Abstract
Over-expression of sirtuins (NAD+-dependent protein deacetylases) has been reported to increase lifespan in budding yeast, Caenorhabditis elegans and Drosophila melanogaster1-3. Studies of gene effects on ageing are vulnerable to confounding effects of genetic background4. We re-examined the reported effects of sirtuin over-expression on ageing and found that standardisation of genetic background and use of appropriate controls abolished the apparent effects in both C. elegans and Drosophila. In C. elegans, outcrossing of a line with high level sir-2.1 over-expression1 abrogated the longevity increase, but not sir-2.1 over-expression. Instead, longevity co-segregated with a second-site mutation affecting sensory neurons. Outcrossing of a line with low copy number sir-2.1 over-expression2 also abrogated longevity. A Drosophila strain with ubiquitous over-expression of dSir2 using the UAS-GAL4 system was long-lived relative to wild-type controls, as previously reported3, but not relative to the appropriate transgenic controls, and nor was a new line with stronger over-expression of dSir2. These findings underscore the importance of controlling for genetic background and the mutagenic effects of transgene insertions in studies of genetic effects on lifespan. The life extending effect of dietary restriction (DR) on ageing in Drosophila has also been reported to be dSir2 dependent3. We found that DR increased fly lifespan independently of dSir2. Our findings do not rule out a role for sirtuins in determination of metazoan lifespan, but they do cast doubt on the robustness of the previously reported effects on lifespan in C. elegans and Drosophila.
Keywords: Ageing, C. elegans, Drosophila, sirtuin, genetic background
The role of sirtuins in ageing was discovered in budding yeast (Saccharomyces cerevisiae), where over-expression of SIR2 increases replicative lifespan5. It was then reported that elevated sirtuin levels increase lifespan in the nematode C. elegans1,2,6 and the fruitfly Drosophila3, suggesting an evolutionarily ancient role of sirtuins in longevity assurance7. Dietary restriction (DR), reduced food intake short of starvation, extends lifespan in organisms ranging from yeast to mammals8, and initial studies suggested that DR increases lifespan by activating sirtuins in yeast9, C. elegans10 and Drosophila3. Pharmacological activation of sirtuins has thus been widely promulgated as a potential means to mimic DR and slow ageing in humans11. However, several aspects of the role of sirtuins in ageing have proved controversial12. Subsequent studies have suggested that sirtuins do not mediate DR effects on ageing, at least in budding yeast and C. elegans13,14. The plant-derived polyphenol resveratrol and other compounds have been reported to activate sirtuins and extend lifespan15,16. More recent findings have challenged both effects17-20. We therefore re-examined the effects of sirtuin over-expression on lifespan in C. elegans and Drosophila. In particular, we wished to exclude the possibility that the increased longevity observed in strains with sirtuin gene over-expression are caused by differences in genetic background, or by the mutagenic effects of transgene insertion, which frequently confound studies of the genetics of ageing4.
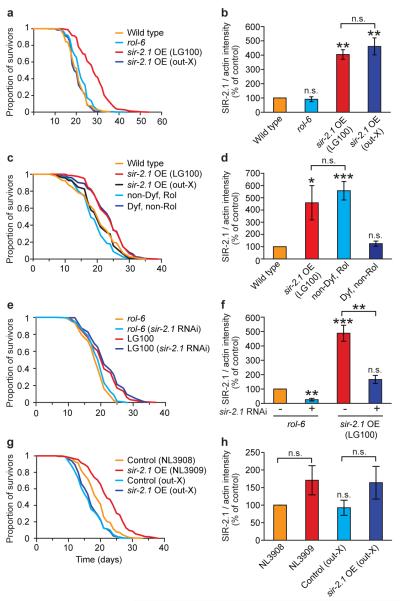
We first examined a high copy number sir-2.1 transgenic C. elegans strain (LG100) carrying the integrated transgene array geIn3 [sir-2.1 rol-6(su1006)] (Ref. 1). As expected, this strain was long lived (Fig. 1a; Table S1). However, outcrossing (x5) of geIn3 to wild type (N2) abrogated the increase in longevity (Fig. 1a, Table S1) without affecting SIR-2.1 protein levels (Fig. 1b). This loss of longevity upon outcrossing was verified by an independent research team (Table S2).
Figure 1. C. elegans: Longevity of LG100 and NL3909 is not attributable to sir-2.1 over-expression.
a, b. Outcrossing of LG100 removes life extension without affecting SIR-2.1 protein levels. Data in b derived from Western blots (mean of three trials each using an independent protein preparation). A representative Western blot is shown in Fig S1a. qRT-PCR showed that sir-2.1 mRNA is also elevated in both strains (data not shown). c, LG100-derived Dyf, non-Rol segregant lines are long-lived while non-Dyf, Rol lines are not. d. Non-Dyf Rol segregant lines have elevated SIR-2.1 levels, while Dyf, non-Rol lines do not. e, f. sir-2.1 RNAi does not suppress LG100 longevity, but reduces SIR-2 protein levels. g, h. Outcrossing of NL3909 removes life extension without affecting SIR-2.1 protein levels. See Tables S1, S3, S4 and S5 for lifespan statistics for a, c, e and g, respectively. Error bars, S.E.M.. *0.01< P < 0.05; ** 0.001 < P < 0.01; ***P < 0.001, n.s., not significant; Student’s t test (two tailed). One remaining possibility is that the outcrossed sir-2.1 strains both contain second site mutations that suppress longevity effects. However, daf-2 RNAi strongly induced longevity in both (data not shown), arguing against the presence of a general suppressor of longevity in each case.
LG100 exhibited a neuronal dye-filling (Dyf) defect22 that did not segregate with the transgene upon outcrossing (Fig. S2A). Dyf mutants often exhibit extended lifespan23. To determine whether the longevity of LG100 might be attributable to a dyf mutation, we derived from it three Dyf, non-Rol lines (lacking geIn3) and three non-Dyf, Rol lines (carrying geIn3). Dyf, non-Rol lines were long-lived and showed wild-type SIR-2.1 protein levels (Fig. 1c,d, Table S3). Non-Dyf, Rol lines showed elevated SIR-2.1 protein levels but had wild-type lifespans. Dyf mutant longevity appeared to be partially daf-16 dependent (Fig. S2B), as seen previously for other Dyf mutants23. The co-segregation of longevity with this dyf mutation but not geIn3 was previously noted by another research team (S.S. Lee, Cornell University, personal communication). Furthermore, knock-down of sir-2.1 expression in LG100 using RNA-mediated interference did not suppress longevity, despite lowering SIR-2.1 protein to wild-type levels (Fig. 1e,f; Table S4). Taken together, these results imply that the longevity of LG100 is attributable to an unidentified dyf mutation (or possibly another mutation closely-linked to the dyf locus), and that high level over-expression of sir-2.1 is not sufficient to increase lifespan in these strains.
A low copy number transgenic strain (NL3909) over-expressing sir-2.1 (Ref. 7) is also long-lived2. We confirmed the increased lifespan of NL3909 (pkIs1642 [sir-2.1 unc-119] unc-119(ed3)) relative to the control strain NL3908 (pkIs1641 [unc-119] unc-119(ed3)) (Fig. 1g, Table S5). We also observed an apparent elevation of SIR-2.1 protein in NL3909 relative to NL3908 (Fig. 1h). Outcrossing (x6) of NL3909 once again abrogated longevity (Fig. 1g, Table S5) without affecting SIR-2.1 protein levels (Fig. 1h, Fig. S1c). This effect of outcrossing was independently verified (Table S6). Thus, the longevity of NL3909 also appears to be attributable to genetic background effects rather than to pkIs1642.
The duplication mDp4 includes the sir-2.1 locus, and the mDp4–containing strain DR1786 is long lived1. We too found that DR1786 is long-lived, and also shows elevated sir-2.1 expression. However, longevity was not suppressed by sir-2.1 RNAi (Fig. S3, Table S7) suggesting causation by factors other than sir-2.1, either on mDp4 or elsewhere in the genome.
In Drosophila over-expression of dSir2 reportedly increases lifespan relative to wild-type controls3. Over-expression was achieved using the GAL4, UAS binary system24, with the largest increases in lifespan produced by combination of EP-UAS-dSir2 (dSir2EP2300) with a ubiquitously-expressed tubulin-GAL4 driver. We outcrossed these two transgenes (x6) into the control white Dahomey (wDah) background. Assayed on a medium similar to that used in the original study, EP-UAS-dSir2/tubulin-GAL4 flies were longer lived than wild-type controls, as previously reported3 (Fig. 2a). However, they were not longer lived than the tubulin-GAL4/+ control flies (Fig. 2a). This implies that life extension is due to transgene-linked genetic effects other than over-expression of dSir2. Lifespan was assayed on a range of food media (see Methods for details) to test for nutrient-dependence of any effect. However, in no case were EP-UAS-dSir2/tubulin-GAL4 flies longer lived than one or both transgenic controls (Fig. S4).
Figure 2. Drosophila: Absence of effects of dSir2 on lifespan.
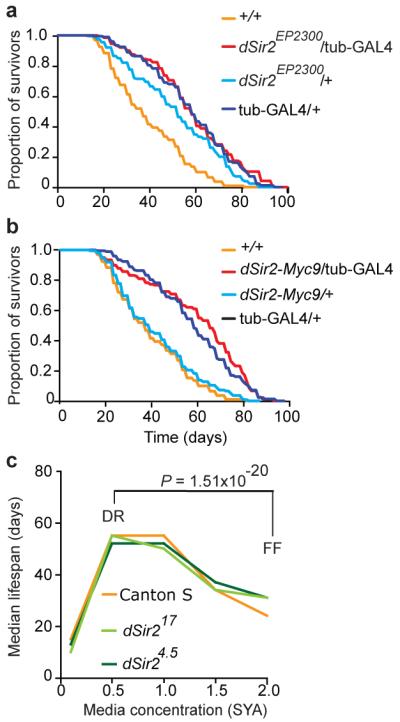
All lines were outcrossed into wDah (+/+) a, Lifespan in flies over-expressing dSir2EP2300 driven via tubulin-GAL4 (tub-GAL4) is longer than wild type, but not than the tubulin-GAL4 /+ genetic control. Median lifespans: +/+ = 39 days, dSir2EP2300/tubulin-GAL4 = 59 days, dSir2EP2300/+ = 53 days, tubulin-GAL4 /+ = 60 days. dSir2EP2300/tubulin-GAL4 vs. dSir2EP2300/+, P = 0.0006; dSir2EP2300/tubulin-GAL4 vs. tubulin-GAL4 /+, P = 0.9295; dSir2EP2300/tubulin-GAL4 vs. +/+, P <0.0001 . b, Lifespan in flies over-expressing dSir2-Myc9 is longer than in wild type, but not than in the tubulin-GAL4 control. Median lifespans: +/+ = 39 days, dSir2-Myc9/tubulin-GAL4 = 67, dSir2-Myc9/+ = 41 days, tubulin-GAL4/+ = 60 days. dSir2-Myc9/tubulin-GAL4 vs. dSir2-Myc9/+, P = 0.0001; dSir2-Myc9/tubulin-GAL4 vs. tubulin-GAL4/+, P = 0.1354; dSir2-Myc9/tubulin-GAL4 vs. +/+, P <0.0001. Compared using log rank test, n=200. c, Effect of dietary restriction on Drosophila lifespan is not dSir2 dependent. Flies were assayed over five concentrations of SYA media and data are presented as the median lifespan on each food concentration. All lines were outcrossed into Canton S (+/+). P values confirm that all flies respond normally to DR when median lifespans are compared for DR vs. fully-fed (FF) conditions30.
Lack of an observable effect on lifespan could reflect the relatively modest increase in dSir2 expression in EP-UAS-dSir2/tubulin-GAL4 flies, both in terms of levels of mRNA (Fig. S5) and protein (+35% relative to wild type, Fig. S6). We therefore created lines with a higher level of over-expression of dSir2 (UAS-dSir2-Myc9/tubulin-GAL4). Here dSir2 mRNA and protein levels were robustly increased relative to wild type (+318% relative to wild-type protein levels; Fig. S5, S6). We examined recombinant protein raised in E. coli to check that the presence of the Myc tag did not interfere with dSir2 histone deacetylase (HDAC) activity, as measured by deacetylation of the fluorophore-containing p53 (Fluor de Lys) or native acetylated histone H4 substrates, and it did not (Fig. S7). We also found that dSir2 HDAC activity was unaffected by addition of resveratrol in either assay (Fig. S7). We saw no increase in lifespan in UAS-dSir2-Myc/tubulin-GAL4 flies relative to tubulin-GAL4/+ controls, on food medium similar to that used in the original study (Fig. 2b) or relative to either control on a range of other media (Fig. S4b,c,f). An independent research team also saw no increase in lifespan in tubulin-GAL4/UAS-dSir2-Myc9 flies (Fig. S8). These results suggest that the previously observed longevity of EP-UAS-dSir2/tubulin-GAL4 flies was not attributable to elevated expression of dSir2, and that stronger, ubiquitous over-expression of dSir2 also does not extend fly lifespan.
The role of sirtuins in the extension of lifespan by DR in yeast and C. elegans is controversial, with multiple groups reporting that sirtuins are not required for life span extension from DR in both organisms8. In Drosophila, it was reported that DR does not increase lifespan in dSir2 deletion mutant flies3. We tested this too, using the deletion alleles dSir24.5 (tested previously3) and dSir217. We first outcrossed these alleles (Fig. S9a) into the Canton S wild type (see Methods), used in the previous DR study3. We then checked the effect of each allele on dSir2 gene expression. dSir217 abrogated dSir2 mRNA, implying that this is a null allele. By contrast, dSir24.5, which contains a relatively small deletion at the 5′ end of the gene, did not reduce dSir2 mRNA levels (Fig. S9b,c).
To reassess the role of dSir2 in DR in Drosophila, we compared lifespans of wild-type (Canton S), dSir24.5 and dSir217 homozygotes. All genotypes responded similarly and normally to DR in trials conducted by two independent research teams (Fig. 2c, Fig. S10), hence the effect of DR on lifespan did not require dSir2.
In this study, we were unable to verify the effect of sirtuin over-expression on lifespan in either C. elegans or Drosophila. Increased lifespan was seen in two C. elegans lines with elevated sir-2.1 expression derived from independent studies, as previously reported, but in each case this was abrogated by outcrossing. sir-2.1 over-expression does exert effects on traits other than lifespan. For example, geIn3 is neuroprotective in a worm model of neuron dysfunction in Huntington’s disease25 and, importantly, this effect is not attributable to the dyf mutation (Fig. S11). Moreover, both NL3909 and its outcrossed derivative are thermotolerant (M. Somogyvári, C. Sőti, unpublished data). In Drosophila, lines over-expressing dSir2 were longer-lived than wild type controls, as previously reported, but they were not longer lived than lines containing the appropriate transgenic controls. That all transgenic lines were longer lived than the Dahomey wild type into which they had been outcrossed could reflect heterosis in the vicinity of the transgene inserts, or a mutagenic effect of the GAL4 insert.
Lifespan was not increased by over-expression either of sir-2.1 from its own promoter in C. elegans, or dSir2 ubiquitously from a heterologous promoter in Drosophila. Our findings call into question the robustness of earlier reports of a role of sirtuins in longevity-assurance based upon over-expression in C. elegans and Drosophila, and also on the role of dSir2 in the response to DR in Drosophila. However, sirtuins can affect lifespan in animals under certain conditions: C. elegans daf-2(e1370) mutants are hypersensitive to genetic effects on lifespan26, and here deletion of sir-2.1 reproducibly increases lifespan6 (Fig. S12).
Our finding that resveratrol does not activate HDAC activity of dSir2 using a native histone H4 peptide is consistent with earlier findings with yeast SIR2 and mammalian SirT1 (Ref. 17,18). Resveratrol increased Drosophila lifespan in one study27 but not another21. In principle, this could reflect sensitivity of resveratrol effects to subtle differences in culture conditions. If this were the case, our findings would imply that such effects are not attributable to direct activation of dSir2 by resveratrol.
METHODS SUMMARY
Nematode strains and maintenance
Nematodes were maintained on nematode growth medium (NGM) agar at 20°C, with Escherichia coli OP50 bacteria as a food source. Nematode strains used included: wild type (N2), GA707 wuEx166 [rol-6(su1006)] (rol-6 control), LG100 geIn3 [sir-2.1 rol-6(su1006)] dyf-?(wu250), NL3909 pkIs1642 [sir-2.1 unc-119] unc-119(ed3), and the control strain NL3908 pkIs1641 [unc-119] unc-119(ed3).
Nematode lifespan measurements
These were performed as described28, at 20°C. To prevent progeny production, 5-fluoro-2′-deoxyuridine (FUdR) was added to seeded plates, to a final concentration of 10, 40 or 50 μM. Before testing effects of RNA-mediated interference (RNAi) on lifespan, worms were kept for 2 generations on the RNAi bacteria. Statistical significance of effects on lifespan was estimated using the log rank test, performed using JMP, Version 7 (SAS Institute).
Fly stocks and maintenance
tubulin-GAL4 and dSir2EP2300 were obtained from the Bloomington Stock Center. dSir2-Myc2 and dSir2-Myc9 lines were generated by germ-line transformation into strain w04. dSir24.5/SM6B, dSir217/Cyo and Canton S were gifts from S. Pletcher, J. Rine and S. Helfand. All lines were outcrossed at least 6 times into the relevant controls. Experiments were performed at 25°C on a 12h : 12 h light-dark cycle at constant humidity.
Fly lifespan assays
Flies were bred at standard density, allowed to mate for 48 hours after emerging then sorted into 10 females per vial. Vials were changed every 48 hours, and deaths per vial scored until all flies were dead. Over-expression studies n=200. dSir2 mutant studies n=100. For statistical methodology, see above.
dSir2 deacetylation assays
We used both the SirT1 Fluorimetric Drug Discovery Kit (Enzo Life Sciences) and an HPLC-based acetyl-histone H4 deacetylation assay29. dSir2 and dSir2-Myc were cloned into pET SUMO (Invitrogen) and purified on HisPur cobalt spin columns (Thermo Scientific).
Supplementary Material
Acknowledgements
We thank A. Gartner for providing an antibody against C. elegans SIR-2.1, D. Chen, P. Kapahi, S. Pletcher and D. Skorupa for providing data, S.S. Lee for permission to cite unpublished results, S. Helfand and J. Rine for providing fly strains, W. Mair for performing preliminary trials, and R. Baumeister for useful discussion. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. We acknowledge funding from the the Drosophila Aging Core of the Nathan Shock Center of Excellence in Basic Biology of Aging at University of Michigan (S.P.), the European Union (FP6-036894 to C.B., D.G., L.P. and S.V. and FP6-518230 to D.G. and C.S.), the Hungarian Science Foundation and Norway Grants (NNF-78794 to C.S.), INSERM and ANR, Paris, France (R.V., A.M.O. and C.N.), the National Institutes of Health (CA129132 to A.B., R01AG031108 to M.K. and T32AG000057 to G.S.), and the Wellcome Trust (Strategic Award to C.A., F.C., D.G., L.P. and M.R.). C.S. is a Bolyai Research Scholar of the Hungarian Academy of Sciences, and M.K. is an Ellison Medical Foundation New Scholar in Aging.
Appendix A
METHODS
C. elegans
Nematode strains and maintenance
C. elegans were cultured under standard monoxenic conditions31,32. Strains used included N2 (wild type), GA707 wuEx166 [rol-6(su1006)], HT1593 unc-119(ed3), LG100 geIn3 [sir-2.1 rol-6(su1006)] dyf-?(wu250), NL3908 pkIs1641 [unc-119] unc-119(ed3), and NL3909 pkIs1642 [sir-2.1 unc-119] unc-119(ed3).
Outcrossing of strains
LG100 was outcrossed with N2, and the Rol trait used to detect the presence of gIn3. NL3908 and NL3909 were outcrossed using HT1593 unc-119(ed3). Rescue of Unc was used to detect the presence of the transgene array.
Isolation of Dyf, non-Rol and non-Dyf, Rol lines
LG100 was crossed with N2, and lines established from individual F2 animals exhibiting Dyf, non-Rol or non-Dyf, Rol phenotypes. The Dyf phenotype was identified by staining with the dye DiI, and looking for absence of dye uptake into the amphid and phasmid neurons. Non-Dyf, Rol F2 animals that were heterozygous for the geIn3 transgene array (the rol-6 marker is dominant) were identified by the presence of non-Rol animals in the F3, and excluded.
RNA-mediated interference (RNAi)
Animals were fed E. coli containing the HT115 vector either with or without a portion of the sir-2.1 gene cloned into it. The sir-2.1 feeding strain was obtained from the Ahringer RNAi library33. Worms were maintained on RNAi feeding strains for two generations prior to lifespan measurements. One day before starting measurements, 5-fluoro-2′-deoxyuridine (FUDR) was applied to seeded plates to 10 μM to prevent progeny production.
Analysis of SIR-2.1 protein levels
Protein was prepared from synchronous nematode cultures (L4 larvae and young adults) raised on E. coli OP50 or RNAi bacteria for two generations. Western blots were performed with anti-actin monoclonal antibodies (Santa-Cruz Biotechnology), anti-SIR-2.1 polyclonal antibody (kindly provided by A. Gartner34). For all assays, 3-5 replicate worm cultures were used.
Neuroprotection assays
To test for sirtuin protection from expanded polyglutamines (polyQs), we crossed GA919 (geIn3 dissociated from dyf-?(wu250)) to strains carrying integrated polyQ arrays. These polyQ strains co-express the first 57 amino acids of human huntingtin (htt) with either 19 or 128 Glns fused to CFP, expressed from the mec-3 promoter, and YFP expressed from the mec-7 promoter in touch receptor neurons25. Response to touch at the tail was tested as described25. Three trials were performed and 150-200 animals/genotype tested.
Lifespan analysis
Lifespans of synchronized population cohorts were measured as previously described28. FUDR was applied to the plates, to 10, 40 or 50 μM (see Supplemental Tables). Lifespan experiments were performed at 20°C. A small proportion of animals were censored, usually due to uterine rupture, which mainly occurred at mid-adulthood (~day 9-11).
Statistical analysis
Statistical significance of effects on lifespan were estimated using the log rank test, performed using JMP, Version 7 (SAS Institute).
Drosophila
Fly stocks and maintenance
tubulin-GAL4 and dSir2EP2300 lines were obtained from the Bloomington Stock Center. dSir2-Myc2 and dSir2-Myc9 lines were generated by germ-line transformation. These were outcrossed into white Dahomey (wDah). dSir24.5/SM6B (Ref. 35), dSir217/Cyo (Ref. 36), kindly provided by S. Pletcher and J. Rine, were outcrossed into Canton S. All lines were out-crossed at least 6 times. The presence of the deletion was detected by PCR using the following primers: 149F (5′-AGATATGACATAAGGCAGTGGC-3′), 1427R (5′-TCCCGTTAGCACAATGATCTTC-3′); 3909R (5′-GAAGGCGGTAGCAATGG TGACAA-3′). Flies were maintained at 25°C on a 12h : 12 h light-dark cycle at constant humidity.
Myc-tagged dSir2
The Myc tag was added to RE27621 from Riken using standard techniques and cloned into pUASP. The construct was microinjected into w04 and the transformant lines dSir2-Myc2 and dSir2-Myc9 recovered. Primers: Sir5’R2 (5′-CAAGAATTCCAACGAGAATTTTACACAGGTCGTGTG-3′), Sir3’Xba (5′-ATC GAGTCTAGACACTGCTGCTAACTGTCCTGGAGG-3′) MYC3’Xba (5′-GAGCT ATCTAGAGGATCCGAGGAGCAGAAGCTGATC-3′).
Lifespan assays
Flies were bred at standard density, allowed to mate for 48 hours after emerging (once mated) then sorted into 10 females per vial (UCL) or 35 per vial on 15% SYA (University of Michigan). Vials were changed every 48 hours, and deaths per vial scored until all flies were dead. Numbers of flies used in lifespan assays: over-expression studies, n, ~200 (UCL) or ~350 (U. Michigan). DR studies, n=100. For the over-expression studies, the fly food recipes were as follows. SYA (100 g yeast, 50 g sugar, 15 g agar, 30 ml nipagin and, in most trials, 3 ml propionic acid per litre food), ASG (20 g yeast, 85 g sugar, 10 g agar, 60 g maize, per litre food), ASG1 (31 g yeast, 124 g sugar, 9 g agar, 53 g cornmeal, 25 ml nipagin per litre food), 15% SYA (150 g yeast, 150 g sugar, 21 g agar, 15 ml tegosept). For the DR trials the food dilutions used were as follows. 15 g agar, 30 ml nipagin, 3 ml propionic acid, with yeast and sugar both altered to final concentrations of 10 g, 50 g, 100 g, 150 g, 200 g per litre food. All food was prepared as previously described21.
Genetic crosses
tubulin-GAL4/TM3 males were crossed to dSir2EP2300, dSir2-Myc2 or dSir2-Myc9 virgin females, and dSir2EP2300/+; tubulin-GAL4/+, dSir2-Myc2/+; tubulin-GAL4/+ or dSir2-Myc9/+; tubulin-GAL4/+ females were selected from the progeny. For the controls, tubulin-GAL4/TM3 males or dSir2EP2300, dSir2-Myc2 or dSir2-Myc9 virgin females were crossed to wDah and dSir2EP2300/+, tubulin-GAL4/+, dSir2-Myc2/+ or dSir2-Myc9/+ females were selected from the progeny.
QRT-PCR
RNA was extracted from ten 10 day old females using standard techniques and transcribed into cDNA. 4 biological replicates were run per genotype, each in triplicate. Samples were normalised to either Actin5C or RP49. Primers: Sir2-4 5′-GCTCTCCACCGTTGTCTGAGGGCC-3′ (Ref. 3), Sir2-5 5′-GGCGGCAGCTGTGCTGCGATGAG-3′ (Ref. 3), Actin5CF 5′-CACACCAAATCTTACAAAATGTGTGA-3′, ActinCR 5′-AATCCGGCCTTGCACATG-3′, RP49F 5′-ATGACCATCCGCCCAGCATCAGG-3′, RP49R 5′-ATCTCGCCGCAGTAAACG-3′.
Analysis of dSir2 protein levels
Protein was extracted from 30 females aged 7 days. Western blots were performed using antibodies c-myc 9E10 (Santa Cruz Biotechnology), p2E2 (Developmental Studies Hybridoma Bank), and Tubulin (Sigma).
dSir2 deacetylation assays
dSir2 (RE27621) and dSir2-Myc were cloned into pET SUMO (Invitrogen) and purified on HisPur cobalt spin columns (Thermo Scientific). For the Fluor de Lys assay, using the SirT1 Fluorimetric Drug Discovery Kit (Enzo Life Sciences), results presented are the mean ± S.E.M. of 3 biological replicates. Within each biological replicate samples were run in triplicate. Final concentrations: Resveratrol and suramin 0.2 mM, NAD+ 0.1 mM. Deacetylation of native acetyl histone H4 peptide was monitored by HPLC. Deacetylation of histone H4 N-terminal peptide (SGRGKGGKGLGKGGA(acetyl-K)RHRC) (Biomatik) was carried out using 500 μM NAD+, 100 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 mM DTT and 0.05% Triton X-100 and monitored by HPLC (Agilent 1100) with an ACE C8-300 150×3.0mm column. The elution profiles were analyzed using Chemstation for LC 3D software.
Statistical analyses
Survivorships and the response to DR were compared using the log-rank test and analyses were performed using JMP, Version 7 (SAS Institute).
Additional references
- 31.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sulston J, Hodgkin J. In: The nematode Caenorhabditis elegans. Wood WB, editor. Cold Spring Harbor; 1988. pp. 587–606. [Google Scholar]
- 33.Kamath R, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 34.Greiss S, Hall J, Ahmed S, Gartner A. C. elegans SIR-2.1 translocation is linked to a proapoptotic pathway parallel to cep-1/p53 during DNA damage-induced apoptosis. Genes Dev. 2008;22:2831–2842. doi: 10.1101/gad.482608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman BL, Lundblad JR, Chen Y, Smolik SM. A Drosophila homologue of Sir2 modifies position-effect variegation but does not affect life span. Genetics. 2002;162:1675–1685. doi: 10.1093/genetics/162.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Astrom SU, Cline TW, Rine J. The Drosophila melanogaster sir2+ gene is nonessential and has only minor effects on position-effect variegation. Genetics. 2003;163:931–937. doi: 10.1093/genetics/163.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
Full Methods and any associated references are available in the online version of the paper.
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends life span in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 2.Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Rogina B, Helfand S. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Partridge L, Gems D. Benchmarks for ageing studies. Nature. 2007;450:165–167. doi: 10.1038/450165a. [DOI] [PubMed] [Google Scholar]
- 5.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 7.Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 8.Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- 9.Lin S, Defossez P, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garber K. A mid-life crisis for aging theory. Nat Biotechnol. 2008;26:371–374. doi: 10.1038/nbt0408-371. [DOI] [PubMed] [Google Scholar]
- 13.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenyon C. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 15.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 16.Milne J, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaeberlein M, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 18.Borra M, Smith B, Denu J. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 19.Beher D, et al. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem Biol Drug Des. 2009;74:619–624. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 20.Pacholec M, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bass T, Weinkove D, Houthoofd K, Gems D, Partridge L. Effects of resveratrol on lifespan in Drosophila melanogaster and Caenorhabditis elegans. Mech Ageing Dev. 2007;128:546–552. doi: 10.1016/j.mad.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Hedgecock E, Culotti J, Thomson J, Perkins L. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- 23.Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- 24.Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 25.Parker JA, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 26.Patel DS, et al. Clustering of genetically defined allele classes in the Caenorhabditis elegans DAF-2 insulin/IGF-1 receptor. Genetics. 2008;178:931–946. doi: 10.1534/genetics.107.070813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood J, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 28.Gems D, et al. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borra MT, Denu JM. Quantitative assays for characterization of the Sir2 family of NAD(+)-dependent deacetylases. Methods Enzymol. 2004;376:171–187. doi: 10.1016/S0076-6879(03)76011-X. [DOI] [PubMed] [Google Scholar]
- 30.Grandison RC, Wong R, Bass TM, Partridge L, Piper MD. Effect of a standardised dietary restriction protocol on multiple laboratory strains of Drosophila melanogaster. PLoS One. 2009;4:e4067. doi: 10.1371/journal.pone.0004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.