and SrtA-L-FLAG3 were the same in the rpsL+/rpsL1 complemented strains as in the rpsL1 background (data not shown). We conclude that the rpsL1 allele did not influence the location patterns of these proteins.
Flotillin family (also designated SPFH or PHB) proteins copurify with SecA in a detergent-resistant membrane (DRM) fraction of S. aureus (31). Therefore, we investigated whether the locations of SecASpn, HtrASpn, and SrtASpn were altered in mutants lacking the pneumococcal homologues of the flotillin family proteins. Two genes in S. pneumoniae strain D39, spd_1962 and spd_1984, encode proteins containing PHB domains, identified by the SMART program (http://smart.embl-heidelberg.de/). Knockout of spd_1962, spd_1984, or both spd_1962 and spd_1984 (see Table S1 in the supplemental material) did not result in any overt growth or cell morphology defects under the conditions tested so far (data not shown). In addition, SecA-L-FLAG3, HtrA-L-FLAG3, and SrtA-L-FLAG3 showed wild-type localization patterns in the single spd_1962 and spd_1984 mutants and the double spd_1962 spd_1984 mutant (data not shown). We conclude that the putative pneumococcal flottilin proteins do not influence cell division or cellular location of SecASpn, HtrASpn, and SrtASpn.
Last, we tested whether SecASpn or HtrASpn localization depended on SrtASpn and whether SecASpn and SrtASpn location depended on HtrASpn. SecA-L-FLAG3 and HtrA-L-FLAG3 showed wild-type localization patterns in an srtASpn mutant (data not shown) (Fig. 1 and 4; see Table S1 in the supplemental material). Likewise, the localization pattern of SecA-L-FLAG3 and the random distribution of SrtA-L-FLAG3 were the same in an htrASpn mutant as in the isogenic htrA+ parent (data not shown) (Fig. 1; see Fig. S5 and Table S1 in the supplemental material). The SecASpn gene is essential and could not be knocked out in these experiments. We conclude that SecASpn and HtrASpn localization does not depend on the presence of SrtASpn, and SecASpn and SrtASpn localization does not depend on HtrASpn.
DISCUSSION
We show herein that the SecA and SecY translocase subunits of S. pneumoniae localize to mid-cell regions of actively dividing cells of S. pneumoniae (Fig. 1). In predivisional cells, SecASpn and SecYSpn are primarily located at equators, which will develop into divisional septa. Some predivisional cells also have SecASpn and SecYSpn at one pole, possibly as remnants of the previous division. As division progresses and septation begins, both Sec subunits cover the new hemispheres of cells in a distinctive “χ” (Chi)-like distribution between the septa and future equators of the dividing cell. This “χ” distribution is maintained in newly divided cells, until SecASpn and SecYSpn relocate to equators. This localization of the Sec translocase subunits overlaps the regions of peptidoglycan (PG) biosynthesis in dividing pneumococcal cells (see references 21 and 24). Localization of SecASpn and SecYSpn to equators and septa is lost as cells stop growing and transition to stationary phase (Fig. 3) (data not shown), which eventually induces autolysis of pneumococcal cultures.
Notably, the localization of SecA in S. pneumoniae reported here (Fig. 1 and 3) is markedly different from the concentration of SecA at a single ExPortal microdomain reported previously for S. pyogenes (5, 6) and E. faecalis (7). There are two possible explanations for this difference. Although all are ovococcus species, S. pneumoniae is evolutionarily distant from both S. pyogenes and E. faecalis (14), and it is possible that different mechanisms of protein export are used, especially by species like S. pyogenes that secrete numerous toxins and superantigens (17). On the other hand, the difference may reflect the methods of detection. This study used IFM to locate Sec subunits in a Streptococcus species for the first time, whereas in previous studies, SecASpy and E. faecalis SecA (SecAEfa) were detected using IG-EM techniques (5, 7, 8). It is possible that the thin sectioning or high levels of fixation used in IG-EM methods did not allow the entire distribution pattern of SecASpy and SecAEfa to be detected (see reference 32). Consistent with this interpretation, other studies using the IG-EM method found SecASpy distributed over the entire surface of cells (10), similar to the pattern reported here for SecASpn in transition-phase cells (Fig. 3). Moreover, the IFM approach used here has now located several pneumococcal cytoplasmic and surface proteins in different patterns, including FtsZ, whose localization was already established (Fig. 1, 3, and 4; see Fig. S2 and S4 to S6 in the supplemental material) (21, 22, 33–35). These diverse localization patterns of different proteins argue that the IFM method is allowing sufficient access of antibody to target proteins fused to the FLAG3 epitope. Taken together, our results do not support the presence of an ExPortal in S. pneumoniae and indicate a dynamic distribution of the Sec translocation apparatus.
To attain their ellipsoid shape, pneumococcal cells use two modes of PG biosynthesis that extend the side walls from mid-cells and close septa between daughter cells, respectively (see references 21 and 24). Consequently, cell division proteins, including FtsZSpn (see Fig. S6 in the supplemental material), MreCSpn(21), penicillin binding proteins (e.g., PBP2bSpn and PBP2xSpn) (24), and PcsBSpn (L.-T. Sham, unpublished data), and regions of active PG biosynthesis revealed by fluorescent vancomycin staining localize to equators, septa, and mid-cell regions (see references 19 and 21), similar to SecASpn and SecYSpn. The SecA motor protein plays roles in exporting proteins from cells and in secreting large hydrophilic domains of membrane-anchored proteins into the extracellular space (reviewed in reference 1). Many proteins that carry out or mediate PG biosynthesis, such as the penicillin binding proteins (PBPs), MreC, DivIB, and carboxypeptidase (see references 24, 33, and 36), contain single transmembrane domains or membrane anchors linked to large extracellular domains, which are likely exported in a SecA-dependent manner. Therefore, an attractive model that emerges is that the Sec translocase of S. pneumoniae directs secretion of the PG biosynthesis apparatus to regions where PG biosynthesis occurs. The protein interactions that direct SecASpn and SecYSpn to equators, septa, and mid-cell regions are currently under investigation.
S. pneumoniae serotype 2 strain D39 used in this study contains only the SecA general Sec translocase pathway for protein export (20). Strain D39 lacks the accessory SecA2 system found in some serotypes of S. pneumoniae (37). In addition, the twin-arginine transporter (38) is absent from S. pneumoniae. Besides PG biosynthetic proteins, 5% of the 1,914 polypeptides encoded by strain D39 are predicted to contain signal peptides according to the LocateP program (http://www.cmbi.ru.nl/locatep-db/cgi-bin/locatepdb.py) (18). These proteins include secreted proteins, lipid-anchored glycoproteins, including 28 substrate binding proteins of ABC transporters, and 12 of the 16 LPXTG motif-containing proteins that are anchored to the PG by sortase. Previous work with S. aureus (11) and S. pyogenes (10) suggests that signal peptides containing a YSIRK/GS amino acid motif direct the proteins to division septa. In contrast, sortase-attached proteins lacking this motif end up at cell poles (10, 11).
Six of the sortase-attached proteins of strain D39 contain sequences related to the YSIRK/GS motif. The localization patterns of SecASpn, SecYSpn, and SrtASpn reported here could accommodate two modalities of localization of sortase-attached proteins. In exponentially growing cells, SecASpn and SecYSpn are located over hemispheres fanning out from division septa (Fig. 1; see Fig. S2 in the supplemental material), whereas SrtASpn is distributed over the entire surface of cells (see Fig. S5 in the supplemental material), similar to a fraction of the SrtA in S. pyogenes (9). However, even in preliminary experiments that localize SrtA-L-FLAG3 by 3D imaging (data not shown), we did not detect any patterns of SrtASpn localization or preferential association with septa, as reported for SrtASpy(9). Nevertheless, the overlap between SecASpn, SecYSpn, and SrtASpn in hemispheres of dividing cells (Fig. 1; see Fig. S2 and S5 in the supplemental material) would allow sortase-attached proteins to be directed to septa. In newly divided cells, SecASpn and SecYSpn remain momentarily at cell poles (Fig. 1; see Fig. S2), and SecASpn and SecYSpn spread over surfaces as cells stop dividing and enter stationary phase (Fig. 3). Either of these configurations would allow sortase-attached proteins lacking the YSIRK/GS motif to be localized to poles. Experiments are in progress to determine the localization of sortase-attached proteins in S. pneumoniae.
Unexpectedly, we found that the HtrASpn protein is predominantly located at the equators and septa of dividing pneumococcal cells (Fig. 4), largely coincident with SecASpn and SecYSpn (Fig. 1; see Fig. S2 in the supplemental material). HtrASpn is a member of a widely conserved family of serine proteases that are crucial for protein homeostasis in extracytoplasmic compartments (39). DegP of E. coli is the most studied member of this family and exhibits antagonistic functions as a chaperone and protease within a single polypeptide (40, 41). The substrates of HtrASpn are unknown, but htrA mutants of S. pneumoniae show increased sensitivity to thermal and oxidative stress (25), decreased nasopharyngeal colonization (42) and virulence (25), and increased bacteriocin activity (43). Additionally, htrASpn appears to influence expression of pneumococcal competence (see reference 44) and is a member of the regulon controlled by the CiaRH two-component system, which responds to antibiotics that inhibit PG biosynthesis (45). The localization of HtrASpn to regions of PG biosynthesis in unstressed, exponentially growing cells, coincident with the localization of SecASpn and SecYSpn, suggests that HtrASpn may play a general role in the quality control of proteins exported by the Sec translocase in S. pneumoniae. The basis for HtrASpn localization to division sites is unknown.
Finally, normal anionic lipid content was required for localization of SecASpn and, to a lesser extent, HtrASpn to cell equators and division septa (Fig. 2; see Fig. S4 in the supplemental material). Previously, the anionic lipids phosphatidylglycerol (PGly) and cardiolipin (CL) were shown to play important roles in protein translocation across the membranes of other bacterial species (26). SecA has a high affinity for anionic lipids (46), and its ATPase activity is enhanced by the presence of acidic phospholipids (47). CL and, to a lesser extent, PGly promote the association of SecA and SecYEG and prime translocation (3). Consistent with these biochemical results in other bacteria, S. pneumoniae ∆cls (CL synthase) mutants, which lack CL and possibly accumulate PGly, showed mislocalization of SecASpn and HtrASpn in growing cells (Fig. 2; see Fig. S4). This mislocalization is reminiscent of the rapid disruption of B. subtilis SecA (SecABsu) spirals in B. subtilis cells depleted of anionic lipids (4). ∆cls mutants of B. subtilis and several other bacterial species do not show growth defects under nonstressed conditions and may contain biosynthetic routes that compensate for loss of CL synthesis (3, 27, 48, 49). In contrast, S. pneumoniae D39 ∆cls mutants are impaired for growth and form chains of misshapen cells that readily lyse (see Fig. S3 and S4 in the supplemental material). The defective growth and SecASpn localization phenotypes of ∆cls mutants were largely complemented by ectopic expression of the cls+ gene (see Fig. S3 and S4). It remains to be determined whether the impaired growth and fragility of pneumococcal ∆cls mutants are caused by decreased function and localization of the Sec translocase.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Unencapsulated derivatives of Streptococcus pneumoniae serotype 2 strain D39 were used in this study (20). Strains containing antibiotic resistance markers were constructed by transforming linear DNA amplicons synthesized by overlapping fusion PCR into competent pneumococcal cells, as described previously (29). All intermediate and final strains synthesized are listed in Table S1 in the supplemental material, and primers used for the generation of the amplicons are listed in Table S2 in the supplemental material. Strains containing markerless secA-L-FLAG3, secY-L-FLAG3, htrA-L-FLAG3, srtA-L-FLAG3, or ∆cls alleles in native gene loci (see Table S1) were generated using the Pc-[Kanr-rpsL+] (Janus cassette [50]) allele replacement method as described in reference 35. Intermediate strains IU4370 (∆htrA::Pc-[Kanr-rpsL+]) and IU4372 (∆srtA::Pc-[Kanr-rpsL+]) were constructed with the deletion of htrA and srtA reading frames, except for the 60 bp at the 5′ and 3′ ends, and replaced with the Pc-[Kanr-rpsL+] cassette. Since secA and secY are essential genes, the intermediate strains IU4404 (secA+ intergenic::Pc-[Kanr-rpsL+]) and IU5445 (secY+ intergenic::Pc-[Kanr-rpsL+]) were constructed by the insertion of the Pc-[Kanr-rpsL+] cassette after the secA and secY coding sequences. Strains were grown on plates containing Trypticase soy agar II (TSAII; modified) (Becton-Dickinson; BD) and 5% (vol/vol) defibrinated sheep blood (TSAII BA) and incubated at 37°C in an atmosphere of 5% CO2 (mixed with 95% of the normal atmospheric composition). For antibiotic selections, TSAII BA plates were supplemented with 250 µg kanamycin per ml, 250 µg streptomycin per ml, or 0.3 µg erythromycin per ml. For liquid cultures, strains were cultured statically in BD brain heart infusion (BHI) broth at 37°C or 41.2°C (to test for temperature sensitivity) in an atmosphere of 5% CO2.
Localization of SecA-L-FLAG3, SecY-L-FLAG3, HtrA-L-FLAG3, SrtA-L-FLAG3, and FtsZ-FLAG3 by immunofluorescence microscopy (IFM).
IFM localization of FLAG3-tagged proteins using anti-FLAG polyclonal antibody (Sigma, F7425) and nucleoid staining with 4′,6-diamidino-2-phenylindole (DAPI) were performed as described previously (35). Control experiments repeatedly showed no detectable labeling of cells that did not express a protein fused to the FLAG epitope tag (data not shown). Strains were cultured in BHI broth to mid-exponential phase (OD620 of 0.13) or to the transition to stationary phase (OD620 of 0.78 15 min before reaching the maximal culture OD620 of 0.82). Stained cells were examined using a Nikon E-400 epifluorescence phase-contrast microscope. Images were captured using a CoolSNAP HQ2 charge-coupled device (CCD) camera (Photometrics) and processed with NIS-Elements AR imaging software (Nikon). For the 300-ms exposure time used, micrographic images of control strain IU1824 (D39 ∆cps not expressing a FLAG3-tagged protein) did not show detectable labeling. The background pixel intensity corresponded to 1,000 arbitrary units (AU) (dynamic range, 0 to 16,383), and there was no difference in background pixel intensities between areas within cells or outside cells. Micrographs of strains expressing SecA-L-FLAG3, SecY-L-FLAG3, HtrA-L-FLAG3, or SrtA-L-FLAG3 showed regions of intense labeling (Fig. 1, 3, and 4; see Fig. S2 and S4 to S6 in the supplemental material), corresponding to maximal pixel intensities ranging from 4,000 to 15,000 AU. In comparison, the pixel intensities in regions of cells not showing labeling in micrographs was close to (i.e., 1,200 AU) the background level. Validation experiments by this IFM approach using a strain expressing FtsZ-FLAG3 showed patterns of FtsZSpn localization during cell division (see Fig. S6) similar to those reported previously using anti-FtsZSpn antibody (22, 34).
Determination of distribution pattern frequencies.
Unencapsulated derivatives of strain D39 grown in BHI broth, which form mainly single cells and diplococci and some short chains of 4 cells (19), were sorted into different stages of division. For each experiment, 50 to 100 cells from multiple phase-contrast microscopic fields of each strain were binned as single cells (oval cells), early divisional cells (slight indentation at mid-cell), or late-divisional cells (width of septum less than half of cell width). Images of immunofluorescently labeled cells using anti-FLAG antibody were then superimposed on phase-contrast images. Cells in each stage of division were then scored for protein localization at equators, septa, poles, hemispheres, or elsewhere. Similar localization patterns were observed in at least five independent experiments, and quantitative data were obtained from two separate experiments.
SUPPLEMENTAL MATERIAL
Supplemental Materials and Methods. Download Text S1, DOC file, 0.1 MB.
Supplemental results from control experiments. Download Text S2, DOC file, 0.1 MB.
Native genetic loci of ftsZ-FLAG3 Pc-erm (A), SecA-L-FLAG3 (B), secY-L-FLAG3 (C), htrA-L-FLAG3 (D), and srtA-L-FLAG3 (E) and their neighboring genes in the Streptococcus pneumoniae D39 chromosome. Pc-erm cassette sequence was added after ftsZ-FLAG3 as an antibiotic resistance marker for cotransformation. Strains containing markerless SecA-L-FLAG3, secY-L-FLAG3, htrA-L-FLAG3, and srtA-L-FLAG3 alleles in the respective native loci (see Fig. S1 and Table S1 in the supplemental material) were generated using the Pc-[Kanr-rpsL+] (Janus cassette) allele replacement method as described in Materials and Methods. Short sequences from bgaA, SecA, secY, htrA, or srtA were added for primer design reasons. Download Figure S1, PDF file, 0.1 MB.
SecY-L-FLAG3 localizes primarily to equators, septa, and hemispheres of exponentially growing S. pneumoniae cells in a pattern similar to that of SecA-L-FLAG3 (Fig. 1). Strain IU5471 (D39 ∆cps rpsL1 secY-L-FLAG3) was grown to mid-exponential phase in BHI broth, and DAPI staining and IFM of cells were performed as described in Materials and Methods. Cells at different stages of division were binned based on phase-contrast microscopy images (column 1), and then localization patterns of DAPI (pseudocolored red; column 2) and SecY-L-FLAG3 (pseudocolored green; column 3) were determined from images obtained with epifluorescent microscopy. Columns 4 and 5 show overlays of columns 1 and 3 or columns 1 and 2, respectively. Images of representative cells are shown at different stages of division. A schematic summary is given at right showing the SecYSpn distribution pattern in ≈75% of the cell population (see Results). The experiment was performed 2 times independently. Multiple fields were observed in each experiment, with similar results. Scale bar = 1 µm. Download Figure S2, PDF file, 0.1 MB.
Typical growth curves of unencapsulated parent strain D39 SecA-L-FLAG3 (IU4472), the isogenic ∆cls::Pc-erm strain (IU5093), and the ∆cls::Pc-erm/CEP::PfcsK-cls+ strain (complementation strain IU5138) in BHI broth at 37°C with limited aeration (static) in an atmosphere of 5% CO2 and 95% atmospheric composition (Materials and Methods). In IU5138, cls is expressed under the control of the PfcsK promoter in the ectopic CEP site. Growth of strain IU5138 was similar in BHI broth containing 0, 0.2, or 0.8% fucose. The growth curve without fucose is shown in this figure. Download Figure S3, PDF file, 0.1 MB.
Distribution of SecA-L-FLAG3 in (A) IU4472 (unencapsulated parent strain D39 SecA-L-FLAG,3) (B) isogenic ∆cls::Pc-erm strain IU5093, and (C) ∆cls::Pc-erm//CEP::PfcsK-cls+ strain IU5138 (complementation strain) grown in BHI broth at 37°C in exponential phase (OD620 of 0.1). Columns 1 and 2 are phase-contrast images of fixed cells and IFM of the FLAG-tagged SecA proteins using anti-FLAG antibody. Column 3 shows overlays of columns 1 and 2, and column 4 shows overlays of IFM (pseudocolored green) and DAPI-stained (pseudocolored red) images. The ∆cls strain (B) is characterized by chaining of cells, and cell lysis (block arrows in column 1) within a chain. Yellow arrows indicate a hemispheric labeling pattern, and white arrows indicate a mid-cell staining pattern with anti-FLAG. Scale bar = 1 µm. Download Figure S4, PDF file, 0.2 MB.
Apparently random punctate distribution of SrtA-L-FLAG3 over surfaces of strain IU4470 (D39 ∆cps srtA-L-FLAG3) cells grown to mid-exponential phase. Carboxyl-terminal fusion of the chromosomally expressed proteins to the linker (L) and FLAG epitope tags (see Table S1 in the supplemental material) did not cause any observable changes in cell morphology, growth, or β-galactosidase distribution compared to the srtA+ parent strain (column 1 and see Text S2 in the supplemental material). The column arrangement is the same as in Fig. 1. Rows 1 to 4 represent predivisional cells, early and late-divisional cells, and postdivisional diplococci, respectively. No readily discernable pattern was recognized by visual inspection of cells in each divisional stage. More than 3 biological replicates were performed, and multiple fields of cells were observed by IFM and analyzed as described in Materials and Methods. Scale bar = 1 µm. Download Figure S5, PDF file, 0.1 MB.
Distribution of FtsZ-FLAG3 in strain IU4368 (D39 ∆cps ftsZ-FLAG3) at different stages of cell division during exponential growth. Columns 1 to 3 are phase-contrast images of fixed cells, DAPI-stained images (pseudocolored red) to locate nucleoids, and IFM (pseudocolored green) of the FLAG-tagged FtsZ proteins using anti-FLAG antibody, respectively. Columns 4 and 5 show overlays of columns 1 and 3 and overlays of columns 1 and 2, respectively. Cells are arranged from rows 1 to 6 to represent the progression through the cell cycle: predivisional cells with undivided and duplicated nucleoids in rows 1 and 2, early and late-divisional cells in rows 3 and 4, a postdivisional diplococcus in row 5, and a diplococcus with two predivisional daughter cells in row 6. Scale bar = 1 µm. Download Figure S6, PDF file, 0.1 MB.
Bacterial strains used in this study
Oligonucleotides used in this study
ACKNOWLEDGMENTS
We thank Krystyna Kazmierczak for discussions and information.
This work was supported in part by grant RO1AI060744 from the National Institute of Allergy and Infectious Diseases to M.E.W. Several pneumococcal mutants were constructed with support from grant RO1GM085697 from the National Institute of General Medical Science to Carol A. Gross. K.J.W. was a predoctoral trainee supported by training grant T32GM007757 from the National Institutes of Health.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the granting agencies.
Footnotes
Citation Tsui H-CT, Keen SK, Sham L-T, Wayne KJ, Winkler ME. 2011. Dynamic distribution of the SecA and SecY translocase subunits and septal localization of the HtrA surface chaperone/protease during Streptococcus pneumoniae D39 cell division. mBio 2(5):e00202-11. doi:10.1128/mBio.00202-11.
REFERENCES
- 1. du Plessis DJ, Nouwen N, Driessen AJ. 2011. The Sec translocase. Biochim. Biophys. Acta 1808:851–865 [DOI] [PubMed] [Google Scholar]
- 2. Sardis MF, Economou A. 2010. SecA: a tale of two protomers. Mol. Microbiol. 76:1070–1081 [DOI] [PubMed] [Google Scholar]
- 3. Gold VA, et al. 2010. The action of cardiolipin on the bacterial translocon. Proc. Natl. Acad. Sci. U. S. A. 107:10044–10049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campo N, et al. 2004. Subcellular sites for bacterial protein export. Mol. Microbiol. 53:1583–1599 [DOI] [PubMed] [Google Scholar]
- 5. Rosch J, Caparon M. 2004. A microdomain for protein secretion in Gram-positive bacteria. Science 304:1513–1515 [DOI] [PubMed] [Google Scholar]
- 6. Rosch JW, Caparon MG. 2005. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol. Microbiol. 58:959–968 [DOI] [PubMed] [Google Scholar]
- 7. Kline KA, et al. 2009. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J. Bacteriol. 191:3237–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu P, Bian Z, Fan M, Huang M, Zhang P. 2008. Sec translocase and sortase A are colocalised in a locus in the cytoplasmic membrane of Streptococcus mutans. Arch. Oral Biol. 53:150–154 [DOI] [PubMed] [Google Scholar]
- 9. Raz A, Fischetti VA. 2008. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc. Natl. Acad. Sci. U. S. A. 105:18549–18554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carlsson F, et al. 2006. Signal sequence directs localized secretion of bacterial surface proteins. Nature 442:943–946 [DOI] [PubMed] [Google Scholar]
- 11. DeDent A, Bae T, Missiakas DM, Schneewind O. 2008. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 27:2656–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Henriques-Normark B, Normark S. 2010. Commensal pathogens, with a focus on Streptococcus pneumoniae, and interactions with the human host. Exp. Cell Res. 316:1408–1414 [DOI] [PubMed] [Google Scholar]
- 13. van der Poll T, Opal SM. 2009. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet 374:1543–1556 [DOI] [PubMed] [Google Scholar]
- 14. Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73:407–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Banas JA. 2004. Virulence properties of Streptococcus mutans. Front. Biosci. 9:1267–1277 [DOI] [PubMed] [Google Scholar]
- 16. Kadioglu A, Weiser JN, Paton JC, Andrew PW. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6:288–301 [DOI] [PubMed] [Google Scholar]
- 17. Musser JM, Shelburne SA., III 2009. A decade of molecular pathogenomic analysis of group A Streptococcus. J. Clin. Invest. 119:2455–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou M, Boekhorst J, Francke C, Siezen RJ. 2008. LocateP: genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinform. 9:173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barendt SM, et al. 2009. Influences of capsule on cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae. J. Bacteriol. 191:3024–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lanie JA, et al. 2007. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Land AD, Winkler ME. 2011. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J. Bacteriol. 193:4166–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morlot C, Zapun A, Dideberg O, Vernet T. 2003. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol. Microbiol. 50:845–855 [DOI] [PubMed] [Google Scholar]
- 23. Pérez-Núñez D, et al. 2011. A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol. Microbiol. 79:759–771 [DOI] [PubMed] [Google Scholar]
- 24. Zapun A, Vernet T, Pinho MG. 2008. The different shapes of cocci. FEMS Microbiol. Rev. 32:345–360 [DOI] [PubMed] [Google Scholar]
- 25. Ibrahim YM, Kerr AR, McCluskey J, Mitchell TJ. 2004. Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect. Immun. 72:3584–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Vrije T, de Swart RL, Dowhan W, Tommassen J, de Kruijff B. 1988. Phosphatidylglycerol is involved in protein translocation across Escherichia coli inner membranes. Nature 334:173–175 [DOI] [PubMed] [Google Scholar]
- 27. Rosch JW, Hsu FF, Caparon MG. 2007. Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J. Bacteriol. 189:801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guiral S, et al. 2006. Construction and evaluation of a chromosomal expression platform (CEP) for ectopic, maltose-driven gene expression in Streptococcus pneumoniae. Microbiology 152:343–349 [DOI] [PubMed] [Google Scholar]
- 29. Tsui HC, et al. 2010. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 192:264–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chan PF, et al. 2003. Characterization of a novel fucose-regulated promoter (PfcsK) suitable for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae. J. Bacteriol. 185:2051–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. López D, Kolter R. 2010. Functional microdomains in bacterial membranes. Genes Dev. 24:1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oliver C, Jamur MC. 2010. Fixation and embedding. Methods Mol. Biol. 588:353–362 [DOI] [PubMed] [Google Scholar]
- 33. Barendt SM, Sham LT, Winkler ME. 2011. Characterization of mutants deficient in the L,D-carboxypeptidase (DacB) and WalRK (VicRK) regulon, involved in peptidoglycan maturation of Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 193:2290–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giefing C, Jelencsics KE, Gelbmann D, Senn BM, Nagy E. 2010. The pneumococcal eukaryotic-type serine/threonine protein kinase StkP co-localizes with the cell division apparatus and interacts with FtsZ in vitro. Microbiology 156:1697–1707 [DOI] [PubMed] [Google Scholar]
- 35. Wayne KJ, et al. 2010. Localization and cellular amounts of the WalRKJ (VicRKX) two-component regulatory system proteins in serotype 2 Streptococcus pneumoniae. J. Bacteriol. 192:4388–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Claessen D, et al. 2008. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 68:1029–1046 [DOI] [PubMed] [Google Scholar]
- 37. Rigel NW, Braunstein M. 2008. A new twist on an old pathway—accessory Sec systems. Mol. Microbiol. 69:291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. HTRA proteases: regulated proteolysis in protein quality control. Nat. Rev. Mol. Cell Biol. 12:152–162 [DOI] [PubMed] [Google Scholar]
- 40. Krojer T, et al. 2008. Structural basis for the regulated protease and chaperone function of DegP. Nature 453:885–890 [DOI] [PubMed] [Google Scholar]
- 41. Spiess C, Beil A, Ehrmann M. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339–347 [DOI] [PubMed] [Google Scholar]
- 42. Sebert ME, Palmer LM, Rosenberg M, Weiser JN. 2002. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70:4059–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dawid S, Sebert ME, Weiser JN. 2009. Bacteriocin activity of Streptococcus pneumoniae is controlled by the serine protease HtrA via posttranscriptional regulation. J. Bacteriol. 191:1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Halfmann A, Kovács M, Hakenbeck R, Brückner R. 2007. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol. 66:110–126 [DOI] [PubMed] [Google Scholar]
- 45. Mascher T, Heintz M, Zähner D, Merai M, Hakenbeck R. 2006. The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and by mutations in pbp2x involved in beta-lactam resistance. J. Bacteriol. 188:1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ulbrandt ND, London E, Oliver DB. 1992. Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding. J. Biol. Chem. 267:15184–15192 [PubMed] [Google Scholar]
- 47. Lill R, Dowhan W, Wickner W. 1990. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell 60:271–280 [DOI] [PubMed] [Google Scholar]
- 48. López CS, Alice AF, Heras H, Rivas EA, Sánchez-Rivas C. 2006. Role of anionic phospholipids in the adaptation of Bacillus subtilis to high salinity. Microbiology 152:605–616 [DOI] [PubMed] [Google Scholar]
- 49. Tsai M, et al. 2011. Staphylococcus aureus requires cardiolipin for survival under conditions of high salinity. BMC Microbiol. 11:13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods. Download Text S1, DOC file, 0.1 MB.
Supplemental results from control experiments. Download Text S2, DOC file, 0.1 MB.
Native genetic loci of ftsZ-FLAG3 Pc-erm (A), SecA-L-FLAG3 (B), secY-L-FLAG3 (C), htrA-L-FLAG3 (D), and srtA-L-FLAG3 (E) and their neighboring genes in the Streptococcus pneumoniae D39 chromosome. Pc-erm cassette sequence was added after ftsZ-FLAG3 as an antibiotic resistance marker for cotransformation. Strains containing markerless SecA-L-FLAG3, secY-L-FLAG3, htrA-L-FLAG3, and srtA-L-FLAG3 alleles in the respective native loci (see Fig. S1 and Table S1 in the supplemental material) were generated using the Pc-[Kanr-rpsL+] (Janus cassette) allele replacement method as described in Materials and Methods. Short sequences from bgaA, SecA, secY, htrA, or srtA were added for primer design reasons. Download Figure S1, PDF file, 0.1 MB.
SecY-L-FLAG3 localizes primarily to equators, septa, and hemispheres of exponentially growing S. pneumoniae cells in a pattern similar to that of SecA-L-FLAG3 (Fig. 1). Strain IU5471 (D39 ∆cps rpsL1 secY-L-FLAG3) was grown to mid-exponential phase in BHI broth, and DAPI staining and IFM of cells were performed as described in Materials and Methods. Cells at different stages of division were binned based on phase-contrast microscopy images (column 1), and then localization patterns of DAPI (pseudocolored red; column 2) and SecY-L-FLAG3 (pseudocolored green; column 3) were determined from images obtained with epifluorescent microscopy. Columns 4 and 5 show overlays of columns 1 and 3 or columns 1 and 2, respectively. Images of representative cells are shown at different stages of division. A schematic summary is given at right showing the SecYSpn distribution pattern in ≈75% of the cell population (see Results). The experiment was performed 2 times independently. Multiple fields were observed in each experiment, with similar results. Scale bar = 1 µm. Download Figure S2, PDF file, 0.1 MB.
Typical growth curves of unencapsulated parent strain D39 SecA-L-FLAG3 (IU4472), the isogenic ∆cls::Pc-erm strain (IU5093), and the ∆cls::Pc-erm/CEP::PfcsK-cls+ strain (complementation strain IU5138) in BHI broth at 37°C with limited aeration (static) in an atmosphere of 5% CO2 and 95% atmospheric composition (Materials and Methods). In IU5138, cls is expressed under the control of the PfcsK promoter in the ectopic CEP site. Growth of strain IU5138 was similar in BHI broth containing 0, 0.2, or 0.8% fucose. The growth curve without fucose is shown in this figure. Download Figure S3, PDF file, 0.1 MB.
Distribution of SecA-L-FLAG3 in (A) IU4472 (unencapsulated parent strain D39 SecA-L-FLAG,3) (B) isogenic ∆cls::Pc-erm strain IU5093, and (C) ∆cls::Pc-erm//CEP::PfcsK-cls+ strain IU5138 (complementation strain) grown in BHI broth at 37°C in exponential phase (OD620 of 0.1). Columns 1 and 2 are phase-contrast images of fixed cells and IFM of the FLAG-tagged SecA proteins using anti-FLAG antibody. Column 3 shows overlays of columns 1 and 2, and column 4 shows overlays of IFM (pseudocolored green) and DAPI-stained (pseudocolored red) images. The ∆cls strain (B) is characterized by chaining of cells, and cell lysis (block arrows in column 1) within a chain. Yellow arrows indicate a hemispheric labeling pattern, and white arrows indicate a mid-cell staining pattern with anti-FLAG. Scale bar = 1 µm. Download Figure S4, PDF file, 0.2 MB.
Apparently random punctate distribution of SrtA-L-FLAG3 over surfaces of strain IU4470 (D39 ∆cps srtA-L-FLAG3) cells grown to mid-exponential phase. Carboxyl-terminal fusion of the chromosomally expressed proteins to the linker (L) and FLAG epitope tags (see Table S1 in the supplemental material) did not cause any observable changes in cell morphology, growth, or β-galactosidase distribution compared to the srtA+ parent strain (column 1 and see Text S2 in the supplemental material). The column arrangement is the same as in Fig. 1. Rows 1 to 4 represent predivisional cells, early and late-divisional cells, and postdivisional diplococci, respectively. No readily discernable pattern was recognized by visual inspection of cells in each divisional stage. More than 3 biological replicates were performed, and multiple fields of cells were observed by IFM and analyzed as described in Materials and Methods. Scale bar = 1 µm. Download Figure S5, PDF file, 0.1 MB.
Distribution of FtsZ-FLAG3 in strain IU4368 (D39 ∆cps ftsZ-FLAG3) at different stages of cell division during exponential growth. Columns 1 to 3 are phase-contrast images of fixed cells, DAPI-stained images (pseudocolored red) to locate nucleoids, and IFM (pseudocolored green) of the FLAG-tagged FtsZ proteins using anti-FLAG antibody, respectively. Columns 4 and 5 show overlays of columns 1 and 3 and overlays of columns 1 and 2, respectively. Cells are arranged from rows 1 to 6 to represent the progression through the cell cycle: predivisional cells with undivided and duplicated nucleoids in rows 1 and 2, early and late-divisional cells in rows 3 and 4, a postdivisional diplococcus in row 5, and a diplococcus with two predivisional daughter cells in row 6. Scale bar = 1 µm. Download Figure S6, PDF file, 0.1 MB.
Bacterial strains used in this study
Oligonucleotides used in this study