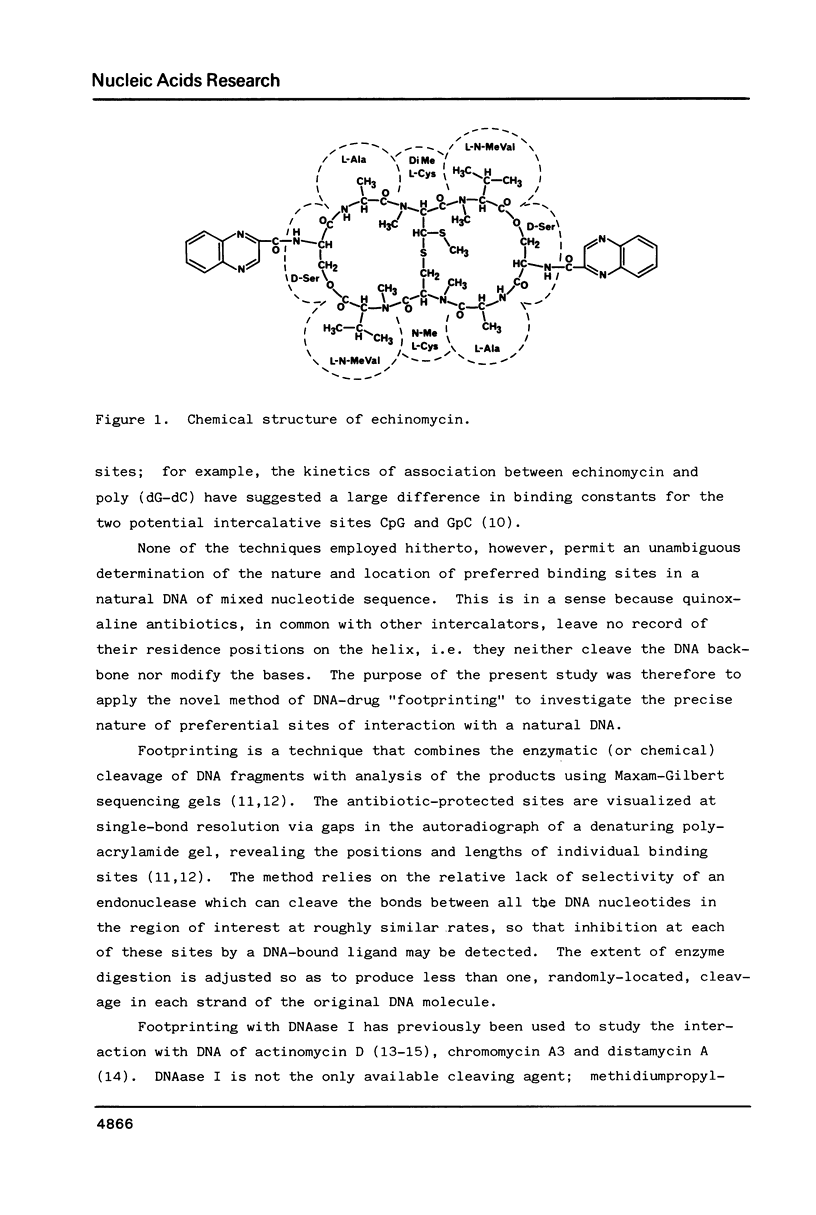
Abstract
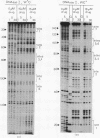
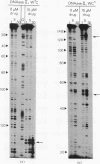
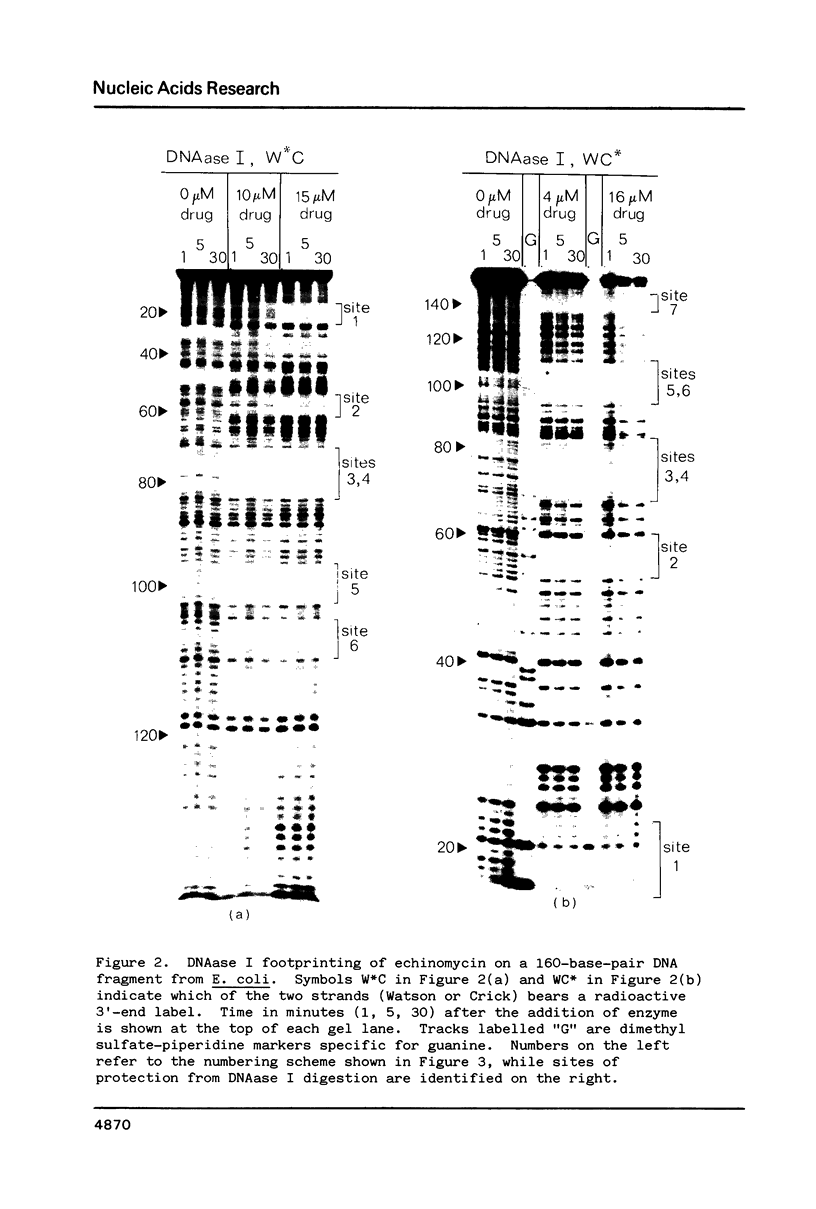
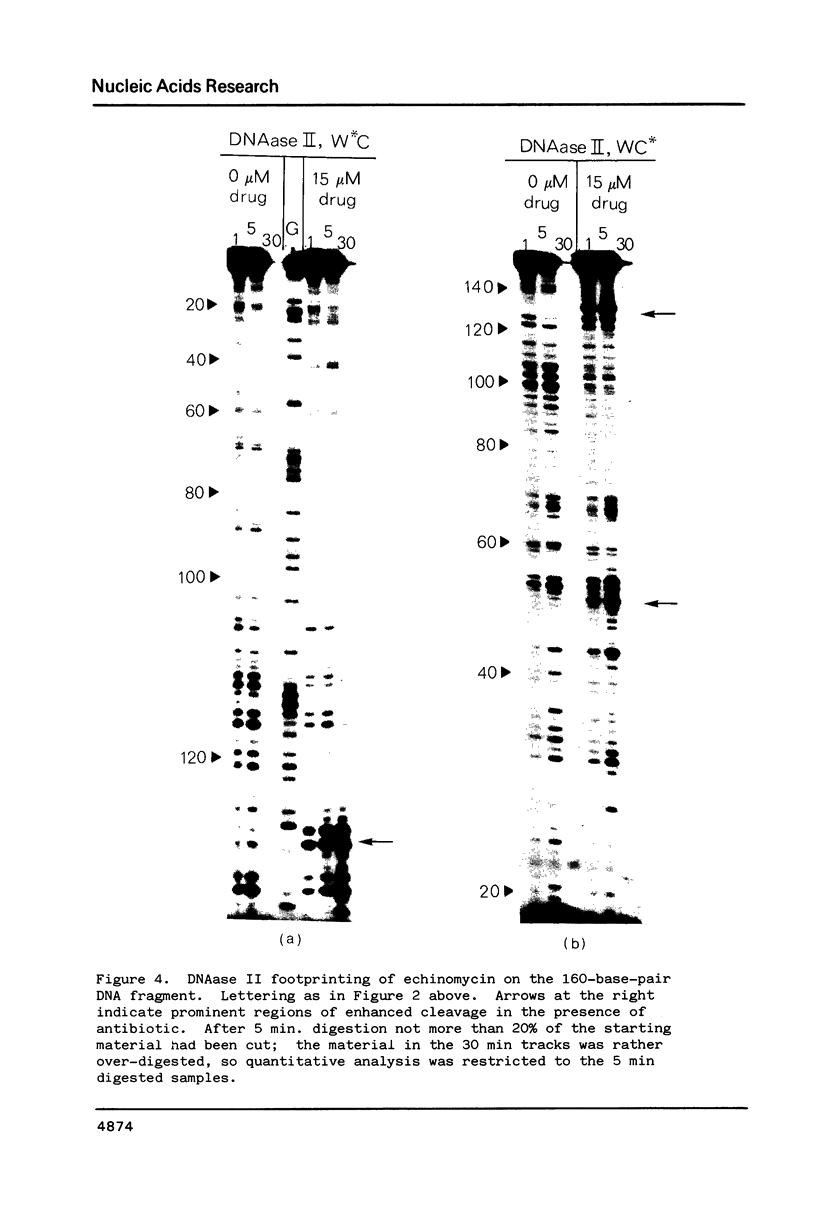
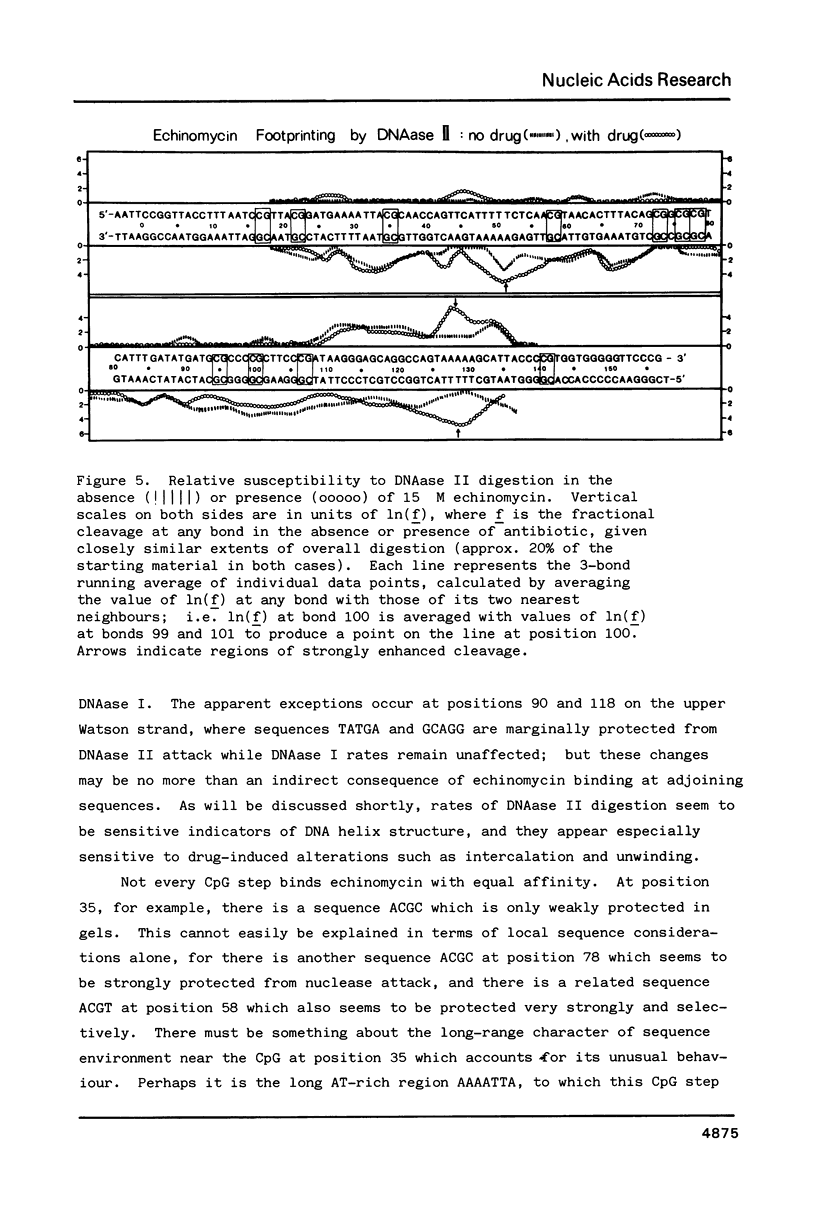
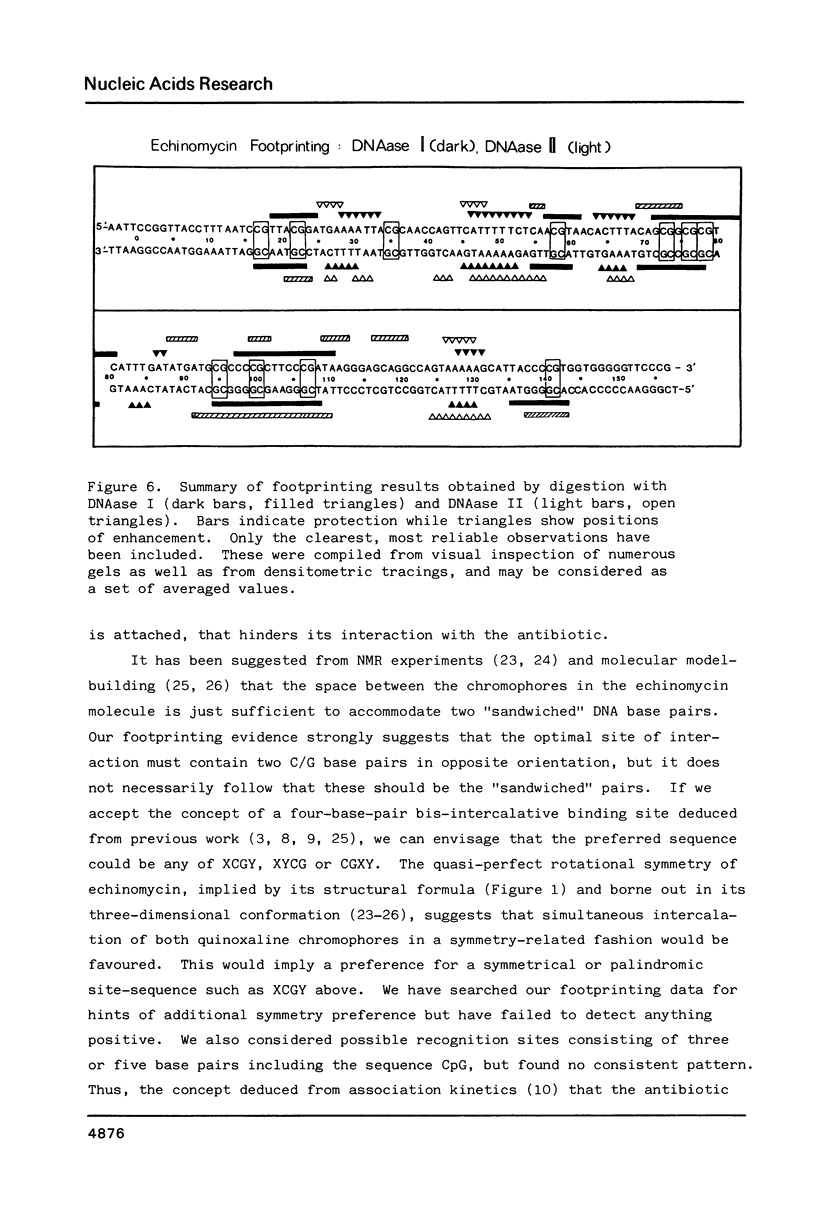
The technique of DNAase I footprinting has been used to investigate preferred binding sites for echinomycin on a 160-base-pair DNA fragment from E. coli containing the tyr T promoter sequence. Six binding sites have been precisely located in the sequence; a seventh has been partially identified. The minimum site-size is six base pairs. All the binding sites contain the dinucleotide sequence CpG but no other regularities can be discerned. When the protected regions on each complementary strand are compared it is evident that they are staggered by 2-3 base-pairs towards the 3' end at each site. Footprinting with DNAase II reports a similar, though less precise, pattern of protection. Cutting by both enzymes is markedly enhanced at AT-rich regions flanking the antibiotic-binding sites. This increased susceptibility to nuclease attack can be attributed to an altered helix conformation in the vicinity of the bis-intercalated echinomycin molecule. It seems that certain sequences, mainly runs of A or runs of T, switch from a nuclease-resistant to a nuclease-sensitive form when echinomycin binds nearby.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Cornish A., Williams R. C., Waring M. J. The use of radiolabelled triostin antibiotics to measure low levels of binding to deoxyribonucleic acid. Biochem J. 1983 Jun 1;211(3):543–551. doi: 10.1042/bj2110543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Gauvreau D., Goodwin D. C., Waring M. J. Binding of quinoline analogues of echinomycin to deoxyribonucleic acid. Role of the chromophores. Biochem J. 1980 Dec 1;191(3):729–742. doi: 10.1042/bj1910729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Olsen R. K., Waring M. J. Interaction between synthetic analogues of quinoxaline antibiotics and nucleic acids: role of the disulphide cross-bridge and D-amino acid centres in des-N-tetramethyl-triostin A. Br J Pharmacol. 1980 Sep;70(1):25–40. doi: 10.1111/j.1476-5381.1980.tb10900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Wakelin L. P., Waring M. J. Kinetics of the interaction between echinomycin and deoxyribonucleic acid. Biochemistry. 1981 Sep 29;20(20):5768–5779. doi: 10.1021/bi00523a020. [DOI] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. J., Dabrowiak J. C., Vournakis J. N. Sequence specificity of actinomycin D and Netropsin binding to pBR322 DNA analyzed by protection from DNase I. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3260–3264. doi: 10.1073/pnas.80.11.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pecq J. B., Le Bret M., Barbet J., Roques B. DNA polyintercalating drugs: DNA binding of diacridine derivatives. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2915–2919. doi: 10.1073/pnas.72.8.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C. DNase II digestion of the nucleosome core: precise locations and relative exposures of sites. Nucleic Acids Res. 1981 Sep 11;9(17):4251–4265. doi: 10.1093/nar/9.17.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcolm A. D., Moffatt J. R., Fox K. R., Waring M. J. Differential inhibition of a restriction enzyme by quinoxaline antibiotics. Biochim Biophys Acta. 1982 Dec 31;699(3):211–216. doi: 10.1016/0167-4781(82)90109-9. [DOI] [PubMed] [Google Scholar]
- Scamrov A. V., Beabealashvilli R. S. Binding of actinomycin D to DNA revealed by DNase I footprinting. FEBS Lett. 1983 Nov 28;164(1):97–101. doi: 10.1016/0014-5793(83)80027-1. [DOI] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979 Jan;6(1):111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A., Lamond A. I., Mace H. A., Berman M. L. RNA polymerase interactions with the upstream region of the E. coli tyrT promoter. Cell. 1983 Nov;35(1):265–273. doi: 10.1016/0092-8674(83)90229-5. [DOI] [PubMed] [Google Scholar]
- Ughetto G., Waring M. J. Conformation of the deoxyribonucleic acid-binding peptide antibiotic echinomycin based on energy calculations. Mol Pharmacol. 1977 May;13(3):579–584. [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Chromomycin, mithramycin, and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA)iron(II). Biochemistry. 1983 May 10;22(10):2373–2377. doi: 10.1021/bi00279a011. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakelin L. P., Romanos M., Chen T. K., Glaubiger D., Canellakis E. S., Waring M. J. Structural limitations on the bifunctional intercalation of diacridines into DNA. Biochemistry. 1978 Nov 14;17(23):5057–5063. doi: 10.1021/bi00616a031. [DOI] [PubMed] [Google Scholar]
- Wakelin S. P., Waring M. J. The binding of echinomycin to deoxyribonucleic acid. Biochem J. 1976 Sep 1;157(3):721–740. doi: 10.1042/bj1570721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M. J., Wakelin L. P. Echinomycin: a bifunctional intercalating antibiotic. Nature. 1974 Dec 20;252(5485):653–657. doi: 10.1038/252653a0. [DOI] [PubMed] [Google Scholar]
- Waring M. J., Wakelin L. P., Lee J. S. A solvent-partition method for measuring the binding of drugs to DNA. Application to the quinoxaline antibiotics echinomycin and triostin A. Biochim Biophys Acta. 1975 Oct 1;407(2):200–212. doi: 10.1016/0005-2787(75)90285-3. [DOI] [PubMed] [Google Scholar]