Abstract
Theoretical studies predict hydrophobic matching between transmembrane domains of proteins and bilayer lipids to be a physical mechanism by which membranes laterally self-organize. We now experimentally study the direct consequences of mismatching of transmembrane peptides of different length with bilayers of different thicknesses at the molecular level. In both model membranes and simulations we show that cholesterol critically constrains structural adaptations at the peptide-lipid interface under mismatch. These constraints translate into a sorting potential and lead to selective lateral segregation of peptides and lipids according to their hydrophobic length.
Keywords: mattress model, membrane domain, protein–lipid interaction, self-assembly, annular lipid
Hydrophobicity is the key criterion for lipids and proteins to integrate into membranes (1). This self-assembly is driven by the minimization of hydrophobic surface area exposed to the aqueous phase (2). Next to sterols, eukaryotic membranes contain a variety of phospholipids with different chain length (3) and proteins with a variety of transmembrane (TM) domain lengths (4). Why cell membranes contain so many lipids is still enigmatic. But one reason could be membrane sorting due to grouping of TM proteins and lipids with similar hydrophobic length.
The “mattress model” predicts that the embedding of a rigid, helical TM protein into a fluid bilayer causes the lipids to adapt locally to a mismatch (5). In this way exposure of hydrophobic surface area is minimized. The adaptive flexing and straightening of the lipids can also be accompanied by a tilting of the protein (6). Because of these strain-causing adaptations of the bilayer, selective association of matching lipids with TM proteins as well as macroscopic sorting processes according to hydrophobic length have been predicted by theory and simulation (7, 8). Membrane properties such as elasticity—modulated by cholesterol—were also predicted as crucial parameters (9). Such mismatch-dependent, cholesterol-induced sorting has indeed been proposed as a retention mechanism for the Golgi-resident proteins in the secretory pathway (10).
Due to the complexity of cell membranes, model membranes have proven a valuable system to investigate hydrophobic matching (11–13). Hydrophobic peptides of the poly-leucine type have been used as generic TM proteins because of the ease of their organic solvent-based reconstitution (14, 15). From these experiments, it has become clear that TM proteins indeed tolerate moderate mismatch with the bilayer (13, 14, 16). However, large mismatch as well as cholesterol have been found to reduce efficiency of TM peptide incorporation into bilayers, suggesting that there are energetic limitations to mismatch buffering (12).
Despite indication for selective protein–lipid interactions and hydrophobic matching (17, 18), actual sorting of TM proteins and accompanying lipid cosorting has not been reconstituted in vitro. Therefore, it remains unclear whether hydrophobic mismatch is a significant parameter in the organization of proteinacious membranes.
Using TM peptides and lipids of defined acyl chain length in atomistic molecular dynamics (MD) simulations and reconstituted proteo-liposomes we have investigated hydrophobic mismatching with respect to (i) the structural impact of mismatched helices on lipid bilayers; and (ii) its consequences on the lateral distribution of peptides and lipids.
Our results suggest that cholesterol severely strains the peptide–lipid interface under mismatch if the bilayer is thicker than the TM segment. Moreover, it enforces a redistribution of TM peptides and lipids according to hydrophobic length. We propose that cholesterol reduces the adaptability of the lipids to mismatched proteins and therefore makes hydrophobic mismatch energetically more relevant. Our data thus provide a molecular basis for the role of TM domain (TMD) length and cholesterol in protein and lipid sorting in the secretory pathway.
Results
First we investigated the structure of the protein–lipid interface in model systems under conditions of hydrophobic mismatch. We carried out atomistic MD simulations pcarallel to organic solvent reconstitution of bilayers containing LW19 (K2W2L7AL7W2K2), LW21 (K2W2L8AL8W2K2), or LW29 (K2W2L12AL12W2K2) peptides (Fig. S1A). To vary hydrophobic mismatch we used a series of monounsaturated phosphatidylcholines (PC), having different fatty acid lengths from C16∶1 to C24∶1 (13, 19) and defined positive mismatch as lengthprotein > thicknessbilayer and negative mismatch as lengthprotein < thicknessbilayer (Table S1).
Atomistic MD Simulations.
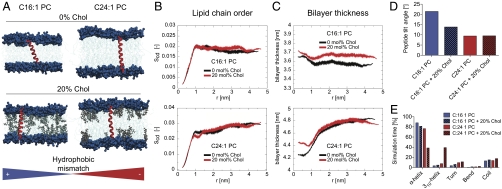
In the 500 ns atomistic MD simulations we used the two extreme conditions of LW21 in C16∶1 PC and C24∶1 PC in the absence and presence of 20 mol% cholesterol (Fig. 1A and Movies S1–S4). In the C16∶1 PC membranes the radial profiles and average maps showed increased acyl chain order and bilayer thickness around the TM helix (Fig. 1 B and C and Fig. S2). This became less pronounced with cholesterol (Fig. 1 B and C and Fig. S2). In addition the peptide tilted in the membrane, being more prominent without cholesterol (Fig. 1D). This suggested that the mismatch adaption (bilayer thickening and tilt) became less in the presence of cholesterol. Peptide length measured from N- to C-terminal Cα remained unaffected, being 3.80 ± 0.12 nm without cholesterol and 3.90 ± 0.14 nm with cholesterol. In the C24∶1 PC system, the bilayer around the peptide was markedly less ordered and thinner. In the presence of cholesterol this region localized closer to the peptide (Fig. 1C and Fig. S2) and was associated with a marked stretching of the helix from 4.08 ± 0.12 nm to 4.42 ± 0.13 nm and a conversion from the α into the longer 310 form (Fig. 1E). These data indicated that the strongest protein-induced bilayer deformation occurred under negative mismatch. Cholesterol counteracted bilayer adaptation by shifting the deformation closer to the peptide surface and by forcing the helix to stretch.
Fig. 1.
Hydrophobic mismatch between transmembrane peptide and lipid determines bilayer structure, helix orientation, and fold. (A) Snapshots from the MD simulations with LW21 TM peptide (red) embedded in C16∶1 and C24∶1 PC (headgroups dark blue, chains light blue) in the presence and absence of cholesterol (gray). Positive hydrophobic mismatch (lengthTMD > thicknessbilayer) and negative mismatch (lengthTMD < thicknessbilayer) is indicated by the blue/red color bar. (B) Distance profiles of lipid chain order (Scd of Sn1) with respect to the helix backbone at 0 nm (t = 500 ns) for C16∶1 and C24∶1. Black trace is 0% cholesterol, red trace is 20% cholesterol. The Scd parameter shown here represents the average value of the Scd profile along the hydrocarbon chain. (C) Membrane thickness (P-P distance in units of nanometers) with respect to the helix backbone at 0 nm (t = 500 ns) for C16∶1 and C24∶1. Black trace is 0% cholesterol, red trace is 20% cholesterol. (D) Bar graph of the average tilt angle of LW21 in the four simulations in degrees (see axis). (E) Fold adopted by LW21 over the simulation time in percent (see legend).
Reconstitution in Large Unilamellar Vesicles.
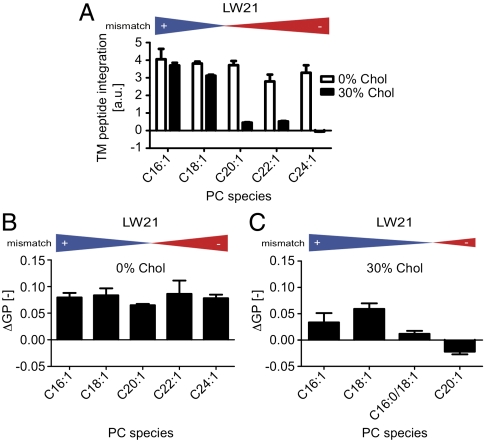
To test predictions from the simulation studies, we reconstituted peptides of two different lengths (3 mol% of LW21 and LW19 peptides) into large unilamellar vesicles (LUVs) by organic solvent reconstitution and extrusion (100 nm pore size). Incorporation of the peptides into LUVs could be measured by intrinsic Trp fluorescence (Fig. 2A and Fig. S3A) or by coomassie staining (Fig. S3B). We also validated integral membrane association by protection from proteinase K digestion (Fig. S1B) and their predominately helical structure (> 85%) by circular dichroism (CD) measurements (Fig. S1C and Table S2). We found that 30 mol% cholesterol greatly reduced incorporation of LW21 into negatively mismatched bilayers (Fig. 2A; full spectra in Fig. S3A). The unincorporated peptide could be recovered from the extruder membrane and did not remain on the vesicles (Fig. S3B). Also the shorter LW19 showed this trend (Fig. S4A). In the absence of cholesterol, incorporation was limited only under extreme mismatch of approximately 9 Å (Fig. S4A). L17, a peptide similar to LW21 that lacked double Trp (but retained double Lys) in the flanks, showed integration behavior very similar to that of LW21 (Fig. S3C). Taken together the data showed that cholesterol induced a selective limitation to the integration of negatively mismatched TM peptides at angstrom precision.
Fig. 2.
Hydrophobic mismatch between transmembrane peptide and lipid affects protein integration and lipid packing. (A) Trp fluorescence emission indicates integration of LW21 peptide into PC LUVs in the absence and presence of 30 mol% cholesterol. (B) Differences in lipid packing (ΔGP) indicate the structural impact of 3 mol% LW21 peptide in the absence of cholesterol on unsaturated PC bilayers from C16∶1 to C24∶1 (error bars = SD, n = 3). (C) Differences in lipid packing (ΔGP) in the presence of 30 mol% cholesterol. All error bars are SD (n = 3).
We next studied the impact of 3 mol% of LW21 (if integrated) on lipid packing. For this we used the fluorescent membrane probe C-laurdan, whose membrane hydration-dependent red shift in fluorescence emission can be translated into a relative index for lipid packing: the generalized polarization (GP) value (20, 21). We found a peptide-mediated increase in lipid packing (ΔGP) for all mismatch conditions in the absence of cholesterol (Fig. 2B). However, with cholesterol present, the lipid packing strongly decreased with increasing negative mismatch (Fig. 2C). In order to bridge between C18∶1 and C20∶1 PC we additionally used C18∶1/C16∶0 PC, which represents an intermediate lipid length (22). The shorter LW19 peptide showed the same trend (Fig. S4 B and C). This implied that the peptide generally rigidified the bilayer but that under negative mismatch in the presence of cholesterol the peptide induced a relative disordering of the bilayer.
Peptide Sorting.
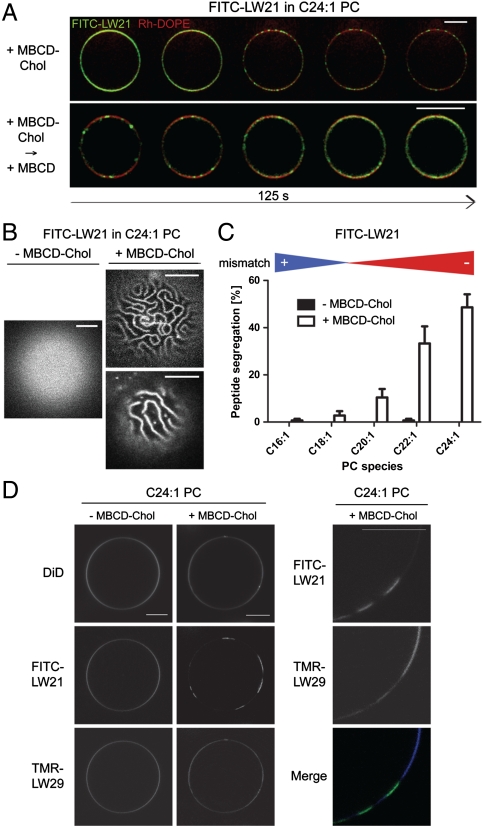
Next we tested the role of cholesterol on the distribution of TM peptides in mismatched vesicles. We prepared giant unilamellar vesicles (GUVs) for microscopy composed of PCs of different chain length, 3 mol% of a fluorescein-isothiocyanate (FITC)-labeled LW21 (FITC-LW21), and the lipid marker lissamine rhodamine-dioleoylphosphoethanolamine (Rh-DOPE) either by swelling LUVs or by swelling the dried lipid-peptide mixture. We always maintained 10 mol% of C18∶1 PC to keep the bilayer fluid and more stable (see SI Text). Then we carefully infused 3.8 μM cholesterol as a methyl-β-cyclodextrin complex (MBCD-Chol) (23). This induced a substantial clustering of FITC-LW21 in the C24∶1 PC membrane into line-shaped domains within minutes (Fig. 3 A and B). This segregation could be reversed by cholesterol extraction with 1 mM uncomplexed MBCD (Fig. 3A). The extent of segregation decreased with shorter PCs and was not observed under positive mismatch (Fig. 3C). These results suggested that cholesterol reversibly induced a segregating activity under negative mismatch. In the course of the cholesterol-loading reaction FITC fluorescence decreased to around 50% just before microscopic segregation and recovered after dissolution of the domains upon cholesterol extraction (Fig. S5 B–D). This implied that the FITC molecules concentrated and underwent a reversible self-quenching, which likely reflected peptide oligomerization during the transition to microscopic segregation. However, we also observed budding of domains from the cholesterol-loaded GUVs representing partial release of the TM peptides from the mother vesicles.
Fig. 3.
Hydrophobic mismatch and cholesterol determine lateral transmembrane peptide distribution. (A) Fluorescence microscopy images of GUVs depicting the segregation of FITC-LW21 peptide (3 mol%) and lipid marker Rh-DOPE (0.05 mol%) C24∶1 PC (87 mol%) with C18∶1 PC (9.95 mol%) during of 3.8 μM MBCD-cholesterol loading (Upper) and subsequent cholesterol extraction with 1 mM MBCD (Lower) (125 s). (B) Fluorescence microscopy images of GUV apexes depicting the patterns of FITC-LW21 peptide before and after 3.8 μM MBCD-cholesterol loading. (C) Quantification of FITC-LW21 segregation in GUVs before and after 3.8 μM cholesterol loading in different PCs (bars = SEM, n = 3). (D) Fluorescence microscopy images of GUVs depicting the localization of FITC-LW21 peptide (1.5 mol%), TMR-LW29 peptide (1.5 mol%) and lipid marker DID (0.05 mol%) in C24∶1 PC (87 mol%) with C18∶1 PC (9.95 mol%) before and after 8.1 μM MBCD-cholesterol loading. All image bars = 10 μm.
Next, we tested a positively mismatched peptide for cholesterol-dependent segregation. To this end we mixed 1.5 mol% of a long tetramethylrhodamine (TMR)-labeled LW peptide (TMR-LW29; Fig. S1A) into GUVs containing 1.5 mol% of FITC-LW21. It integrated best into C22∶1 PC to C24∶1 PC but not into C16∶1 PC membranes (Fig. S6A). When we infused cholesterol at 8.1 μM into the C24∶1 PC membrane, FITC-LW21 was selectively clustered into domains from which the longer TMR-LW29 was partially depleted (Fig. 3D). Results were independent of the preparation (swelling crude peptide/lipid mixture vs. LUVs; see SI Text). The behavior implied that the segregation force was acting selectively on FITC-LW21 and not on TMR-LW29. This correlated well with LUV experiments, where LW21 was excluded from the C24∶1 PC-cholesterol bilayer, whereas TMR-LW29 was well integrated (Fig. S6B).
Lipid Sorting.
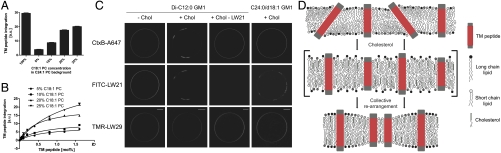
The previous LUV experiments had shown that negative mismatch prevented peptide integration in the presence of cholesterol. Thus we tested whether TMD-matched lipids would “rescue” peptide integration. We reconstituted LW21 in the C24∶1 PC bilayer and replaced 10, 20, and 25 mol% C24∶1 PC with an equal amount of C18∶1 PC. This led to an increase of Trp fluorescence, reflecting increased peptide integration through matching lipids (Fig. 4A). Next we added increasing amounts of LW21 to preparations containing 5, 10, 20, or 25 mol% of C18∶1 PC in a C24∶1 PC-cholesterol background. Here, peptide integration showed a saturation behavior (Fig. 4B). This indicated that the matching C18∶1 PC became a limiting factor for the integration of LW21 into the mismatching C24∶1 PC-cholesterol bilayer. It was suggestive of a lateral association between C18∶1 PC and LW21.
Fig. 4.
Hydrophobic mismatch and cholesterol determine lateral lipid distribution. (A) Trp fluorescence emission indicates integration of LW21 peptide into LUVs with 100, 0, 10, 20, and 25 mol% C18∶1 PC in a C24∶1 PC background containing 30 mol% cholesterol (bars = SD, n = 3). (B) Trp fluorescence emission indicates integration of the titrated LW21 peptide into LUVs with 5, 10, 20, and 25 mol% C18∶1 PC in a C24∶1 PC background containing 30 mol% cholesterol (bars = SD, n = 3). (C) Fluorescence microscopy images of GUVs depicting the localization of FITC-LW21 (1.5 mol%), TMR-LW29 (1.5 mol%), and CtxB-A647-labeled Di-C12∶0 GM1 (0.1 mol%) before and after MBCD-cholesterol loading and in the absence of FITC-LW21. Last column shows CtxB-A647-labeled C24∶0/dC18∶1 GM1 (0.1 mol%) after MBCD-cholesterol loading. bar = 10 μm. (D) Scheme of cholesterol’s function to rearrange TM peptide and lipid distribution according to hydrophobic length. It extends the acyl chains of the lipids making them less adaptable to mismatching TM peptides.
Next, we tested whether short chain lipids would laterally associate with the clustered FITC-LW21 domain as implied by the previous result. We doped C24∶1 PC GUVs containing FITC-LW21 and TMR-LW29 with the (neo)glycolipid di-C12∶0-GM1 (24) containing short acyl chains or the long chain C24∶0/dC18∶1-GM1 at 0.1 mol%. Addition of Alexa647-cholera toxin B (A647-CtxB) caused a homogeneous staining of GM1 in the membrane (Fig. 3C). After infusion of 8.1 μM MBCD-Chol di-C12∶0-GM1 redistributed and colocalized with the FITC-LW21 domain (Fig. 3C). Redistribution was not seen in the absence of FITC-LW21. In contrast to di-C12∶0-GM, C24∶0/dC18∶1-GM1 remained homogenously distributed in the presence of FITC-LW21 domains (Fig. 3C). These results showed that upon cholesterol loading the glycolipid with the short lipid chains became attracted to the FITC-LW21 domain, whereas the glycolipid with long chains did not. GM1s with intermediate chain length exhibited partial enrichment in the LW21 domain (Fig. S7).
Discussion
In this study we investigated (i) how hydrophobic mismatch between the lipid bilayer and a helical TM segment influences the structure of the interface, (ii) how cholesterol alters the lipid–peptide configuration, and (iii) whether structural alterations de-randomize the long-range distribution of the molecules. We chose the common series of monounsaturated PCs from di-16∶1 to di-C24∶1 as well described model membranes in which thickness can be varied (13, 14, 19). These membranes are fluid at room temperature. We selected characterized poly-leucine-type hydrophobic peptides with Trps at the water-bilayer boundary and flanking lysines that readily form stable TM helices when reconstituted from organic solvent (6, 11, 15). We confirmed integral membrane association by protection from proteinase K digestion and helical structure by CD spectroscopy for LW19, LW21, and LW29 (Fig. S1 and Table S2). The peptides only differed in their amount of leucines in the TM segment (14 vs. 16 vs. 24).
First we performed atomistic MD simulations over 500 ns of LW21 in C16∶1 PC and C24∶1 PC in the absence and presence of 20 mol% cholesterol. These conditions covered similar positive and negative mismatch (Fig. 1A and Table S1). The data indicated local, peptide-induced acyl chain straightening and TM segment tilting as response to positive mismatch and acyl chain flexing as a result of negative mismatch (Fig. 1 B, C and D). This is in line with predictions of the mattress model and experimental results by others (14, 22). Related membrane protein induced changes in the structure and also the dynamics of lipids around the protein have been observed in recent atomistic simulations (25). The most striking effect was a considerable membrane thinning around the peptide under negative mismatch (Fig. 1C). Cholesterol counteracted this effect presumably by straightening lipid acyl chains. This concentrated the deformation in direct vicinity of the helix surface (Fig. 1 B and C) leading to a marked stretching of the α-helix into a 310 conformation by more than 3 Å (Fig. 1E). We interpret this as a strain exerted by the stiffer lipid environment and a destabilization toward unfolding of the peptide (26, 27).
When we reconstituted LW21 in a series of PCs of different length, we indeed observed that efficiency of membrane integration decreased sharply under negative mismatch in the presence of 30 mol% cholesterol (Fig. 2A). Similar effects have also been reported by other groups (12–14, 28). This indicated that the activity of cholesterol made the peptide unstable in the membrane in agreement with our simulation data. A direct measurement of the increased 310 helix content predicted by the simulation could not be realized due to the strong reduction of peptide content in the bilayer. A similar trend of reduced incorporation was observed for LW19 albeit shifted toward thinner bilayers as expected (Fig. S4A). Moreover, integration was also compromised under large negative mismatch (9 Å) in the absence of cholesterol, again suggesting that cholesterol increased the energy penalty for membrane deformation under negative mismatch. Importantly, L17 a peptide similar to LW21 but lacking the flanking Trps, recapitulated the reduced membrane integration under negative mismatch with cholesterol seen with LW21 (Fig. S3C). This suggested that a minimal TM architecture of the type “charge-hydrophobic stretch-charge” is sufficient for the behavior.
When we monitored lipid packing with C-laurdan we found that the peptide generally increased average membrane order except under growing negative mismatch with cholesterol (Fig. 2 B and C). Thus the rigid structure of the peptide reduces the conformational freedom of the lipids (29) whereas in thicker bilayers it caused a relative disordering. Here the peptide may obstruct the cholesterol-mediated straightening of the chains in agreement with the stretching of the helix observed in the simulations. Our results corroborate NMR data that indicate similar ordering effects (22). However, in our system we cannot fully rule out that the helix partially converts from a TM into a peripheral helix conformation as a result of the strain (12).
Our data clearly indicated that the lipid–peptide interface experienced a deformation strain under negative mismatch with cholesterol. We predicted that this strain represented a potential for lateral segregation because clustering of TM peptides into a domain should minimize the interface area (7, 9). Moreover, this reaction should be inducible by cholesterol and reversible. We reconstituted the fluorescent FITC-LW21 at 3 mol% into C24∶1 PC GUVs and infused cholesterol by a recently developed MBCD-Chol exchange protocol. This allowed careful loading of membranes below saturation levels (23). Using confocal microscopy we observed the segregation of FITC-LW21 into microscaled, line-shaped domains within a fluid bilayer. Extraction of cholesterol reversed the reaction as predicted (Fig. 3A). Because this selective TM peptide segregation does not require or induce lipid phase immiscibility, it is fundamentally different from the packing-related exclusion of peptides and proteins from liquid ordered lipid phases (20, 30). Reversible self-quenching of the FITC-moieties along the reaction from dispersed to segregated TM peptides indicated peptide oligomerization as a transition state as expected (Fig. S5 B–D). We also noticed budding of domains, which represents an irreversible reaction beyond segregation.
Peptide segregation in GUVs was a function of negative mismatch (Fig. 3C) in line with the conclusions from our LUV experiments. Line-shaped domains (Fig. 3B) have also been observed for solid lipid domains in a cholesterol-PC phase (31). They require repelling forces that prevent coalescence. We speculate that the line shape observed here is a product of the rigid helices and the repulsion arising from the net positive charge due to the four lysines. Given the results from our LUV system, we predicted that a positively mismatched TM peptide should resist the segregating force. For this we reconstituted the very long TMR-LW29, which integrated only into the thick PC bilayer suggesting mismatch-limited TM integration as discussed above (Fig. S6A). When it was introduced at the same molar ratio as FITC-LW21 (1.5%) TMR-LW29 did not get sequestered upon cholesterol addition as did the shorter peptide. Instead it became depleted from the FITC-LW21 domains (the extent of which eluded further quantification due to the irregular shape of the domains) (Fig. 3B). We also validated the selective activity on the negatively mismatched peptide by the exclusion from LUVs. Here we observed a failure of integration for FITC-LW21, whereas TMR-LW29 was not affected (Fig. S6B). The data suggest that the failure of peptide incorporation at a fixed cholesterol level under negative mismatch translates into a propensity for TM length-based lateral segregation of peptides upon gradual cholesterol loading (13).
How then does the distribution of matching/mismatching lipids correlate? In our LUV system addition of LW21-matching C18∶1 PC to a C24∶1 PC bilayer increased the amount of incorporated peptide (Fig. 4A). Moreover, the peptide titration showed saturation trends implying the matching lipid to be a limiting factor for peptide incorporation (Fig. 4B). We interpret this behavior as a marked reduction of the mismatch-related strain by the matching lipid. Accordingly it should localize to the surface of the helix like an annular lipid. We tested this by labeling the short chain neoglycolipid di-C12∶0-GM1 (0.1 mol%) with A647-CtxB. Upon segregation the lipid indeed colocalized with FITC-LW21 but remained homogeneous in the absence of the peptide, suggesting that the lipid sorting is dependent on the peptide domain. As a negative control we used the glycolipid C24∶0/dC18∶1-GM1 with long chains, which did not experience attraction by the peptide domain. Exclusion was not readily detectable due to the domain shape (Fig. 4C). A native and a neo-GM1 of intermediate chain length both exhibited partial enrichment in the peptide domain as expected. CtxB is pentavalent for GM1; therefore clustering may reinforce the sorting behavior of the individual lipid. The experiments showed that the model set up from the LUV system holds for the GUVs and that cholesterol induces the requirement and cosegregation of short chain lipids with the peptide.
In summary, we have demonstrated that cholesterol forces a membrane containing high amounts of TM peptide to undergo a collective rearrangement according to hydrophobic length (Fig. 4D). Differences of two amino acids (LW19 vs. LW21) or two methylene groups (C22∶1 vs C24∶1 PC) were significant. We propose that cholesterol sterically constrains acyl chain rearrangements required for mismatch adaptation. This could be an essential, physiological function of cholesterol at the molecular level.
In the eukaryotic secretory pathway, TM proteins of different lengths get integrated in the cholesterol-poor—therefore adaptable—membranes of the endoplasmic reticulum (1). Indeed, cholesterol loading blocks this integration (32). In the Golgi apparatus the concentration of cholesterol increases and promotes the sorting of short TM domain-containing Golgi proteins and short lipids for recycling out of the forward membrane flow (33) that carries significantly longer TMD proteins together with long phospholipids and cholesterol to the plasma membrane (10).
More work is required to clarify how hydrophobic matching relates to (i) sphingolipid-cholesterol(raft)-based lateral heterogeneity and (ii) chemically specific protein–lipid interactions (34). Another open issue is whether moderate length differences between lipids and proteins paired with 40 mol% cholesterol in the plasma membrane influence protein and lipid distribution as well as the conformation of receptor proteins and channels (35). Because the segregation potential also scales with the square of the radius of the TM segment there are interesting implications for multihelix proteins/complexes as well as for ligand-induced lipid and protein clustering (9).
Altogether, our data provide a structural perspective on TM protein–lipid interactions in sterol-rich membranes with interesting biological implications for the function of cholesterol and the organization of the secretory pathway.
Material and Methods
Reagents.
LW21 (K2W2L8AL8W2K2), FITC-LW21 (FITC- (CH2)2-K2W2L8AL8W2K2), LW19 peptide (K2W2L7AL7W2K2), L17 (AK2L8AL8K2A), and TMR-LW29 (TMR-K2W2L12AL12W2K2) peptides (> 98% purity) were obtained from Genscript. Phosphatidylcholines (PC), cholesterol (Chol), and lissamine rhodamine B-dioleoylphosphatidylethanolamine (Rh-DOPE) were from Avanti polar lipids Inc. C24∶0/dC18∶1-GM1 was purchased from Sandro Sonnino, University of Milan, Italy. Di-C12∶0-GM1 was synthesized in the lab of Ten Feizi, Imperial College London. Methyl-beta-cyclodextrin (MBCD), Proteinase K, D-(+)-Trehalose, SDS, and silica TLC plates, and organic solvents were from Sigma-Aldrich. Dioctadecyl-tetramethyl-indodicarbocyanine (DiD), and Alexa-647-cholera toxin B (A647-CtxB) was purchased from Invitrogen. 6-dodecanoyl-2-N-methyl-N-carboxymethyl-aminonaphthalene (C-laurdan) was a kind gift from Bong-Rae Cho, Korea University of Seoul. For more details see SI Text.
Molecular Dynamics Simulations.
We performed 500-ns atomistic molecular dynamics simulations for four systems consisting of a lipid bilayer with a transmembrane peptide. Two systems were single-component bilayers comprised of either di-16∶1 or di-24∶1 PC lipids, and two other systems were mixtures of these PC systems with 20 mol% cholesterol. The transmembrane peptide used in the studies was LW21 (K2W2L8AL8W2K2). Simulations were performed with GROMACS (36) using the OPLS force field (37). The model for the membrane part is based on earlier work (38), which also describes the simulation protocol. Details of the model and its validation, data analysis, and additional results are described in SI Text.
Preparation and Analysis of Large Unilamellar Vesicles (LUVs).
LUVs were prepared according to Kalvodova et al. (39) with 3.0 mol% of the respective peptide in ethanol being added to the lipid mixture before drying, hydration, and extrusion. Proteinase K protection, Trp fluorescence readings, peptide thin layer chromatography (TLC), and C-laurdan spectroscopy was performed as described in SI Text.
Preparation and Cholesterol-Loading of Giant Unilamellar Vesicles (GUVs).
GUVs were prepared according to Bacia et al. (40) with peptides in ethanol being added to the lipid mixture before drying and swelling as described in SI Text. Alternatively GUVs were swelled from LUVs as described in Kahya et al. (41). For cholesterol loading, Chol was complexed to MBCD and afterwards mixed with uncomplexed MBCD in 300 mM sucrose according to Mahammad et al. (23) as specified in SI Text. Extraction was performed with 1 mM MBCD. Fluorescent CtxB was added to GUVs at 1 μg/mL after preparation.
Fluorescence Microscopy and Image Analysis.
For confocal microscopy of GUVs, a Zeiss LSM 510 inverted setup with the appropriate filters and a 63× and 100× OI objective was used. Imaging was performed at room temperature. All images were recorded in 8 bit format, normalized, and background corrected and contrast enhanced. Quantification of peptide segregation was performed as described in SI Text.
Supplementary Material
Acknowledgments.
We thank Sebastian Günther for preparation of the Pymol LW models, Erwin London for the initial donation of LW21 peptide, Bong-Rae Cho for the C-laurdan sample, and the Simons Lab for helpful discussions. Computational resources were provided by the Finnish IT Centre for Science (CSC). The Academy of Finland is thanked for funding. This work was also supported by Deutsche Forschungsgemeinschaft (DFG) “Schwerpunktprogramm1175” Grant SI459/2-1, DFG “Transregio 83” Grant TRR83 TP02, Bundesministerium für Bildung und Forschung “ForMaT” Grant 03FO1212, European Science Foundation “LIPIDPROD” Grant SI459/3-1, and the Klaus Tschira Foundation (to K.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.H.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103742108/-/DCSupplemental.
References
- 1.Hessa T, et al. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 2.Tanford C. The hydrophobic effect and the organization of living matter. Science. 1978;200:1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- 3.Ejsing CS, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharpe HJ, Stevens TJ, Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mouritsen OG, Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys J. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huschilt JC, Millman BM, Davis JH. Orientation of alpha-helical peptides in a lipid bilayer. Biochim Biophys Acta. 1989;979:139–141. doi: 10.1016/0005-2736(89)90534-8. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt U, Weiss M. Hydrophobic mismatch-induced clustering as a primer for protein sorting in the secretory pathway. Biophys Chem. 2010;151:34–38. doi: 10.1016/j.bpc.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Sperotto MM, Ipsen JH, Mouritsen OG. Theory of protein-induced lateral phase separation in lipid membranes. Cell Biophys. 1989;14:79–95. doi: 10.1007/BF02797393. [DOI] [PubMed] [Google Scholar]
- 9.Lundbaek JA, Andersen OS, Werge T, Nielsen C. Cholesterol-induced protein sorting: an analysis of energetic feasibility. Biophys J. 2003;84:2080–2089. doi: 10.1016/S0006-3495(03)75015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 11.de Planque MR, et al. Influence of lipid/peptide hydrophobic mismatch on the thickness of diacylphosphatidylcholine bilayers. A 2H NMR and ESR study using designed transmembrane alpha-helical peptides and gramicidin A. Biochemistry. 1998;37:9333–9345. doi: 10.1021/bi980233r. [DOI] [PubMed] [Google Scholar]
- 12.Ren J, Lew S, Wang Z, London E. Transmembrane orientation of hydrophobic alpha-helices is regulated both by the relationship of helix length to bilayer thickness and by the cholesterol concentration. Biochemistry. 1997;36:10213–10220. doi: 10.1021/bi9709295. [DOI] [PubMed] [Google Scholar]
- 13.Webb RJ, East JM, Sharma RP, Lee AG. Hydrophobic mismatch and the incorporation of peptides into lipid bilayers: A possible mechanism for retention in the Golgi. Biochemistry. 1998;37:673–679. doi: 10.1021/bi972441+. [DOI] [PubMed] [Google Scholar]
- 14.de Planque MR, et al. Sensitivity of single membrane-spanning alpha-helical peptides to hydrophobic mismatch with a lipid bilayer: Effects on backbone structure, orientation, and extent of membrane incorporation. Biochemistry. 2001;40:5000–5010. doi: 10.1021/bi000804r. [DOI] [PubMed] [Google Scholar]
- 15.Fastenberg ME, Shogomori H, Xu X, Brown DA, London E. Exclusion of a transmembrane-type peptide from ordered-lipid domains (rafts) detected by fluorescence quenching: Extension of quenching analysis to account for the effects of domain size and domain boundaries. Biochemistry. 2003;42:12376–12390. doi: 10.1021/bi034718d. [DOI] [PubMed] [Google Scholar]
- 16.Lewis BA, Engelman DM. Bacteriorhodopsin remains dispersed in fluid phospholipid bilayers over a wide range of bilayer thicknesses. J Mol Biol. 1983;166:203–210. doi: 10.1016/s0022-2836(83)80006-0. [DOI] [PubMed] [Google Scholar]
- 17.Killian JA, Nyholm TK. Peptides in lipid bilayers: The power of simple models. Curr Opin Struct Biol. 2006;16:473–479. doi: 10.1016/j.sbi.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Marsh D, Horvath LI. Structure, dynamics and composition of the lipid-protein interface. Perspectives from spin-labelling. Biochim Biophys Acta. 1998;1376:267–296. doi: 10.1016/s0304-4157(98)00009-4. [DOI] [PubMed] [Google Scholar]
- 19.Lewis BA, Engelman DM. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol. 1983;166:211–217. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser HJ, et al. Order of lipid phases in model and plasma membranes. Proc Natl Acad Sci USA. 2009;106:16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HM, et al. A Two-Photon Fluorescent Probe for Lipid Raft Imaging: C-Laurdan. Chembiochem. 2007;8:553–559. doi: 10.1002/cbic.200700003. [DOI] [PubMed] [Google Scholar]
- 22.Nezil FA, Bloom M. Combined influence of cholesterol and synthetic amphiphillic peptides upon bilayer thickness in model membranes. Biophys J. 1992;61:1176–1183. doi: 10.1016/S0006-3495(92)81926-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahammad S, Dinic J, Adler J, Parmryd I. Limited cholesterol depletion causes aggregation of plasma membrane lipid rafts inducing T cell activation. Biochim Biophys Acta. 2010;1801:625–634. doi: 10.1016/j.bbalip.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Chai W, Stoll MS, Galustian C, Lawson AM, Feizi T. Neoglycolipid technology: Deciphering information content of glycome. Methods Enzymol. 2003;362:160–195. doi: 10.1016/S0076-6879(03)01012-7. [DOI] [PubMed] [Google Scholar]
- 25.Niemela PS, et al. Membrane proteins diffuse as dynamic complexes with lipids. J Am Chem Soc. 2010;132:7574–7575. doi: 10.1021/ja101481b. [DOI] [PubMed] [Google Scholar]
- 26.Millhauser GL. Views of helical peptides: A proposal for the position of 3(10)-helix along the thermodynamic folding pathway. Biochemistry. 1995;34:3873–3877. doi: 10.1021/bi00012a001. [DOI] [PubMed] [Google Scholar]
- 27.Popot JL, Engelman DM. Membrane protein folding and oligomerization: The two-stage model. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 28.Ren J, Lew S, Wang J, London E. Control of the transmembrane orientation and interhelical interactions within membranes by hydrophobic helix length. Biochemistry. 1999;38:5905–5912. doi: 10.1021/bi982942a. [DOI] [PubMed] [Google Scholar]
- 29.Jahnig F. Critical effects from lipid-protein interaction in membranes. II. Interpretation of experimental results. Biophys J. 1981;36:347–357. doi: 10.1016/S0006-3495(81)84736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schafer LV, et al. Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc Natl Acad Sci USA. 2011;108:1343–1348. doi: 10.1073/pnas.1009362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weis RM, McConnell HM. Cholesterol stabilizes the crystal-liquid interface in phospholipid monolayers. J Phys Chem. 1985;89:4453–4459. [Google Scholar]
- 32.Nilsson I, Ohvo-Rekila H, Slotte JP, Johnson AE, von Heijne G. Inhibition of protein translocation across the endoplasmic reticulum membrane by sterols. J Biol Chem. 2001;276:41748–41754. doi: 10.1074/jbc.M105823200. [DOI] [PubMed] [Google Scholar]
- 33.Brugger B, et al. Evidence for segregation of sphingomyelin and cholesterol during formation of COPI-coated vesicles. J Cell Biol. 2000;151:507–518. doi: 10.1083/jcb.151.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 35.Jensen MO, Mouritsen OG. Lipids do influence protein function-the hydrophobic matching hypothesis revisited. Biochim Biophys Acta. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 38.Bjorkbom A, et al. Effect of sphingomyelin headgroup size on molecular properties and interactions with cholesterol. Biophys J. 2010;99:3300–3308. doi: 10.1016/j.bpj.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalvodova L, et al. The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J Virol. 2009;83:7996–8003. doi: 10.1128/JVI.00635-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bacia K, Schwille P, Kurzchalia T. Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc Natl Acad Sci USA. 2005;102:3272–3277. doi: 10.1073/pnas.0408215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahya N, Brown DA, Schwille P. Raft partitioning and dynamic behavior of human placental alkaline phosphatase in giant unilamellar vesicles. Biochemistry. 2005;44:7479–7489. doi: 10.1021/bi047429d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.