Abstract
To investigate the properties of metazoan replication origins, recent studies in cell culture have adopted the strategy of identifying origins using genome-wide approaches and assessing correlations with such features as transcription and histone modifications. Drosophila amplicon in follicle cells (DAFCs), genomic regions that undergo repeated rounds of DNA replication to increase DNA copy number, serve as powerful in vivo model replicons. Because there are six DAFCs, compared with thousands of origins activated in the typical S phase, close molecular characterization of all DAFCs is possible. To determine the extent to which the six DAFCs are different or similar, we investigated the developmental and replication properties of the newly identified DAFC-34B. DAFC-34B contains two genes expressed in follicle cells, although the timing and spatial patterns of expression suggest that amplification is not a strategy to promote high expression at this locus. Like the previously characterized DAFC-62D, DAFC-34B displays origin activation at two separate stages of development. However, unlike DAFC-62D, amplification at the later stage is not transcription-dependent. We mapped the DAFC-34B amplification origin to 1 kb by nascent strand analysis and delineated cis requirements for origin activation, finding that a 6-kb region, but not the 1-kb origin alone, is sufficient for amplification. We analyzed the developmental localization of the origin recognition complex (ORC) and the minichromosome maintenance (MCM)2-7 complex, the replicative helicase. Intriguingly, the final round of origin activation at DAFC-34B occurs in the absence of detectable ORC, although MCMs are present, suggesting a new amplification initiation mechanism.
Keywords: gene amplification, oogenesis, chorion, developmental gene expression
Initiation of DNA replication is a critical regulatory step in complete duplication of the genome. In eukaryotes, DNA replication initiates from hundreds to thousands of sites, called replication origins, across the genome. Bidirectional replication proceeds from these initiation sites until replication forks from adjacent origins converge and the genome is fully replicated. Eukaryotic cells use diverse mechanisms to ensure that origin licensing and origin activation occur in distinct stages of the cell cycle such that each sequence is replicated precisely once per cell division (1). Failure to regulate these processes properly can result in rereplication from a single origin or unreplicated portions of the genome. Various studies have reported the inappropriate expression of proteins involved in origin licensing, such as minichromosome maintenance (MCM)2-7 and CDT1, in human cancers, implicating loss of replication control as a potential cause of genomic instability and an enabling characteristic in the development and progression of cancer (2, 3).
In most eukaryotes apart from budding yeast, there is no sequence-specific motif for localization of origin recognition complex (ORC), the essential replication initiation factor that is conserved in all eukaryotes (4, 5). However, there has been a tremendous increase in the number of metazoan origins identified recently, due to the application of methods to select origin-centered DNA and detection by microarray and high-throughput sequencing technologies (6–12). These origin datasets have enabled correlative analyses to genome-wide features such as transcription, epigenetic modifications, and replication timing. The picture emerging from these studies is that origin activation and ORC binding are significantly influenced by an open chromatin structure, showing a strong correlation with transcription start sites, particularly active promoters. These studies have a number of limitations, however. First, despite the general consensus in conclusions, different origin mapping techniques generated datasets with modest overlap in studies using HeLa cells on a common microrarray platform, suggesting that none was fully comprehensive (7, 10, 12). In addition, the large number of identified origins precluded careful analysis of any individual origin. Finally, these studies relied on cell culture, and studying events directly at the time of origin activation was not possible.
Gene amplification in Drosophila follicle cells is an excellent in vivo model for investigating the regulation of DNA replication initiation (13). It is an example of developmentally programmed DNA rereplication in which repeated origin activation is followed by bidirectional fork progression, resulting in a gradient of replicated DNA spanning ∼100 kb. Importantly for its use as a replication model, gene amplification relies on the same replication machinery and cell cycle kinase regulation used in the typical eukaryotic S phase. Genes that encode structural components of the eggshell and eggshell cross-linking enzymes are located in the peak of amplification at several amplicons (14, 15), and this increase in genomic template enables construction of the eggshell in a short developmental period.
Recently, we identified all Drosophila amplicon in follicle cells (DAFCs) using an array-based comparative genomic hybridization approach (aCGH), which uncovered two new amplicons in addition to the four previously known amplicons (16). That study examined the relationship of gene amplification to transcription, ORC localization, and histone H4 acetylation on a genome-wide scale. Although all six amplicons mapped to regions containing genes, not all contained highly expressed genes. Furthermore, all six amplicons showed ORC localization, but only the two amplicons containing the chorion gene clusters showed significant enrichment of histone H4 acetylation. This work highlighted the diversity of the six amplification origins with respect to transcription and possible factors that might influence localization of prereplicative complex proteins.
Detailed analyses of DAFC-66D, the third chromosome chorion amplicon, and DAFC-62D have revealed major differences beyond those uncovered by our genome-wide studies. At DAFC-66D, replication initiation and exclusive elongation occur in distinct phases of egg chamber development, with origin activation events confined to stages 10B and 11, followed by elongation of existing replication forks (17). In contrast, DAFC-62D displays two stages of replication initiation, one at stage 10B and another at stage 13, with elongation occurring in the intervening stages (15). Furthermore, whereas DAFC-66D amplification can be delineated to the cis interaction of a 320-bp amplification enhancer ACE3 on a major 884-bp replication origin, Oriβ (18), ectopic amplification of DAFC-62D cannot be narrowed down to a region smaller than 10 kb (19). In addition, the second phase of replication initiation at DAFC-62D is transcription-dependent, requiring active transcription for loading of MCMs, but not ORC localization (19).
The differences in developmental and regulatory strategies of DAFC-66D and DAFC-62D, despite their being amplicons of the same cell type and developmental stage, suggested that close molecular analysis of uncharacterized follicle cell amplicons would uncover new developmental programs of gene amplification or mechanisms regulating DNA replication initiation. In the present study, we analyzed DAFC-34B and found distinct examples of developmental and regulatory control strategies from DAFC-66D and DAFC-62D.
Results
Two Genes in DAFC-34B Are Expressed in Follicle Cells.
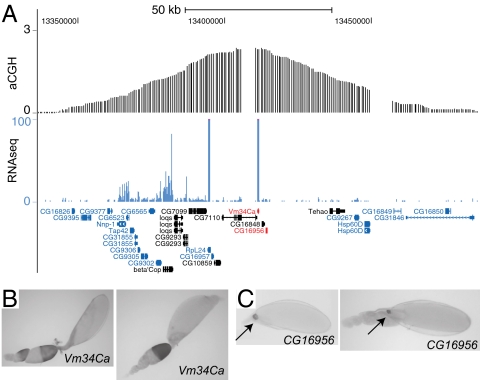
In recent work, we identified DAFC-34B using an aCGH strategy to uncover all follicle cell amplicons (16). Additionally, Illumina RNA sequencing of amplification stage follicle cells showed expression of several genes in the amplified region, including Vm34Ca, a vitelline membrane gene located in the peak of amplification (Fig. 1A). To assess the timing and spatial expression pattern of genes in the amplified region more precisely, we performed in situ hybridization of 10 genes in the central amplified region. Consistent with the RNA-seq data, Vm34Ca was highly expressed in follicle cells. However, this gene was expressed beginning in stage 8, before the onset of synchronous gene amplification at stage 10B, and continued until stage 10B (Fig. 1B), recapitulating previous expression analysis by Northern blot analysis (20).
Fig. 1.
Two genes located in DAFC-34B are expressed in follicle cells. (A) A 100-kb amplified region of DAFC-34B with aCGH in log2 ratio and RNA-seq data in number of mapped reads. The gap space reflects repetitive DNA or nonunique sequences in the Drosophila reference genome sequence not present on the microarray. Ten genes were tested by in situ hybridization for expression in follicle cells. The genes in black were tested but did not show any signal in follicle cells. (B) Vm34Ca is expressed broadly in all follicle cells from stage 8, before the onset of synchronous gene amplification, through stage 10B. (C) CG16956 is expressed exclusively in a small population of cells at the anterior region (arrows). (Left) Stage 12. (Right) Stage 13.
Of the remaining genes that we examined, we found CG16956 expression in stages 12 and 13, but only in a small subset of follicle cells at the anterior region (Fig. 1C), similar to the late-stage expression patterns of yellow-g and yellow-g2 in DAFC-62D (15). Gene amplification generally is considered a strategy for promoting high expression levels. Because the expression patterns of Vm34Ca and CG16956 did not conform to this simple model, showing high expression before the start of synchronous amplification and being expressed in only a limited number of cells, we investigated the precise timing and possible cell population specificity of replication events.
DAFC-34B Shows Two Distinct Stages of Replication Initiation.
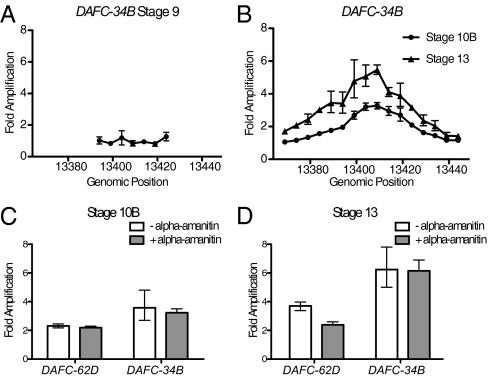
To determine the replication profile at DAFC-34B, we hand-sorted individual egg chamber stages to isolate genomic DNA and assessed DNA copy levels by quantitative real-time PCR (qPCR) compared with a nonamplified locus. Because Vm34Ca is expressed beginning in stage 8, one possibility was that gene amplification begins at an earlier stage at DAFC-34B compared with the other amplicons. However, at stage 9, no amplification was detected at DAFC-34B (Fig. 2A), confirming that Vm34Ca expression occurs before amplification. At stage 10B, we observed an approximate threefold increase in copy number, indicating replication initiation at this stage (Fig. 2B). At stages 11 and 12, we found no further increase in copy number, but noted a slight widening of the replication profile, indicating the process of replication elongation (Fig. S1). At stage 13, we observed a further doubling of DNA copy number, revealing a second phase of DNA replication initiation (Fig. 2B). Thus, DAFC-34B has two stages of replication initiation in the same region separated by a period of elongation, similar to the previously characterized DAFC-62D (19).
Fig. 2.
DAFC-34B exhibits two stages of replication initiation, the second of which is transcription-independent. Genomic DNA was isolated from hand-sorted staged egg chambers, and DNA copy levels were quantified by qPCR compared with a nonamplified locus at 62C5. Error bars show SD for triplicate reactions. Genomic position is shown on the x-axis (13380 is Chr2L:13,380,000). (A) Gene amplification is not first observed at stage 9, despite expression of Vm34Ca. (B) Replication initiation occurs in stage 10B to reach threefold amplification, followed by a second round of replication initiation at stage 13. (C and D) Whole ovaries were in vitro- cultured with α-amanitin for 5 h, after which staged egg chambers were hand-dissected and analyzed as in A and B. qPCR quantification of stage 10 and stage 13 are shown for DAFC-34B and DAFC-62D. (C) At stage 10, α-amanitin treatment has no effect on amplification at either amplicon. (D) At stage 13, α-amanitin treatment specifically inhibits late amplification at DAFC-62D, but has no effect at DAFC-34B.
CG16956 expression in stages 12 and 13 was consistent with the timing of gene amplification, but the expression pattern in a small number of cells raised the possibility that DAFC-34B might be amplified differently in different follicle cell populations. To address whether CG16956-expressing cells amplify this genomic region to higher levels than other follicle cell populations, we used a cell-sorting approach to enrich for CG16956-expressing cells. The expression pattern of CG16956 was reminiscent of slow borders (slbo), a gene expressed in and required for the migration of border cells (21). We found that CG16956 expression colocalized with the slbo marker (Fig. S2A). We isolated slbo-expressing cells by FACS, using the GAL4-UAS system to drive GFP expression with the slbo regulatory region (slbo-GAL4) (22). Compared with DNA recovered using a ubiquitous follicle cell driver (c323a), slbo-positive cells did not display higher gene amplification levels (Fig. S2B).
We next addressed whether transcription influences DAFC-34B amplification. At DAFC-62D, the final round of initiation is dependent on transcription, as demonstrated by the finding that culturing egg chambers in the presence of α-amanitin, which blocks RNAPII translocation, for 5 h specifically blocked stage 13 origin activation but had no effect on stage 10 amplification (19). Given that both DAFC-34B and DAFC-62D exhibit two stages of replication initiation, we tested the effect of transcription inhibition on DAFC-34B amplification to see if these late-stage amplification events occur by the same mechanism. As for DAFC-62D, 5 h of α-amanitin treatment had no effect on stage 10B origin activation at DAFC-34B (Fig. 2C). However, whereas α-amanitin specifically blocked stage 13 origin activation at DAFC-62D, as reported previously (19), it had no effect on late-stage origin activation at DAFC-34B (Fig. 2D), indicating a different mode of regulation.
DAFC-34B Amplification Origin Corresponds to Vm34Ca Transcription Unit.
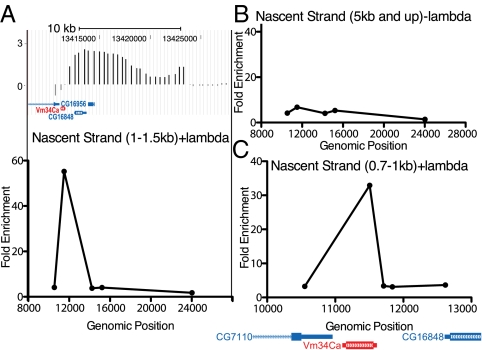
Mapping of the amplification origins of DAFC-66D and DAFC-62D has demonstrated distinct origin positions with respect to the surrounding genes. The major origin at the chorion amplicon DAFC-66D is intergenic, although still residing in a gene-rich region (23), whereas the origin at DAFC-62 coincides with the yellow-g2 gene (19). We used nascent strand analysis to map the stage 10 origin at DAFC-34B and found that the DAFC-34B amplification origin coincides with the transcription unit of Vm34Ca (Fig. 3A). We found that this gene region was highly enriched in the 1.0- to 1.5-kb fraction of DNA. Because of the small size of the Vm34ca transcription unit (455 bp) and the larger size fraction of the nascent DNA that we tested, we could not precisely distinguish whether the origin mapped to the gene region or to the transcription start site. As a control for the efficiency of nuclease digestion and the recovery of short nascent strands, we found that DNA larger than 5 kb showed uniform low enrichment levels across the 10 kb most-amplified region of DAFC-34B (Fig. 3B). We used primers to detect nascent strand abundance in the other gene regions located in DAFC-34B, but found no enrichment (Fig. 3C).
Fig. 3.
The DAFC-34B replication origin maps to the Vm34Ca gene. (A) Size-fractionated nascent DNA was isolated from stage 10 egg chambers, and the abundance of short nascent strands from the 1- to 1.5-kb fraction (plus λ-exonuclease treatment) was quantified by qPCR compared with a nonamplified locus. Genomic position is shown on the x-axis (8000 is Chr2L:13,408,000). The top of the figure shows the ORC2 ChIP on chip data for the region. There is a peak of nascent strand enrichment corresponding to the Vm34Ca transcription unit. (B) This enrichment is absent in the 5-kb and up fraction (minus λ-exonuclease treatment). (C) The region was assessed using additional primers in the 0.7- to 1-kb fraction (plus λ-exonuclease treatment). The enrichment was specific to the Vm34Ca gene and not to adjacent gene regions.
Developmental Control of ORC and MCM Localization at DAFC-34B.
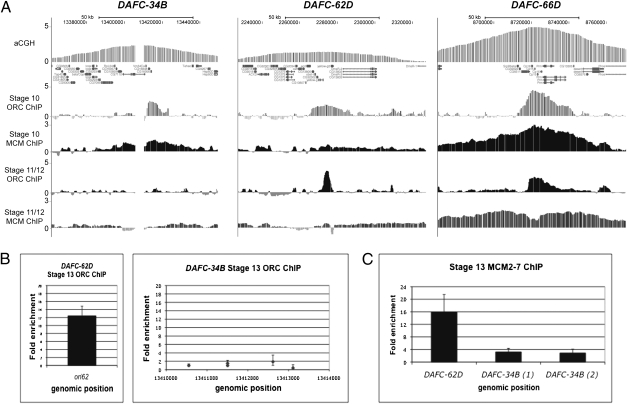
Because DAFC-34B exhibits two rounds of replication initiation, we examined the stage-specific localization of ORC at this locus to see how it corresponds to origin firing. Genome-wide localization using chromatin immunoprecipitation followed by hybridization to a microarray (ChIP-chip) with an ORC2 antibody in stage 10 egg chambers showed that ORC localizes to all follicle cell amplicons in a large zone centered at the peak of amplification (Fig. 4A) (16). We confirmed this result at DAFC-34B by ChIP-qPCR and observed the same 10-kb zone of ORC localization as seen with ChIP-chip (Fig. S3). By both methods, we observed a sharp boundary of ORC binding that coincided with Vm34Ca, corresponding to the origin that we identified by nascent strand analysis. Strikingly, when we examined subsequent stages for ORC localization at DAFC-34B, we found that ORC enrichment was absent despite the second round of replication initiation. ORC2 ChIP-chip for pooled stage 11 and 12 egg chambers revealed no detectable ORC at DAFC-34B (Fig. 4A). In contrast, ORC was present at DAFC-62D, as was previously demonstrated by ChIP-qPCR (19) (Fig. 4A). In addition, examination of ORC localization at DAFC-34B and DAFC-62D at stage 13 by ChIP-qPCR revealed no ORC enrichment at DAFC-34B, but ORC enrichment at DAFC-62D (Fig. 4B).
Fig. 4.
ORC is not detectable at DAFC-34B after stage 10 despite a late stage of replication initiation. ChIP-chip experiments using antibodies specific for ORC2 and MCM2-7 were performed on stage 10 and pooled stages 11 and 12 egg chambers. (A) The amplified regions for DAFC-34B, DAFC-62D, and DAFC-66D. ORC localizes to DAFC-34B in a 10-kb zone with a boundary of ORC binding corresponding to the Vm34Ca transcription unit. After stage 10, there is no localization of ORC at DAFC-34B despite enrichment at DAFC-62D, which also exhibits late amplification. DAFC-66D, which shows replication initiation at stages 10 and 11, is shown for comparison. (B) ChIP-qPCR shows ORC enrichment at DAFC-62D in stage 13 egg chambers, but not at DAFC-34B. (C) Although ORC is not detected at DAFC-34B after stage 10, MCM enrichment is observed at stage 13 by ChIP-qPCR.
We examined the localization of MCMs by ChIP and found that the complex localizes to DAFC-34B, coincident with both stages of replication initiation. At stage 10, we observed MCM enrichment at DAFC-34B by ChIP-chip (Fig. 4A). At stage 13, we observed MCM enrichment by ChIP-qPCR at levels comparable to those found by ChIP-chip at stage 10 (Fig. 4C). This indicates that the late-stage initiation at DAFC-34B does not occur in the complete absence of all prereplicative components. We did not find MCM enrichment in pooled stages 11 and 12 egg chambers at DAFC-34B, indicating that preloaded MCMs are not responsible for stage 13 origin activation (Fig. 4A). In contrast, at DAFC-66D, which undergoes replication initiation at stage 11, we observed significant MCM enrichment in pooled stages 11 and 12 (Fig. 4A). DAFC-34B is an example of replication initiation occurring in the absence of detectable ORC despite the presence of MCM complexes.
ORC Mutant Blocks Both Rounds of Initiation at DAFC-34B.
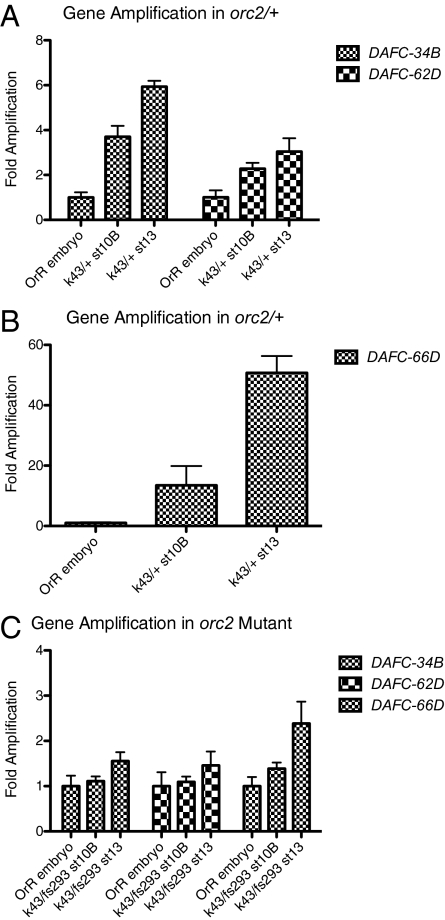
Given the finding of replication initiation in the absence of ORC, we genetically tested whether each period of origin activation is dependent on functional ORC. orc2fs293 is a female-sterile allele that specifically reduces follicle gene amplification, and orc2k43 is a null allele (24). We reasoned that because ORC is absent in late-stage amplification, its function might not be required for this second round of replication initiation, and thus we would observe amplification specifically at stage 13. On examination of DNA copy levels for stages 10B and 13 egg chambers by qPCR, however, we found that the mutant effect on gene amplification at DAFC-34B was comparable to that of the other two amplicons examined (Fig. 5C). All three amplicons showed 1.5- to 2-fold enrichment at stage 13, likely due to low levels of ORC activity and delayed replication. These results indicate that both rounds of replication initiation require ORC, although initiation at stage 13 in the absence of ORC might be explained by some earlier action of ORC that promotes replication initiation; for example, the first round of replication initiation at DAFC-34B might set up conditions that permit the final round of origin firing in the absence of ORC function.
Fig. 5.
ORC function is required to allow both stages of replication initiation at DAFC-34B. DNA copy number was examined in a female-sterile allele of ORC, orc2fs293, which reduces gene amplification. orc2k43 is a null allele. Amplification in the sibling heterozygotes shows typical amplification levels. In orc2fs293/orc2k43, there was no specific increase in gene copy number by stage 10B at DAFC-34B. There was a 1.5- to 2-fold amplification by stage 13 at DAFC-34B, DAFC-62D, and DAFC-66D, likely due to low levels of ORC activity and delayed replication.
Delineation of cis Control Elements for Replication at DAFC-34B.
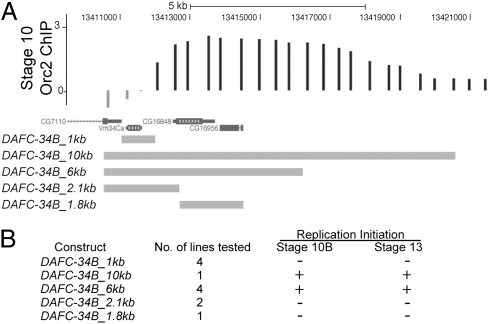
To delineate the cis requirements for amplification at DAFC-34B and determine whether distinct control elements for early- and late-stage initiation can be identified, we used P element-mediated transformation to test the sufficiency for amplification at an ectopic site. We used suppressor of Hairy-wing insulator binding sites to flank the test sequences and minimize inhibitory effects on gene amplification due to integration position (25). We tested the 1-kb origin sequence alone and found that it was insufficient for amplification (Fig. 6 and Fig. S4). Additional sequences, possibly conferring replication enhancer activity, are needed for replication initiation. qPCR analysis revealed a transgenic sequence of 10 kb spanning the stage 10 ORC binding zone is sufficient for gene amplification. This transformant line showed the same timing and magnitude of amplification as the endogenous DAFC-34B locus, indicating that this region contains all of the information necessary for amplification (Fig. 6 and Fig. S4).
Fig. 6.
A 6-kb ORC binding region is sufficient for amplification of DAFC-34B. (A) DNA sequences tested by P element-mediated transformation. (B) Results of ectopic amplification. Neither the 1-kb origin alone nor a 2.1-kb region containing the origin is sufficient for amplification. The 6-kb region containing the 1-kb origin as well as the peak of ORC binding is the minimal sequence that conferred amplification activity.
Previous analyses of DAFC-66D and DAFC-62D have shown that not all follicle cell amplicons can be reduced to equivalent replication control elements. At DAFC-66D, the interaction of ACE3 and Oriβ is sufficient for amplification (18), whereas at DAFC-62D, a region sufficient for amplification cannot be limited to <10 kb (19). DAFC-34B displays an intermediate result. We found that a 6-kb region that spans the peak of ORC binding is sufficient for amplification; however, neither a 2.1-kb fragment containing the DAFC-34B origin nor a 1.8-kb fragment spanning the peak of ORC binding is sufficient for amplification. We identified no distinct replication control elements for the two developmental stages of gene amplification with the sequences that we tested.
Discussion
We have investigated the developmental and regulatory strategies of DAFC-34B and found unique characteristics that make it a valuable new replication model for delineating the relationship between transcription and gene amplification as well as the requirements of ORC function for gene amplification. Although DAFC-34B contains genes expressed in follicle cells, the timing and spatial pattern of expression are previously unobserved compared with the other amplicons. Despite two rounds of replication initiation, we found that the second round of origin firing is not dependent on transcription, as it is in DAFC-62D. Strikingly, we found that the second round of replication initiation occurs in the absence of detectable ORC, even though MCMs are loaded. Finally, we delineated the cis requirements for replication and found that a 6-kb minimal region is sufficient for origin function.
Despite their diverse approaches, recent origin mapping studies have all found that replication origins and ORC binding sites closely correspond to gene regions, specifically the transcription start sites of actively transcribed genes (8, 10, 11). The relationship between active transcription and DNA replication initiation remains unclear, however. Consistent with the majority of origins identified in cell cultures, we found that the DAFC-34B amplification origin corresponds to the transcription unit of Vm34Ca. Nevertheless, the DAFC-34B 2.1-kb construct does not amplify, even though it contains the full-length Vm34Ca gene and at least 1 kb 5′ and 3′ to the gene. Although transcription inhibition by 5-h α-amanitin treatment had no effect on stage 10 and stage 13 amplification, we cannot exclude the possibility that transcription is necessary for stage 10 amplification, because Vm34Ca is expressed for more than 12 h, and inhibiting expression for the duration of stages 8–10 is not possible in culture. Thus, DAFC-34B can be used as a model replication origin to identify the DNA regulatory elements responsible for origin activity and explore how these elements are related to transcriptional regulation.
DAFC-34B is also a distinctive replication model, because the second round of replication initiation occurs in the absence of detectable ORC. ORC binds to the region in a 10-kb zone during stage 10, but is not detectable by ChIP in subsequent stages. In contrast, DAFC-62D (which also exhibits late-stage replication initiation) demonstrates ORC binding throughout the later stages. Despite the absence of ORC, MCMs are enriched at DAFC-34B by stage 13. There is no pool of MCMs in this region during stages 11 and 12, indicating that ORC activity at stage 10 does not help load MCMs that will be activated at a later stage. What, then, can explain the absence of ORC at DAFC-34B after stage 10? One possible model is that ORC is required for late initiation, but failure to detect enrichment by ChIP is due to masking of the epitope, possibly from a significant structural rearrangement uniquely at DAFC-34B after stage 10. Another possibility is that ORC is distributed in a broad initiation zone after stage 10, making specific intervals of ORC localization undetectable, although the levels would have to be quite low. Conversely, ORC might not be required for late initiation at DAFC-34B, and this amplicon may demonstrate ORC-independent MCM loading and replication initiation.
There are at least three mechanistic explanations for ORC-independent replication initiation. First, Park and Asano (26) reported that ORC is dispensable for endoreplication in Drosophila, demonstrating that nuclei in homozygous mutant clones of an orc1 null allele reach the same size as nuclei in WT clones. However, cell proliferation and gene amplification, assessed by loss of BrdU foci, were disrupted in these mutants. Asano (27) proposed the existence of a protein or complex X that can recruit Cdc6 and promote replication initiation. Such a factor, if it exists, also might play a role in late initiation at DAFC-34B. Second, DUP/CDT1 persists in late-stage egg chambers and is present at elongating replication forks (17). A pool of DUP/CDT1 at the origin might recruit MCMs for late amplification at DAFC-34B. Finally, Lydeard et al. (28) reported that break-induced replication in yeast does not require ORC or Cdc6. The replication profile at DAFC-34B shows that late initiation occurs at a specific developmental stage and from the same region as early initiation. Thus, it seems unlikely that double-stranded breaks generated by collapsed replication forks could result in the symmetrical replication profile that we observed. However, if the first round of replication initiation led to increased susceptibility of the DAFC-34B origin to incur a double-stranded break, then random strand invasion occurring from both sides of the break could result in a symmetric replication gradient. The need for the first round of replication initiation to create a double-stranded break also would explain the need for ORC function in both stages of amplification. The ORC mutant does not show late-stage–specific replication initiation, consistent with the second and third models and the need for generation of replication forks in the first round of replication initiation to set up some type of competence for the second round of initiation in the absence of ORC.
Our investigation of the developmental and replication profiles of DAFC-34B shows that it is a powerful model for gaining mechanistic insight into metazoan replication initiation. DAFC-34B has unique properties of developmental gene expression and replication initiation in the absence of detectable ORC, making it a tractable experimental model for studying the influence of transcription on replication initiation and possible ORC-independent means of replication initiation. Furthermore, analysis of multiple follicle cell amplicons highlights the diversity of amplification control mechanisms within the same cell type and is likely representative of similar regulatory diversity during S-phase DNA replication. Finally, because follicle cell gene amplification is developmentally programmed and occurs in cells that will not undergo additional cell division, it is a useful model for investigating the properties that allow DNA to undergo rereplication. Sequencing of cancer genomes has revealed that neoplastic cells display significant levels of chromosomally integrated gene amplification, although the mechanism of this amplification is unknown (29). One model implicates a causal role of rereplication on this process, and recent work in budding yeast has shown that loss of replication control and rereplication from a single origin results in chromosomally integrated gene amplification (30). Understanding how follicle cell amplification origins are regulated is likely to shed light on why certain genomic sequences and not others are capable of repeated origin activation and how these properties might contribute to genomic instability in cancer.
Materials and Methods
Quantitative PCR.
Absolute qPCR was performed as described previously (15). Standard curves were generated from four 10-fold dilutions of stage 1–8 egg chamber DNA. The endogenous control was a nonamplified locus at 62C5 (17). Absolute quantification was used for the DAFC-34B replication profile stage 9 and stage 10A samples as well as for the cell population experiments. Relative quantification was used for all other experiments as described previously (19).
Nascent Strand Analysis.
Nascent strand abundance analysis was performed for stage 10 egg chambers as described previously (19). We hand-sorted stage 10 egg chambers, enriched for replication intermediates, and size-fractionated DNA fragments by gel electrophoresis. Short nascent strands were further enriched using λ-exonuclease treatment to remove nicked DNA lacking 5′ RNA primers. Each fraction was analyzed for the abundance of specific sequences by relative qPCR using 0–4 h embryonic DNA as the calibrator and a nonamplified locus at 62C5 as the endogenous control.
Chromatin Immunoprecipitation.
ChIP-qPCR was performed on 300 staged egg chambers per experiment as described previously (19). ChIP-chip was performed as described by Kim et al. (16). All experiments were compared with input DNA. For hybridization to arrays, DNA was labeled using Invitrogen's BioPrime Total for Agilent aCGH labeling kit. Array intensities were median normalized across channels and smoothed by genomic windows of 1 kb using the Ringo package in R (31). All data have been deposited at Gene Expression Omnibus: ORC2 and MCM2-7 ChIP-chip from Drosophila egg chambers (GEO accession no. GSE31891).
See SI Materials and Methods for additional information.
Supplementary Material
Acknowledgments
We thank Steve Bell for the ORC2 and MCM antibodies, Mary-Lou Pardue for the GFP antibodies, and the Bloomington Stock Center for the flies. We also thank Steve Bell and Peter Reddien for helpful comments on a manuscript draft, Jared Nordman for helpful comments and assistance with the ChIP-chip analysis, and Fang Xie for technical assistance with the α-amanitin experiments. This work was supported by National Institutes of Health Grant GM57960 and by an American Cancer Society Research Professorship (to T.L.O.-W.).
Footnotes
The authors declare no conflict of interest.
Database deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE31891).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114209108/-/DCSupplemental.
References
- 1.Arias EE, Walter JC. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 2.Blow JJ, Gillespie PJ. Replication licensing and cancer—a fatal entanglement? Nat Rev Cancer. 2008;8:799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Cvetic C, Walter JC. Eukaryotic origins of DNA replication: Could you please be more specific? Semin Cell Dev Biol. 2005;16:343–353. doi: 10.1016/j.semcdb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert DM. In search of the holy replicator. Nat Rev Mol Cell Biol. 2004;5:848–855. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas I, et al. High-throughput mapping of origins of replication in human cells. EMBO Rep. 2007;8:770–777. doi: 10.1038/sj.embor.7401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadoret JC, et al. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci USA. 2008;105:15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sequeira-Mendes J, et al. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet. 2009;5:e1000446. doi: 10.1371/journal.pgen.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci USA. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karnani N, Taylor CM, Malhotra A, Dutta A. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell. 2010;21:393–404. doi: 10.1091/mbc.E09-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mesner LD, et al. Bubble-chip analysis of human origin distributions demonstrates on a genomic scale significant clustering into zones and significant association with transcription. Genome Res. 2011;21:377–389. doi: 10.1101/gr.111328.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claycomb JM, Orr-Weaver TL. Developmental gene amplification: Insights into DNA replication and gene expression. Trends Genet. 2005;21:149–162. doi: 10.1016/j.tig.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Spradling AC. The organization and amplification of two chromosomal domains containing Drosophila chorion genes. Cell. 1981;27:193–201. doi: 10.1016/0092-8674(81)90373-1. [DOI] [PubMed] [Google Scholar]
- 15.Claycomb JM, Benasutti M, Bosco G, Fenger DD, Orr-Weaver TL. Gene amplification as a developmental strategy: Isolation of two developmental amplicons in Drosophila. Dev Cell. 2004;6:145–155. doi: 10.1016/s1534-5807(03)00398-8. [DOI] [PubMed] [Google Scholar]
- 16.Kim JC, et al. Integrative analysis of gene amplification in Drosophila follicle cells: Parameters of origin activation and repression. Genes Dev. 2011;25:1384–1398. doi: 10.1101/gad.2043111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claycomb JM, MacAlpine DM, Evans JG, Bell SP, Orr-Weaver TL. Visualization of replication initiation and elongation in Drosophila. J Cell Biol. 2002;159:225–236. doi: 10.1083/jcb.200207046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu L, Zhang H, Tower J. Functionally distinct, sequence-specific replicator and origin elements are required for Drosophila chorion gene amplification. Genes Dev. 2001;15:134–146. doi: 10.1101/gad.822101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie F, Orr-Weaver TL. Isolation of a Drosophila amplification origin developmentally activated by transcription. Proc Natl Acad Sci USA. 2008;105:9651–9656. doi: 10.1073/pnas.0804146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mindrinos MN, et al. Isolation and chromosomal location of putative vitelline membrane genes in Drosophila melanogaster. EMBO J. 1985;4:147–153. doi: 10.1002/j.1460-2075.1985.tb02329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montell DJ, Rorth P, Spradling AC. slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, et al. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev Cell. 2006;10:483–495. doi: 10.1016/j.devcel.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Heck MM, Spradling AC. Multiple replication origins are used during Drosophila chorion gene amplification. J Cell Biol. 1990;110:903–914. doi: 10.1083/jcb.110.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landis G, Kelley R, Spradling AC, Tower J. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc Natl Acad Sci USA. 1997;94:3888–3892. doi: 10.1073/pnas.94.8.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu L, Tower J. A transcriptional insulator element, the su(Hw) binding site, protects a chromosomal DNA replication origin from position effects. Mol Cell Biol. 1997;17:2202–2206. doi: 10.1128/mcb.17.4.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SY, Asano M. The origin recognition complex is dispensable for endoreplication in Drosophila. Proc Natl Acad Sci USA. 2008;105:12343–12348. doi: 10.1073/pnas.0805189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asano M. Endoreplication: The advantage to initiating DNA replication without the ORC? Fly (Austin) 2009;3:173–175. doi: 10.4161/fly.8087. [DOI] [PubMed] [Google Scholar]
- 28.Lydeard JR, et al. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24:1133–1144. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 30.Green BM, Finn KJ, Li JJ. Loss of DNA replication control is a potent inducer of gene amplification. Science. 2010;329:943–946. doi: 10.1126/science.1190966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toedling J, et al. Ringo—an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics. 2007;8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.