Abstract
Next-generation sequencing (NGS) has not been applied to protein-protein interactome network mapping so far because the association between the members of each interacting pair would not be maintained in en masse sequencing. We describe a massively parallel interactome-mapping pipeline, “Stitch-Seq”, that combines PCR stitching with NGS. We use Stitch-Seq to generate a new human interactome dataset. Stitch-Seq is applicable to various interaction assays and should help expand interactome network mapping.
At any time hundreds of thousands of macromolecular interactions occur in a cell, mediating functions that maintain normal cellular activities. High-throughput approaches have been developed to determine interactions in many organisms at large scale. Current high-throughput protein-protein “interactome” datasets are of high quality, but have low coverage1,2. For humans, more than 95% of the interactome remains to be mapped1.
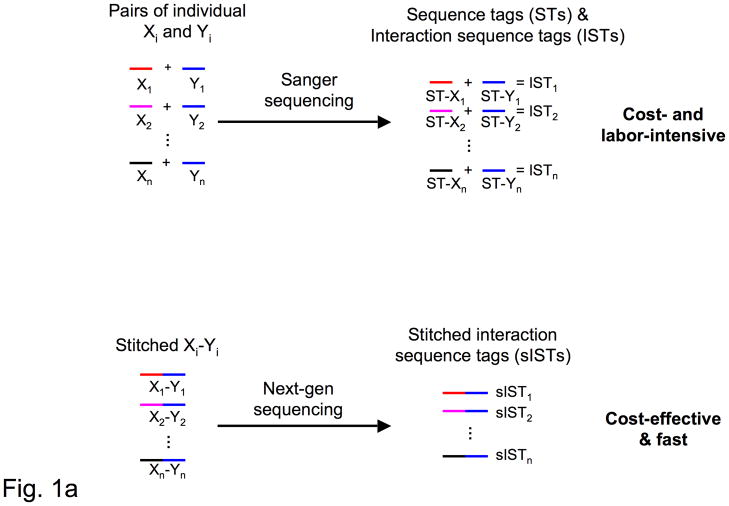
A bottleneck for high-throughput interactome mapping methods, such as yeast one-3, two-4, and three-hybrid5 systems is determining the identities of the interacting protein, DNA, or RNA molecules. Implementation of next-generation DNA sequencing (NGS) technologies6–8, as opposed to Sanger technology, would substantially increase throughput and decrease cost. Although highly effective for genome and transcriptome “shotgun” sequencing, next-generation DNA sequencing technologies are not readily applicable for identifying interacting pairs. The necessary pooling of PCR amplicons in the preparation of interacting sequence tags (ISTs) (Fig. 1a) would inevitably eliminate the association within each pair of DNA sequences coding for interacting molecules.
Figure 1.
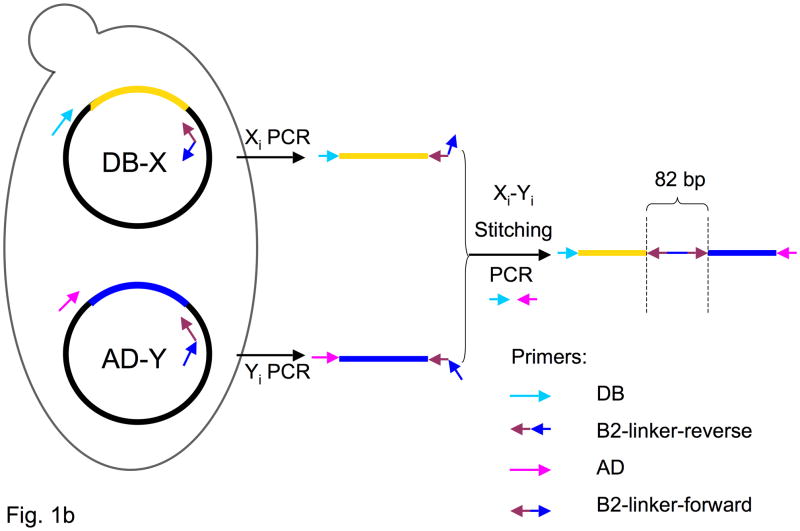
Stitch-Seq interactome mapping. (a) Interactome mapping using different sequencing technologies. Above, each DNA fragment within each interacting pair is PCR-amplified individually and Sanger sequenced. The association is tracked via position on the plate. Below, each pair of DNA fragments is placed on the same PCR amplicon by PCR-stitching. The amplicons are then collected and subjected to next-generation sequencing. (b) Outline of a PCR stitching experiment.
Here we describe a massively parallel interactome mapping strategy that incorporates NGS (Fig. 1a), and test the strategy in a high-throughput yeast two-hybrid (Y2H) system. This general scheme can be readily extended to increase throughput and decrease cost for other interactome mapping methods, particularly other binary protein-protein interaction assays1, yeast one-hybrid3, or genetic screens where pairs of DNA molecules are selected and identified9.
In current protocols of high-throughput Y2H screens, the open reading frames (ORFs) or cDNAs encoding selected pairs of interacting hybrid proteins (X fused to a DNA binding domain (DB-X) and Y fused to an activation domain (AD-Y)) are amplified directly from yeast transformants and subsequently identified by Sanger DNA sequencing (Supplementary Fig. 1)4. Since X and Y originate from recorded positions in paired PCR plates, they can be computationally re-assembled to form pairs of ISTs10.
The first step of our methodology, termed “Stitch-Seq”, is PCR stitching, which places a pair of sequences encoding interacting proteins on the same PCR amplicon, and which has previously been used to link genes encoding interacting pairs20. PCR stitching consists of two rounds of PCR (Fig. 1b). In the first round, X and Y (present on the Y2H DB-X and AD-Y) vectors are amplified with DB- and AD-vector-specific upstream primers, respectively (Supplementary Table 1a). A common sequence on the downstream primers is complementary to the Gateway-specific attB2 site immediately following the ORFs. We tested the PCR stitching concept for Y2H experiments using Gateway clones, though the approach can be generalized to other interactome mapping assays with different vectors. In the second round of PCR (Supplementary Table 1b), X and Y amplicons from the first round are used as templates to produce a concatenated PCR product composed of X and Y ORFs connected by an 82 bp linker sequence (Fig. 1b). All PCR products are then pooled and sequenced by NGS to produce stitched ISTs or “sISTs”.
Concatenated PCR products should, on average, be twice the length of single ORFs (Fig. 1b). To test the length limit of PCR stitching, we chose four DB-X and four AD-Y constructs of various ORF lengths, 500 bp, 1 kb, 2 kb, and 3 kb (Supplementary Fig. 2a). As expected the first-step colony PCR reactions succeeded at amplifying all eight ORFs (Supplementary Fig. 2b). Second-step PCR reactions tested all 16 possible combinations, with the longest combination (A4–D4) over 6 kb. Concatenated ORF pairs up to 6 kb in total length were generated efficiently and accurately (Supplementary Fig. 2c).
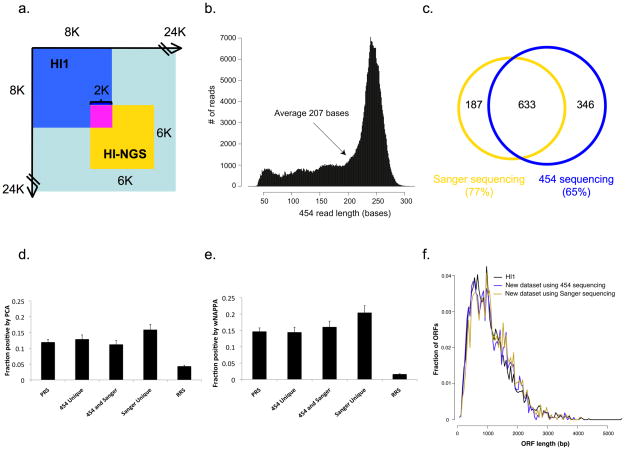
We next applied PCR stitching to pairs of ORFs identified from a Y2H screen aimed at expanding the human interactome map11. After Y2H screening of a 6K by 6K search space within the ORFeome 3.1 set of human ORFs12 (Fig. 2a) with two rounds of phenotype testing, we selected ~5,200 positive colonies. PCR stitching applied to these colonies produced ~5,000 stitched PCR amplicons. We sequenced stitched amplicons with the 454 FLX platform7, producing ~400,000 reads (Table 1). The average read length was 207 bases (Fig. 2b), which is 125 bases longer than the 82 bp linker sequence, so that many reads could unambiguously identify pairs of unique X and Y ORFs, thereby generating sISTs. To identify ORFs encoding pairs of interacting proteins, we selected reads that contain the linker sequence (~10%) and also covered at least 15 bases of ORF specific sequences on both ends of the linker. After matching these sequences to human ORFeome v3.1 (ref. 12) we identified 2,089 unique sISTs.
Figure 2.
Human interactome (“HI-NGS”) produced by massively parallel interactome mapping. (a) ORF Search spaces within the human ORFeome 3.1 space12 of HI1 (8K × 8K11 and HI-NGS (6K × 6K). (b) Length distribution of 454 reads for HI-NGS. (c) Overlaps between interactions identified by 454 FLX and Sanger sequencing. (d, e) Fraction of protein pairs in the indicated datasets that test positive by PCA (d) and wNAPPA (e). (f) Length distribution of the ORFs in HI1 and HI-NGS with 454 sequencing or Sanger sequencing.
Table 1.
Comparison of interactome mapping and IST-sIST identification numbers at key pipeline steps for Sanger sequencing and Stitch-Seq protocols.
| Sanger | 454 | |
|---|---|---|
| Search space | 1.8 * 107 | |
| Colonies | ~5200 | |
| PCR reactions | ~10,400 | ~15,600 |
| Reads | ~8,840 | 395,873 |
| Reads with linker | na | 39,211 |
| ISTs/sISTs | ~8,840 | 18,853 |
| Candidate Y2H interactions (ORF) | 1,602 | 2,089 |
| Verified Y2H interactions (ORF) | 1,032 | 1,318 |
| Verified Y2H interactions (Genes) | 820 | 979 |
| Total verified Y2H interactions | 1,166 | |
We experimentally retested by pairwise Y2H all sISTs starting from fresh yeast transformants stored in our collection and confirmed 1,318 pairs of ORFs as demonstrably encoding Y2H interacting proteins (Table 1). Because the collection contains multiple ORFs for some genes (e.g., splice isoforms), the final tally was 979 interactions among proteins encoded by 997 genes (Table 1). This confirmation rate is virtually identical to that previously described for Y2H screens using Sanger sequencing11. Furthermore, the confirmation rate does not vary between sISTs discovered uniquely and sISTs discovered multiple times (Supplementary Note 1 and Supplementary Fig. 3).
For comparison we also sequenced all of the ~5,200 positive colonies individually by Sanger sequencing, and identified 820 interactions among proteins encoded by 914 genes (Fig. 2c and Table 1). Of these, 633 interactions were also identified by 454 FLX sequencing. This overlap is higher than the expected overlap of ~70% (Supplementary Fig. 4), even taking into account a ~5% failure rate of PCR and Sanger sequencing reactions. We did detect 19% more interactions using our “Stitch-Seq” strategy, but that was probably because of the higher coverage of the 454 FLX sequencing and the inherent failure rate of Sanger sequencing.
We next quantitatively evaluated the quality of this interactome dataset based on orthogonal interaction assays1,13. We selected 94 protein pairs at random from all verified interactions that were identified by: 1) only 454 FLX sequencing (“454 Unique”); 2) only Sanger sequencing (“Sanger Unique”); or 3) both (“454 and Sanger”) (Fig. 2c). We combined these 282 interactions with positive and random reference set interactions (PRS/RRS) consisting of 92 interactions each2,13, which serve to benchmark assay performance13. We tested the 466 pairs by two assays orthogonal to Y2H: a protein complementation assay (PCA)13 and a modified version of the nucleic acid programmable protein array (wNAPPA)13. In all three groups the detection rate of new interactions was statistically indistinguishable from the PRS detection rate of both PCA and wNAPPA (all P values > 0.2), and significantly higher than that of the RRS pairs (all P values < 0.001) (Fig. 2d, e). PRS interactions in the search space were recovered at the expected rate and no RRS pair was found (Supplementary Note 2). Because shorter products can amplify more efficiently than longer ones by PCR, our stitching scheme might have favored identification of shorter ORFs, but the size distributions of ORFs, as determined by both 454 FLX and Sanger sequencing, were identical to that of the ORFs in the previous human interactome version 1 (HI1)11 (Fig. 2f). Thus, large numbers of high-quality sISTs can be identified in a single next-generation sequencing reaction.
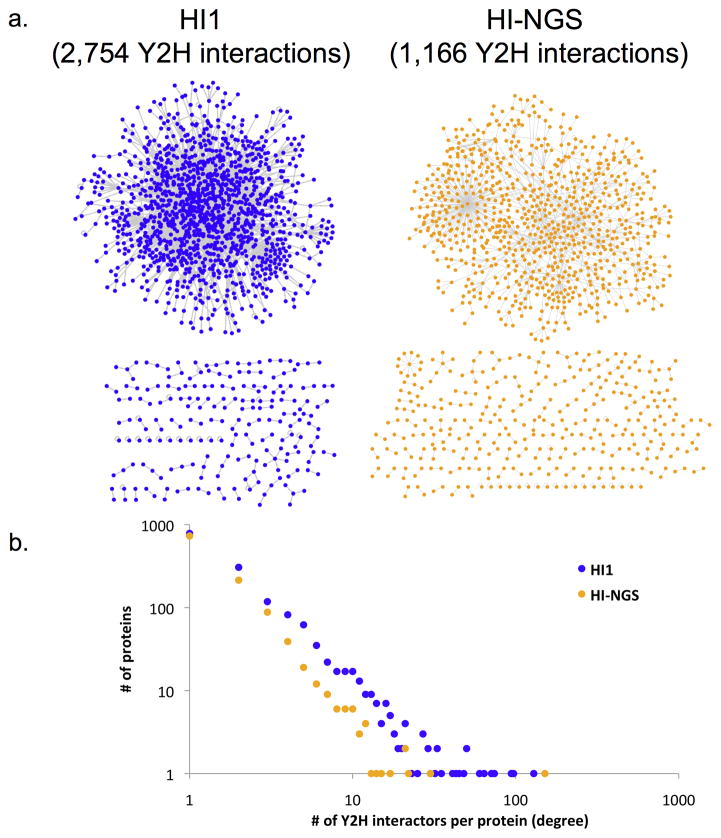
Combining 454 and Sanger sequencing results produced a high-quality human interactome dataset, HI-NGS (Human Interactome produced with Next-Generation Sequencing) containing 1,166 interactions among proteins encoded by 1,147 human genes (Fig. 3a and Supplementary Table 2). This represents a 42% (1,149 novel interactions) increase over HI1 (ref. 11). The overlap of 127 interactions between the two datasets matches the expected overlap of 138 pairs (Supplementary Note 3)1. The distribution of numbers of interactors per protein in HI-NGS is similar to that of previous datasets (Fig. 3b and Supplementary Fig. 5).
Figure 3.
HI-NGS network. (a) Network view (main connected component above the unconnected components) of HI-NGS (gold) produced with PCR stitching compared to HI1 (blue). (b) Degree distribution of HI-NGS compared to HI1.
Despite the PCR stitching protocol involving one additional PCR reaction for each ORF pair compared to the traditional Y2H method, our strategy reduces the overall cost by at least ~40%, and should therefore allow increased throughput (Supplementary Fig. 6 and Supplementary Note 4). With continued improvement of NGS technologies, the cost of sequencing should keep diminishing14. Because 454 sequencing can accommodate lower capacity runs and because samples can be combined with other sequencing samples there is no lower size limit for screens to which this method can be applied. The 82 bp linker has no identical sequence in all of GenBank, so sISTs can in principle be sequenced in combination with other samples (Supplementary Note 5). The approach would be equally effective with cDNA library screens as it was here for an ORFeome library screen.
The linker length of 82 bp requires that the average read length be >100 bp for reliable identification of sISTs. Among existing NGS technologies, the 454 technology is to our knowledge the only one that reliably produces reads of more than 100 bp on average7. The application of paired-end sequencing15 to stitched PCR products would extend the approach to NGS platforms which have average read-lengths less than 100 bp (Supplementary Note 6).
The Stitch-Seq strategy implemented here for Y2H can be readily implemented to other types of interaction assays, leading to improved capacity and expanded scope of interactome network mapping.
ONLINE METHODS
Yeast two-hybrid (Y2H)
High-throughput Y2H screens were carried by published protocols16. Briefly, ~6,000 Entry clones contained in the Human ORFeome version 3.1 (ref. 12) were transferred into pDEST-AD and pDEST-DB vectors (Supplementary Note 7) by Gateway LR reactions17 encoding the Activation domain (AD) and DNA binding domain (DB), respectively. The vectors are available upon request. These LR recombination products were used directly to transform Escherichia coli (DH5α-T1R). Transformed cells were selected on LB media containing ampicillin, and plasmid DNA was extracted and purified. All AD-Y and DB-X plasmids were transformed into Y2H strains MATa Y8800 and MATα Y8930 (Genotype: leu2-3, 112 trp1-901 his3Δ200 ura3-52 gal4Δ gal80Δ GAL2::ADE2 GAL1::HIS3@LYS2 GAL7::lacZ@MET2 cyh2R), respectively. To identify auto-activators18 all DB-X constructs were screened for growth on synthetic complete media lacking leucine and histidine (SC-Leu-His) supplemented with 1 mM of 3-amino-1,2,4-triazole (3-AT). All auto-activators were removed.
The AD-Y containing yeast cells were combined into “minipools” of 188 AD-Y strains. To create these, first 5 μL from glycerol stocks of 94 individual AD-Y yeast strains were each inoculated into 500 μL of liquid SC-tryptophan (Trp) media in 96-well deep-well plates and grown for four days on a shaker at 30°C. Settled yeast were resuspended by thoroughly vortexing the culture plates and the OD600 of every well was measured to verify homogenous yeast growth and hence equal representation of each AD-Y yeast strain in the minipool. The contents of two 96-well culture plates (188 different AD-Y strains) were transferred into a sterile trough and mixed thoroughly to ensure equal representation of all AD-Y yeast strains in the pool. Archival glycerol stocks for storage at −80°C were then prepared by combining 80 μL of the pooled yeast cultures with 80 μL of 40% autoclaved glycerol in round-bottom 96-well microtiter plates.
Proteome-wide Y2H screens were carried out as described16, including inoculation of AD-minipool and DB-X yeast cultures, mating onto rich YEPD media16, and replica-plating onto selective SC-Leu-Trp-His+1 mM 3-AT (SC-His) and SC-Leu-His+1 mM 3-AT plates containing 1 mg/l cycloheximide (SC-His+CHX). The latter control plates select for cells that do not have the AD plasmid due to plasmid shuffling. Growth on selective media thus identifies spontaneous auto-activators18. Only pairs that activated at least one reporter gene and were CHX-sensitive on both control plates were included in the final map. All pairs that exhibited CHX resistance on selective plates or failed to pass the retest were excluded. Six Y2H controls with known phenotypes were included on all Y2H screening plates16. The plates were incubated overnight at 30°C and ‘replica-cleaned’ the following day by placing each plate on a piece of velvet stretched over a replica plating block and pushing evenly on the plate to remove excess yeast cells. Plates were then incubated for another three days, after which positive colonies were picked and used to inoculate liquid cultures in SC-Leu-Trp media. After overnight growth at 30°C, a 5 μL aliquot was spotted onto each of the four plates for secondary phenotype confirmation (Phenotyping II) [SC-His, SC-His+CHX, SC-Leu-Trp-Adenine (SC-Ade), SC-Leu-Ade+CHX (SC-Ade+CHX)] to test for CHX-sensitive expression of the LYS2::GAL1-HIS3 and GAL2-ADE2 reporter genes. All plates were replica-cleaned the following day and scored after three additional days to identify colonies that grow on SC-His or on SC-Ade but not on SC-His+CHX or on SC-Ade+CHX.
For colonies that scored positive the identities of DB-X and AD-Y were determined using PCR stitching followed by massively parallel 454 FLX sequencing. Independently all pairs were identified by Sanger sequencing as described16. Before PCR, yeast cells from positive colonies were lysed in 15 μL of lysis buffer [2.5 mg/ml zymolase 20T (21100 U/g, Seikagaku Corp.) dissolved in 0.1 M sodium phosphate buffer (pH 7.4)] in each well of a 96-well PCR plate. A small amount of yeast cells (not more than what fits on the end of a standard 200 μL tip) were picked and resuspended in lysis buffer in soft shell, V-bottom 96-well microtiter plate (hereafter: PCR plate). PCR plates were put on a thermocycler to run the following lysis program:
-
Step 1
37°C for 15 min
-
Step 2
95°C for 5 min
-
Step 3
Hold at 10°C
Afterwards 100 μL of filter-sterilized water were added to each well. The PCR plates were centrifuged for 10 minutes at 800 × g and stored at −20°C. Conditions and primers for the two rounds of PCR reactions in PCR stitching are given in Supplementary Table 1. From each PCR reaction 5 μl were combined. A 1 ml aliquot of the pooled stitched PCR products was purified using QIAquick PCR Purification Kit (Qiagen catalog number: 28104). A 200 μl aliquot of the purified stitched PCR products was sent to the University of Pennsylvania DNA Sequencing Facility for 454 FLX sequencing.
At the sequencing facility PCR products were processed using Roche kits [GS Standard DNA Library Preparation kit, Catalog number: 04852265001; GS FLX Standard emPCR kit (Shotgun), Catalog number: 0482290001; GS FLX PicoTiterPlate Kit (70×75), Catalog number: 04852427001; and GS FLX Standard LR70 Sequencing Kit, Catalog number: 04932315001] according to instructions of the manufacturer. Briefly, 3–5 μg of the pooled PCR products were fragmented by nebulization for one minute under nitrogen gas pressure of 30 psi (2.1 bar), the DNA fragments were size-selected and subjected to end polishing and adaptor ligation. The library was then immobilized onto streptavidin-coated beads followed by the fill-in reaction to repair the gaps generated by the ligation of non-phosphorylated adaptors to the fragments. A single-stranded library was created by melting off the non-biotinylated strand of bead-bound fragments. Subsequent quality assessment and quantitation were done by 96-well plate fluorometry and analysis on a Bioanalyzer with Agilent RNA Pico 6000 LabChip Kit (Catalog number: 5067-1513). The amount of library DNA needed for optimal results in the emulsion-based clonal amplification (emPCR) procedure was determined by emulsion titration assay according to the instructions of the manufacturer. The library of DNA fragments was amplified from a single bead-bound copy to millions of copies per bead using water-in-oil emulsion PCR. Subsequently, emulsions were broken and the beads carrying the amplified library were recovered using biotinylated amplification primers and streptavidin-coated magnetic beads with manufacturer-provided protocols. Beads were counted, the enrichment ratio calculated and the recommended amount of sequencing primer was added to bead-bound amplified fragments. After annealing and mixing of DNA loaded beads with packing beads, the wells of a GS FLX Standard PicoTiterPlate (PTP) were loaded according to manufacturer provided protocols, i.e., subsequent layers of enzyme beads, the mix of DNA and packing beads, another layer of enzyme beads followed by a layer of apyrase beads. The loaded PTP was inserted into the FLX instrument and run using the standard protocol.
From the 454 sequencing data, we first identified all usable sequencing reads containing the 82 bp linker using “cross_match”19. Then sISTs were identified by mapping both ends of the usable reads to the screened ~6,000 ORFs in human ORFeome 3.1, using BLASTN (mismatches allowed) with an E-value cutoff of 10−3.
From the set of successfully sequenced DB-X and AD-Y pairs, all interacting protein pairs were verified in a single-pass pairwise retesting to ensure the robustness of the His+ or Ade+ phenotypes and to exclude the possibility that physiologic and genetic changes that occurred during the course of the experiment gave rise to experimental artifacts16. For retesting, liquid cultures of individual yeast strains with corresponding AD and DB constructs were inoculated from archival glycerol stocks, grown overnight, and arrayed for pair-wise Y2H analysis using the same procedure outlined above. Briefly, from the YEPD mating plates, yeast colonies were replica-plated onto SC-Leu-Trp plates to select diploid yeast cells containing both AD and DB constructs. Diploid yeast cells were subsequently replica-plated onto four Y2H assay plates identical to the ones used for Phenotyping II. All interactions so verified have been deposited with the International Molecular Exchange (IMEx) consortium.
Protein complementation assay (PCA)
For PCA16 human ORFs available in Gateway Entry vectors were transferred by Gateway LR reactions into vectors encoding the two fragments of YFP (Venus variant) fused to the N-terminus of the tested proteins. Baits were fused to the F1 fragment (amino acids 1-158 of YFP) and preys to the F2 fragment (amino acids 159–239 of YFP). After bacterial transformation, minipreps were prepared on a Qiagen BioRobot, and DNA concentrations were determined by PicoGreen assay (Invitrogen catalog number: P7589) in 96-well format according to protocols of the manufacturer. A 30 ng aliquot of each vector encoding the two proteins was added to 140 ng of a CFP control plasmid for transfection into CHO-K1 cells in 96-well plates, using Lipofectamine2000 (Invitrogen) reagent according to the instructions of the manufacturer. At approximately 18 hrs post-transfection, cells were washed twice with PBS, trypsinized with 15 μl cell culture grade trypsin, suspended in 130 μl PBS and analyzed by fluorescence-activated cell sorting on a Canto II FACS (Becton Dickinson) equipped with a 96-well autosampler. Viable CFP positive cells, i.e. transfected cells, were selected and analyzed for YFP signal. A pair was considered interacting if at least 30% of CFP positive cells were YFP positive, and the YFP/CFP ratio was at least twice as high as the ratio of the average YFP signal across the entire plate over the average CFP ratio on that plate.
Nucleic Acid Programmable Protein Array in Wells (wNAPPA)
For the wNAPPA assay16 ORFs encoding the interacting proteins were cloned into Gateway-compatible pCITE-HA and pCITE-GST vectors by LR reaction. After transformation, growth, DNA minipreps and determination of DNA concentration, ~0.5 μg of each plasmid were added to Promega TnT coupled transcription-translation mix (catalog number: L4610) and incubated for 1.5 hrs to express proteins. During this time anti-GST antibody-coated 96-well plates (Amersham 96-well GST detection module, catalog number: 27-4592-01) were blocked at room temperature with PBS containing 5% dry milk powder. After protein expression, the expression mix was diluted in 100 μl blocking solution, added to the emptied pre-blocked 96-well plates. Binding was for 2 hrs at 16°C with agitation. After capture, plates were washed three times and developed by incubation with primary and secondary antibody. Signal was visualized using enhanced chemiluminescence (Pierce PicoWest ECL reagent, catalog number: 34095) with a Bio-Rad ChemiDoc. Signal was manually assigned a score between 0 and 5 (‘0’ corresponding to background in empty controls and ‘5’ being a saturated signal). Wells that scored ≥2 in either configuration were deemed to contain positively interacting pairs.
P value calculations
All P values were calculated by the following equation:
where x1 is the number of positives in dataset 1 detected by a given assay;
n1 is the total number of pairs in dataset 1;
x2 is the number of positives in dataset 2 detected by the assay;
n2 is the total number of pairs in dataset 2;
p1 = x1/n1
p2 = x2/n2
-
P = 1 − Probability (−z < Z < z)
Supplementary Material
Supplementary Figure 1. The conventional approach for sequencing large numbers of ORF pairs resulting from high-throughput Y2H screens.
Supplementary Figure 2. Robust stitching PCR amplifications over a wide ORF size range.
Supplementary Figure 3. Statistical analysis of the distribution of useful 454 reads for sISTs.
Supplementary Figure 4. Statistical calculation of the overall success rate for PCR stitching with three PCR reactions for each pair of ORFs that encode interacting proteins.
Supplementary Figure 5. Comparison of degree distributions of HI-NGS, HI1, and HI-REPEAT.
Supplementary Figure 6. Calculation and comparison of costs for each pair of ORFs that encode interacting proteins.
Supplementary Table 1. PCR reaction mix setup and PCR conditions of the three PCR reactions for each pair of ORFs that encode interacting proteins.
Supplementary Table 2. Interactions in HI-NGS.
Supplementary Note 1. Distribution of 454 reads for unique sISTs.
Supplementary Note 2. Data quality assessment of HI-NGS.
Supplementary Note 3. Comprehensive comparison of HI1 and HI-NGS.
Supplementary Note 4. Extrapolation to mapping the whole human interactome.
Supplementary Note 5. Size cutoff for adapting Stitch-Seq protocol.
Supplementary Note 6. Useful read length and implementation of paired-end sequencing.
Supplementary Note 7. Vector sequences for pDEST-ADCYH and pDEST-DB.
Acknowledgments
We thank members of the Dana-Farber Cancer Institute (DFCI) Center for Cancer Systems Biology (CCSB) and the Roth Laboratory for helpful discussions. We thank the University of Pennsylvania DNA Sequencing Facility in the Department of Genetics for the 454 FLX sequencing. This work was funded in part by grants R01 HG001715 (awarded to D.E.H., F.P.R. and M.V.) and R21 HG004756 (awarded to F.P.R.) from the National Human Genome Research Institute of the National Institutes of Health, a grant from the Ellison Foundation, Boston, MA (awarded to M.V.), a Canadian Institute for Advanced Research Fellowship and Canada Excellence Research Chair (F.P.R.) and in part by Institute Sponsored Research funds from the DFCI Strategic Initiative in support of CCSB. M.V. is a Chercheur Qualifié Honoraire from the Fonds de la Recherche Scientifique (FRS-FNRS, French Community of Belgium).
Footnotes
AUTHOR CONTRIBUTIONS
M.V. conceived of the project and the PCR-stitching methodology for next-generation sequencing and oversaw all aspects including writing and editing of the manuscript; H.Y. developed the PCR stitching process, did validations, analyzed data and co-wrote manuscript; P.B. oversaw human interactome screening and co-wrote the manuscript; F.P.R oversaw aspects of computational work, helped develop the next-generation sequencing strategy and contributed to editing the manuscript; D.E.H helped with developing experimental protocols and editing the manuscript; T.H. handled database implementation and analysis of raw sequence data; L.T., S.T., E.W., F.G., N.Z., T.H.-K., and J.S. carried out experimental processes for interactome mapping, PCR-stitching, and validation tests; X.Y. carried out processing of PCR products and subsequent 454 sequencing reactions; C.F. and E.R. did computational and network graphing tasks; K.S.A. contributed to troubleshooting the PCR-stitching methods; M.E.C. contributed to writing and editing of the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interest.
References
- 1.Venkatesan K, et al. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu H, et al. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deplancke B, Dupuy D, Vidal M, Walhout AJ. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walhout AJ, Vidal M. Methods. 2001;24:297–306. doi: 10.1006/meth.2001.1190. [DOI] [PubMed] [Google Scholar]
- 5.Hook B, Bernstein D, Zhang B, Wickens M. RNA. 2005;11:227–233. doi: 10.1261/rna.7202705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett ST, et al. Pharmacogenomics. 2005;6:373–382. doi: 10.1517/14622416.6.4.373. [DOI] [PubMed] [Google Scholar]
- 7.Margulies M, et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shendure J, et al. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 9.Tong AH, et al. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 10.Walhout AJ, et al. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 11.Rual JF, et al. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 12.Lamesch P, et al. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun P, et al. Nat Methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coombs A. Nat Biotechnol. 2008;26:1109–1112. doi: 10.1038/nbt1008-1109. [DOI] [PubMed] [Google Scholar]
- 15.Maher CA, et al. Proc Natl Acad Sci USA. 2009;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dreze M, et al. Methods Enzymol. 2010;470:281–315. doi: 10.1016/S0076-6879(10)70012-4. [DOI] [PubMed] [Google Scholar]
- 17.Walhout AJ, et al. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- 18.Walhout AJ, Vidal M. Genome Res. 1999;9:1128–1134. doi: 10.1101/gr.9.11.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ewing B, Hillier L, Wendl MC, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 20.Nirantar SR, Ghadessy FJ. Proteomics. 2011;11:1–5. doi: 10.1002/pmic.201000643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. The conventional approach for sequencing large numbers of ORF pairs resulting from high-throughput Y2H screens.
Supplementary Figure 2. Robust stitching PCR amplifications over a wide ORF size range.
Supplementary Figure 3. Statistical analysis of the distribution of useful 454 reads for sISTs.
Supplementary Figure 4. Statistical calculation of the overall success rate for PCR stitching with three PCR reactions for each pair of ORFs that encode interacting proteins.
Supplementary Figure 5. Comparison of degree distributions of HI-NGS, HI1, and HI-REPEAT.
Supplementary Figure 6. Calculation and comparison of costs for each pair of ORFs that encode interacting proteins.
Supplementary Table 1. PCR reaction mix setup and PCR conditions of the three PCR reactions for each pair of ORFs that encode interacting proteins.
Supplementary Table 2. Interactions in HI-NGS.
Supplementary Note 1. Distribution of 454 reads for unique sISTs.
Supplementary Note 2. Data quality assessment of HI-NGS.
Supplementary Note 3. Comprehensive comparison of HI1 and HI-NGS.
Supplementary Note 4. Extrapolation to mapping the whole human interactome.
Supplementary Note 5. Size cutoff for adapting Stitch-Seq protocol.
Supplementary Note 6. Useful read length and implementation of paired-end sequencing.
Supplementary Note 7. Vector sequences for pDEST-ADCYH and pDEST-DB.