Abstract
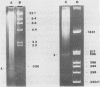
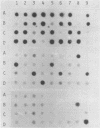
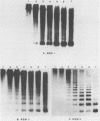
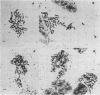
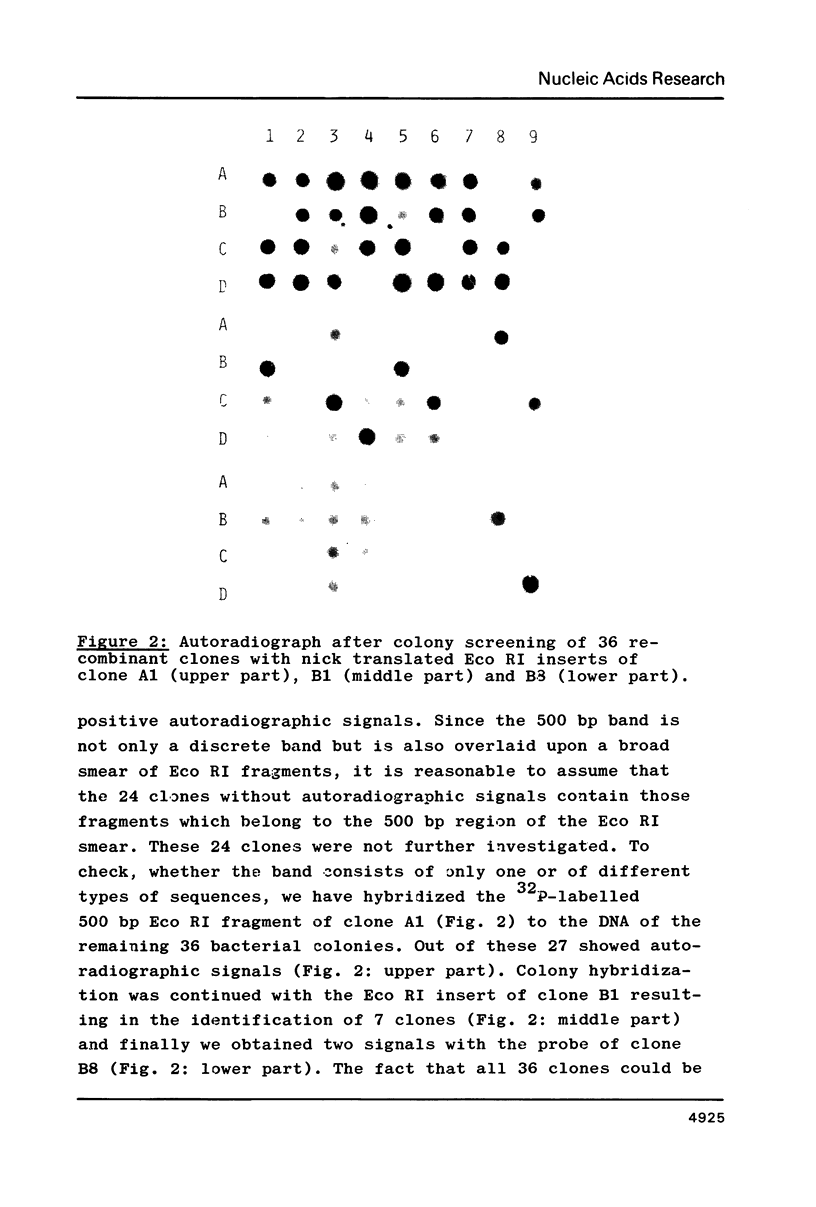
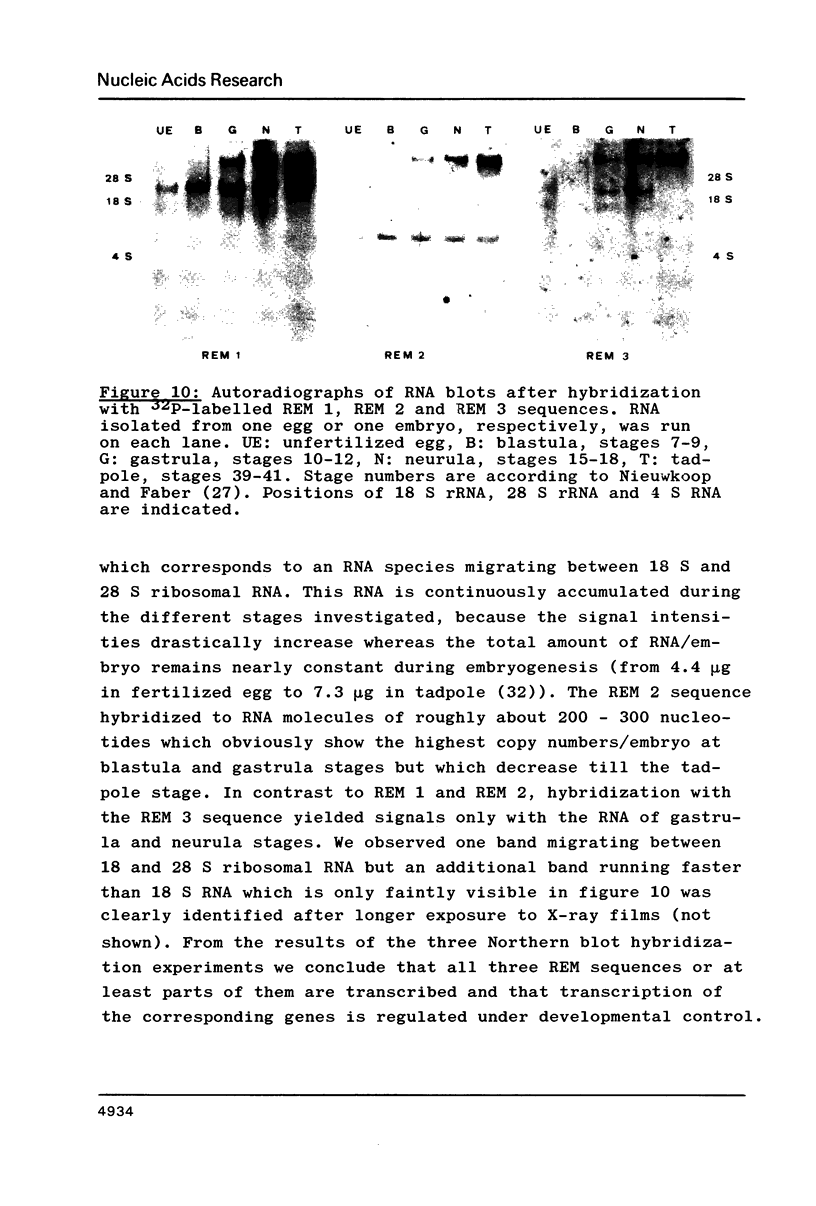
Three different types of repetitive Eco RI fragments, which comigrate within a visible band of approximately 500 bp at gel electrophoresis of Xenopus laevis DNA Eco RI digests have been cloned and sequenced. These sequences are designated as Repetitive Eco RI Monomers: REM 1, REM 2 and REM 3. The sequences contain direct repeats, inverted repeats and palindromic elements. Genomic organization of the most abundant sequence (REM 1; 0.4% of total DNA) is that of an interspersed sequence. REM 2 (0.08%) is partly organized as an interspersed element and partly found in tandem arrangement, whereas REM 3 (0.02%) represents the tandemly repeated monomeric unit of a satellite DNA. In situ hybridization has shown that REM 1 and REM 2 sequences are found on most chromosomes, REM 1 being preferentially located on specific chromosomal loci. REM 3 is located near the centromere region of only one chromosome pair (presumably number 1). Hybridization of Northern blots from RNAs of different developmental stages revealed that REM 1, REM 2 and REM 3 sequences are transcribed and that transcription is under developmental control.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman E. J. Molecular cloning and sequencing of OAX DNA: an abundant gene family transcribed and activated in Xenopus oocytes. EMBO J. 1983;2(8):1417–1422. doi: 10.1002/j.1460-2075.1983.tb01600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRISTOW D. A., DEUCHAR E. M. CHANGES IN NUCLEIC ACID CONCENTRATION DURING THE DEVELOPMENT OF XENOPUS LAEVIS EMBRYOS. Exp Cell Res. 1964 Sep;35:580–589. doi: 10.1016/0014-4827(64)90145-4. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickell P. M., Latchman D. S., Murphy D., Willison K., Rigby P. W. Activation of a Qa/Tla class I major histocompatibility antigen gene is a general feature of oncogenesis in the mouse. Nature. 1983 Dec 22;306(5945):756–760. doi: 10.1038/306756a0. [DOI] [PubMed] [Google Scholar]
- Carmichael G. G., McMaster G. K. The analysis of nucleic acids in gels using glyoxal and acridine orange. Methods Enzymol. 1980;65(1):380–391. doi: 10.1016/s0076-6879(80)65049-6. [DOI] [PubMed] [Google Scholar]
- Chambers J. C., Watanabe S., Taylor J. H. Dissection of a replication origin of Xenopus DNA. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5572–5576. doi: 10.1073/pnas.79.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., Britten R. J. Regulation of gene expression: possible role of repetitive sequences. Science. 1979 Jun 8;204(4397):1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Dawid I. B. Deoxyribonucleic acid in amphibian eggs. J Mol Biol. 1965 Jul;12(3):581–599. doi: 10.1016/s0022-2836(65)80313-8. [DOI] [PubMed] [Google Scholar]
- Fedoroff N. V., Brown D. D. The nucleotide sequence of oocyte 5S DNA in Xenopus laevis. I. The AT-rich spacer. Cell. 1978 Apr;13(4):701–716. doi: 10.1016/0092-8674(78)90220-9. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig W. Molecular hybridization of DNA and RNA in situ. Int Rev Cytol. 1973;36:1–44. doi: 10.1016/s0074-7696(08)60214-4. [DOI] [PubMed] [Google Scholar]
- Jamrich M., Warrior R., Steele R., Gall J. G. Transcription of repetitive sequences on Xenopus lampbrush chromosomes. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3364–3367. doi: 10.1073/pnas.80.11.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay B. K., Dawid I. B. The 1723 element: a long, homogeneous, highly repeated DNA unit interspersed in the genome of Xenopus laevis. J Mol Biol. 1983 Nov 5;170(3):583–596. doi: 10.1016/s0022-2836(83)80122-3. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Lam B. S., Carroll D. Tandemly repeated DNA sequences from Xenopus laevis. I. Studies on sequence organization and variation in satellite 1 DNA (741 base-pair repeat). J Mol Biol. 1983 Apr 25;165(4):567–585. doi: 10.1016/s0022-2836(83)80267-8. [DOI] [PubMed] [Google Scholar]
- Lam B. S., Carroll D. Tandemly repeated DNA sequences from Xenopus laevis. II. Dispersed clusters of a 388 base-pair repeating unit. J Mol Biol. 1983 Apr 25;165(4):587–597. doi: 10.1016/s0022-2836(83)80268-x. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Zeller R. Xenopus laevis U2 snRNA genes: tandemly repeated transcription units sharing 5' and 3' flanking homology with other RNA polymerase II transcribed genes. EMBO J. 1983;2(11):1883–1891. doi: 10.1002/j.1460-2075.1983.tb01675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Meyerhof W., Tappeser B., Korge E., Knöchel W. Satellite DNA from Xenopus laevis: comparative analysis of 745 and 1037 base pair Hind III tandem repeats. Nucleic Acids Res. 1983 Oct 25;11(20):6997–7009. doi: 10.1093/nar/11.20.6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. R., Cartwright E. M., Brownlee G. G., Fedoroff N. V., Brown D. D. The nucleotide sequence of oocyte 5S DNA in Xenopus laevis. II. The GC-rich region. Cell. 1978 Apr;13(4):717–725. doi: 10.1016/0092-8674(78)90221-0. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Gall J. G. Nucleic acid hybridization to the DNA of cytological preparations. Methods Cell Biol. 1975;10:1–16. doi: 10.1016/s0091-679x(08)60727-x. [DOI] [PubMed] [Google Scholar]
- Perlman S., Phillips C., Bishop J. O. A study of foldback DNA. Cell. 1976 May;8(1):33–42. doi: 10.1016/0092-8674(76)90182-3. [DOI] [PubMed] [Google Scholar]
- Peterson R. C., Doering J. L., Brown D. D. Characterization of two xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980 May;20(1):131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- Probst E., Kressmann A., Birnstiel M. L. Expression of sea urchin histone genes in the oocyte of Xenopus laevis. J Mol Biol. 1979 Dec 15;135(3):709–732. doi: 10.1016/0022-2836(79)90173-6. [DOI] [PubMed] [Google Scholar]
- Rosenthal D. S., Doering J. L. The genomic organization of dispersed tRNA and 5 S RNA genes in Xenopus laevis. J Biol Chem. 1983 Jun 25;258(12):7402–7410. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spohr G., Reith W., Crippa M. Structural analysis of a cDNA clone from Xenopus laevis containing a repetitive sequence element. Dev Biol. 1982 Nov;94(1):71–78. doi: 10.1016/0012-1606(82)90069-0. [DOI] [PubMed] [Google Scholar]
- Spohr G., Reith W., Sures I. Organization and sequence analysis of a cluster of repetitive DNA elements from Xenopus laevis. J Mol Biol. 1981 Oct 5;151(4):573–592. doi: 10.1016/0022-2836(81)90424-1. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traboni C., Ciliberto G., Cortese R. A novel method for site-directed mutagenesis: its application to an eukaryotic tRNAPro gene promoter. EMBO J. 1982;1(4):415–420. doi: 10.1002/j.1460-2075.1982.tb01184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett M. A., Jones R. S., Potter S. S. Unusual structure of the FB family of transposable elements in Drosophila. Cell. 1981 Jun;24(3):753–763. doi: 10.1016/0092-8674(81)90101-x. [DOI] [PubMed] [Google Scholar]
- Tymowska J., Kobel H. R. Karyotype analysis of Xenopus muelleri (Peters) and Xenopus laevis (Daudin), Pipidae. Cytogenetics. 1972;11(4):270–278. [PubMed] [Google Scholar]
- Varley J. M., Macgregor H. C., Erba H. P. Satellite DNA is transcribed on lampbrush chromosomes. Nature. 1980 Feb 14;283(5748):686–688. doi: 10.1038/283686a0. [DOI] [PubMed] [Google Scholar]
- Wakefield L., Ackerman E., Gurdon J. B. The activation of RNA synthesis by somatic nuclei injected into amphibian oocytes. Dev Biol. 1983 Feb;95(2):468–475. doi: 10.1016/0012-1606(83)90048-9. [DOI] [PubMed] [Google Scholar]
- Zuker C., Lodish H. F. Repetitive DNA sequences cotranscribed with developmentally regulated Dictyostelium discoideum mRNAs. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5386–5390. doi: 10.1073/pnas.78.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]