Abstract
A number of pathogenic, Gram-negative bacteria are able to secrete specific proteins across three membranes: the inner and outer bacterial membrane and the eukaryotic plasma membrane. In the pathogen Yersinia enterocolitica, the primary structure of the secreted proteins as well as of the components of the secretion machinery, both plasmid-encoded, is known. However, the mechanism of protein translocation is largely unknown. Here we show that Y. enterocolitica polymerizes a 6-kDa protein of the secretion machinery into needles that are able to puncture the eukaryotic plasma membrane. These needles form a conduit for the transport of specific proteins from the bacterial to the eukaryotic cytoplasm, where they exert their cytotoxic activity. In negatively stained electron micrographs, the isolated needles were 60–80 nm long and 6–7 nm wide and contained a hollow center of about 2 nm. Our data indicate that it is the polymerization of the 6-kDa protein into these needles that provides the force to perforate the eukaryotic plasma membrane.
Keywords: electron microscopy, bacterial pathogenicity, cell contact assays, protein translocation
In various Yersinia species, the secretion machinery is formed by about 25 different proteins (1), of which nine are broadly conserved and are found in other pathogens such as Salmonella, Shigella, Pseudomonas, Erwinia, Xanthomonas, and Ralstonia (2, 3). Interestingly, the secretion apparatus of these bacteria is also functionally conserved, so that one system can successfully translocate proteins usually secreted by another system (4–6). In Yersinia there are about 12 proteins called Yops (Yersinia outer proteins) that are secreted by the apparatus (7, 8).
As Yersinia is not engulfed by the eukaryotic cell and as the exported proteins are not detected in the surrounding medium, the translocation process appears to be protected (9–11). In Salmonella typhimurium and enteropathogenic Escherichia coli, filamentous surface appendages have been identified that might be involved in the translocation process (12, 13). In E. coli, the surface structures containing the protein EspA are required for translocation of the protein EspB (13). Apart from these filamentous appendages, needle-like surface structures have been described for S. typhimurium and Shigella flexneri (14–17). These needles contain the homologous proteins PrgI and MxiH, respectively, and are part of a cylindrical organelle that consists of at least three more proteins of the type III secretion system (14, 16–18). However, it is not clear whether these structures are directly involved in the delivery of proteins into the eukaryotic cell or, alternatively, only mediate contact between the bacteria and the eukaryotic cell (19).
So far none of these structures have been described for Yersinia enterocolitica. Two secreted proteins, YopB and YopD, have been suggested to be directly involved in the translocation process (20–22). Indeed, both YopB and YopD have been reported to insert into liposomes and to form channels with high conductance (23). However, the idea of a YopB/YopD import pore in the host cell membrane has been challenged by studies of Y. enterocolitica strains carrying mutations in these genes. These strains showed reduced virulence in the mouse model but were not affected in their ability to transport proteins into HeLa cells (24).
In this study we show that Y. enterocolitica, like S. typhimurium and S. flexneri, assembles many needle-like surface structures during protein secretion. Analyses of the isolated needles indicate that they are composed of a single 6-kDa protein, with homologs in all type III secretion systems of animal pathogens. Our data indicate that the needles are hydrophobic. We propose that their assembly produces the necessary force for the perforation of the host membrane and that they form a conduit for the delivery of specific proteins from the bacterial to the eukaryotic host cell cytosol.
Materials and Methods
Bacterial Strains, Growth Conditions, and Preparation of Secreted Proteins and Antibodies.
The Y. enterocolitica strains used in this study are (i) WA-314 (serotype O:8); (ii) WA-314ΔyopD, which contains a nonpolar mutation of yopD; and (iii) WA-C, a plasmid-less derivative. The induction and preparation of secreted proteins were performed as described (25, 26). To induce needle formation, bacteria were grown in Brain Heart Infusion (BHI) (Difco), Tryptic Soy Broth (TSB) (Difco), or LB medium (GIBCO). No needles were observed after growth in RPMI medium 1640 or DMEM (GIBCO). Bacteria without needles or the plasmid-cured WA-C strain were used as negative controls. Wild-type virulence was tested by passage through mice and reisolation of the bacteria from the spleen. Antibodies against the synthetic YscF peptide NFSGFAKGTDITDLDAVAQTLK were prepared by Cocalico (Reamstown, PA).
Electron Microscopy.
Samples were applied to glow-discharged, carbon-coated grids and negatively stained with 2% (wt/vol) uranyl acetate or 1.5% (wt/vol) sodium phosphotungstate at pH 6.8. The samples were viewed in a Philips CM 12 electron microscope at a nominal magnification of ×39,000 and an operating voltage of 100 kV. The number of needles per bacterium was assessed as described (18).
Purification of the Needles.
For the purification of the needles, bacteria were harvested by centrifugation (10 min at 10,000 × g), washed twice in 20 mM Tris⋅HCl (pH 7.5), and sheared for 5 min on ice with a Polytron blender (Kinematica, Lucerne, Switzerland) at full speed. Unbroken cells and debris were removed (10 min at 10,000 × g), and the supernatant was subjected to ultracentrifugation (1 h at 100,000 × g). The presence of needles in this supernatant was detected by electron microscopy. One-milliliter aliquots of the supernatant were loaded onto 30-ml gradients of 10–30% sucrose (wt/wt) in 20 mM Tris⋅HCl (pH 7.5) and centrifuged at 141,000 × g for 20 h with a Beckman SW 28 rotor. Twelve fractions were collected and dialyzed against 20 mM Tris⋅HCl (pH 7.5). The top four fractions contained needles by electron microscopic analyses. They were combined, and needles were precipitated by the addition of 3 mM CaCl2 (27) and centrifuged at 50,000 × g for 30 min.
Assays for Contact Hemolysis and HeLa Cell Cytotoxicity.
Contact hemolysis assays were performed essentially as described (20). In some experiments, bacteria and sheep erythrocytes were not preincubated for 2 h at 37°C but were immediately resuspended in ice-cold PBS. The released hemoglobin was measured with a spectrophotometer (SmartSpec 3000; Bio-Rad) at 545 nm.
HeLa cells were grown in 6-well plates at 37°C in the presence of 5% CO2 with DMEM supplemented with 5% FCS. At 80% confluency the cells were washed once with DMEM without supplements and then incubated with 100% DMEM, 100% TSB, or one of the following DMEM–TSB mixtures: 75%/25%, 50%/50%, or 25%/75%. Three microliters of bacterial overnight cultures (OD600 = 0.5) was added to each 2-ml well. The plates were briefly centrifuged to bring the bacteria into contact with the HeLa cells and incubated at 37°C for 4 h. Pictures were taken hourly with a Zeiss light microscope equipped with a digital camera (Hamamatsu, Ichinocho, Japan) and phase-contrast optics. To check for the presence of needles, the medium was removed and the HeLa cells were incubated at room temperature with 0.2% digitonin per well. The digitonin extract was centrifuged for 10 min at 5,000 × g, and the pellet was resuspended in a drop of water. The resuspended bacteria were deposited on a grid and examined by negative staining in the electron microscope. Finally, the contact assays were repeated with HeLa cells grown on Formvar/carbon-coated 75-mesh nickel grids. After bacteria were added, the grids were incubated for up to 2 h at 37°C, before negative staining and examination in the electron microscope.
Hydrophobicity Assays.
Needle preparations (3 ml) were overlaid with 500 μl of n-hexadecane and briefly mixed, and the phases were allowed to separate at room temperature. To evaluate partitioning, 1 ml of the aqueous phase before and after the extraction was precipitated with trichloroacetic acid and analyzed by SDS/PAGE. The hydrophobicity of the needles was also assayed by their attachment to latex beads and their aggregation upon salt addition.
SDS/PAGE and Protein Analysis.
Gel electrophoresis was performed with 16% Tris-glycine gels. To identify possible posttranslational modifications, YscF was enzymatically digested, and the resulting peptides were both sequenced and analyzed by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (28).
Results
Y. enterocolitica Forms Needle-Like Structures That Are Composed of a Single Protein, YscF.
Upon induction in BHI medium Y. enterocolitica formed many short, needle-like structures on their cell surface (Fig. 1A). Individual needles were 60–80 nm long and 6–7 nm wide and contained a hollow center of about 2 nm (Fig. 1 B and E). Although the dimensions of the needles were quite constant, longer needles with a length of up to 210 nm were occasionally observed. The number of needles per cell varied greatly and depended strongly on the medium used for cultivation. There were 50–100 needles per cell in BHI and TSB (Fig. 1A), about 30 needles per cell in LB, and no needles at all in DMEM. Compared with wild type, no differences were found in the morphology of the needles of the yopD mutant (data not shown). Overall, the needles of Y. enterocolitica resembled those described for S. typhimurium and S. flexneri (14–17).
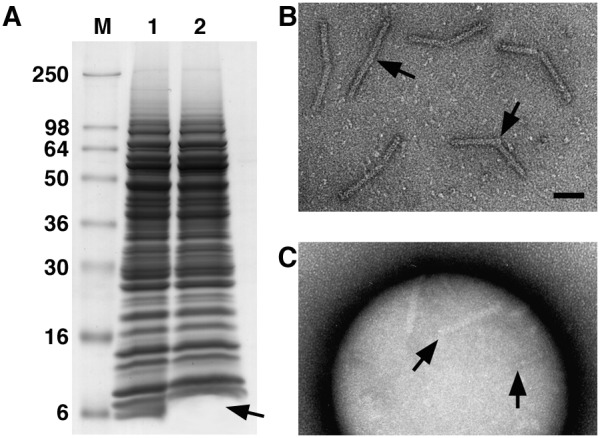
Figure 1.
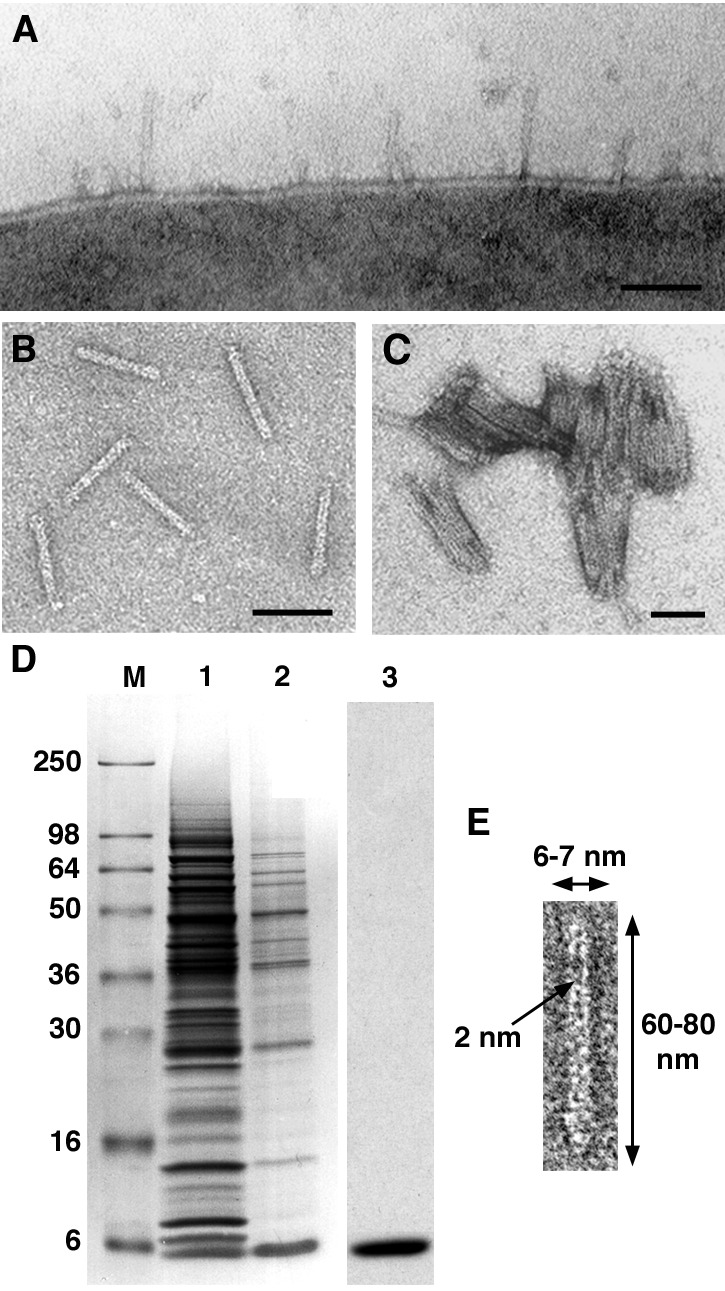
Structural (A–C and E) and biochemical (D) analyses of the needles of Y. enterocolitica. The needles can be visualized on the cell surface (A), after isolation (B), and aggregated upon Ca2+ addition (C). All scale bars are 50 nm. (D) SDS/PAGE of the needle purification. Lane 1 shows the crude extract after mechanical removal of the needles. After sucrose gradient centrifugation, a 6-kDa protein, called YscF, is enriched (lane 2). After calcium aggregation the needles contain only YscF (lane 3, C). (E) At higher magnification, the 2-nm-wide hollow center of the needles is visible. Occasionally one end of the needles is thickened. As this thickening is not observed with needles on the bacterial surface it might be generated by their mechanical removal.
The purification of the needles was monitored by negative-stain electron microscopy and SDS/PAGE. The needles were mechanically sheared from the cells in a blender and could be detected by negative-stain electron microscopy in a 100,000 × g supernatant. This supernatant contained many proteins (Fig. 1D, lane 1). To further enrich the needles, we subjected the supernatant to sucrose density gradient centrifugation and found the needles by negative-stain electron microscopy (Fig. 1B) in the top four fractions. SDS/PAGE analysis of the proteins of the combined fractions still showed a rather complex pattern, although a 6-kDa protein was enriched (Fig. 1D, lane 2). The amino-terminal sequence of this protein was established. The protein was identified as YscF, which is essential for secretion (29). The needles were further purified by calcium-induced aggregation (Fig. 1C). SDS/PAGE analysis of the centrifuged aggregate showed the presence of only one protein, YscF (Fig. 1D, lane 3). Furthermore, immunoblot analysis of the proteins in Lane 3 with antibodies specific to YopB and YopD showed no reactive bands (data not shown). Finally, matrix-assisted laser desorption ionization–time-of-flight analyses indicated that YscF, like PrgI of S. typhimurium (18), was not posttranslationally modified.
The YscF Needle, Not Protein Secretion, Is Responsible for the Hemolytic Activity of Y. enterocolitica.
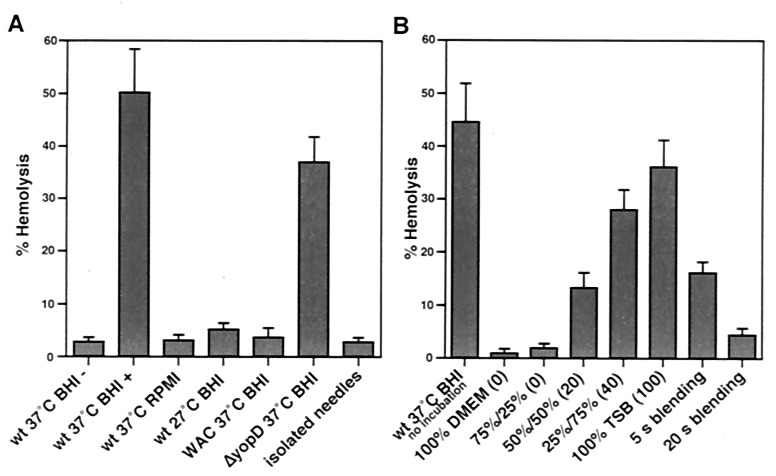
To investigate the function of the YscF needles, we used the contact hemolytic activity of the bacteria as a simple model system. As with S. flexneri (15), contact hemolysis by Yersinia did not occur without centrifugation (Fig. 2A). A comparison of Yersinia wild-type cells grown under different culture conditions with plasmid-cured cells and with the yopD mutant strain showed that all bacteria with needles displayed hemolytic activity; whereas bacteria without needles were nonhemolytic (Fig. 2A). Isolated needles did not show any hemolytic activity in this assay (Fig. 2A).
Figure 2.
Analysis of the contact-dependent hemolytic activity of Y. enterocolitica. (A) The hemolytic activity of the bacteria depends on the presence of the needles and an initial centrifugation step (wt 37°C BHI−, without centrifugation vs. wt 37°C BHI+, with centrifugation). Isolated needles and bacteria that did not form needles (growth in RPMI medium 1640, at 27°C, or the plasmid-cured WA-C strain) were nonhemolytic, whereas strains possessing needles (growth at 37°C in BHI, ΔyopD mutant) induced hemolysis. (B) Yop secretion is not necessary for the induction of contact hemolysis. No difference in hemolysis was found when bacteria and RBCs were immediately resuspended without incubation at 37°C (wt 37°C BHI no incubation vs. wt 37°C BHI+). Hemolysis correlated with the number of needles present on the cells. Bacteria grown in media inducing higher numbers of needles showed higher hemolytic activity (see Fig. 6). The first number is % DMEM; the second number is % TSB; and the numbers in brackets are the numbers of needles per cell. Mechanical removal of the needles from the bacterial surface reduced or completely abolished hemolytic activity. All data show means and standard deviation of assays performed in quadruplicate.
We then attempted to determine whether hemolysis was caused by the needles or by the secretion of proteins during close contact with the erythrocytes. Bacteria were mixed with sheep RBCs, centrifuged, and either incubated at 37°C for 2 h and then resuspended or immediately resuspended without prior incubation. No difference could be detected in the hemolysis resulting from these different protocols (Fig. 2). As secretion is strongly inhibited at room temperature, it was unlikely that hemolysis was due to secreted proteins (8).
We then compared the hemolysis efficiency of bacteria with different numbers of needles, using wild-type cells that were cultivated in various DMEM–TSB mixtures. As can be seen in Fig. 2B, the cells with the highest number of needles induced the strongest response, whereas cells with a lower number showed reduced hemolysis (see also Fig. 6). To further demonstrate the link between needles and contact hemolysis, bacteria were treated with a blender to remove the needles. This short exposure to shear forces had no effect on the integrity and viability of the bacteria, as indicated by microscope and plating experiments (data not shown). However, the ability of the cells to induce hemolysis was reduced to 18% after blending for 5 s and was abolished after 20 s (Fig. 2B). Examination of the cells after blending confirmed that after 5 s some needles could still be found, but that these were undetectable after the 20-s treatment (data not shown).
Figure 6.
Infection of HeLa cells by Y. enterocolitica. HeLa cells were incubated with overnight-grown bacteria for 4 h at 37°C in the following media: 100% DMEM (A), 50% DMEM/50% TSB (B), 25% DMEM/75% TSB (C), or 100% TSB (D). Every hour, the cells were inspected with a light microscope, and electron microscopy was used to detect whether the bacteria had formed needles under these conditions. (A) No needles and no damage to the HeLa cell could be detected in 100% DMEM. (B) At 50% TSB, very few needles could be detected on the bacteria, and only a few HeLa cells showed cellular damage. (C) At 75% TSB, the number of needles per cell increased (up to 40), accompanied by severe damage of many HeLa cells. (D) At 100% TSB each bacterium possessed up to 100 needles, and damage to many HeLa cells could already be seen within 2 h.
To further show that secretion was not involved in hemolysis, we repeated the centrifugation experiments with slight modifications. The centrifugation time was shortened from 10 min to 5, 2, 1, and 0.5 min, and the centrifugal force was doubled at each step. Under these conditions even the shortest centrifugation was able to induce about 60% of the maximal hemolysis (data not shown). Together, these results confirmed that an extremely brief interaction is sufficient to induce hemolysis and that the centrifugation of needle-bearing bacteria, not the secretion of proteins, is likely to account for the phenomenon.
Ultrastructural and Biochemical Analyses of the Bacterium–Erythrocyte Interaction.
To study the interaction between the bacteria and the RBCs during contact-dependent hemolysis in greater detail, we examined the interacting cells by electron microscopy. Bacteria and RBCs were mixed and centrifuged, and the supernatant was carefully removed. The cell pellet was gently resuspended with a drop of water, causing rapid and complete hemolysis of the RBCs. The released hemoglobin was removed by centrifugation, and the RBC ghosts and bacteria were negatively stained. With this approach it was observed that the interaction of many bacteria with the RBCs was stable enough to withstand water-induced hemolysis (Fig. 3A). Closer examination of the cell contacts showed needles piercing the RBCs membranes (Fig. 3 B and C). As each bacterium contained up to 100 needles, this multiple piercing could account for the observed contact-dependent hemolysis.
Figure 3.
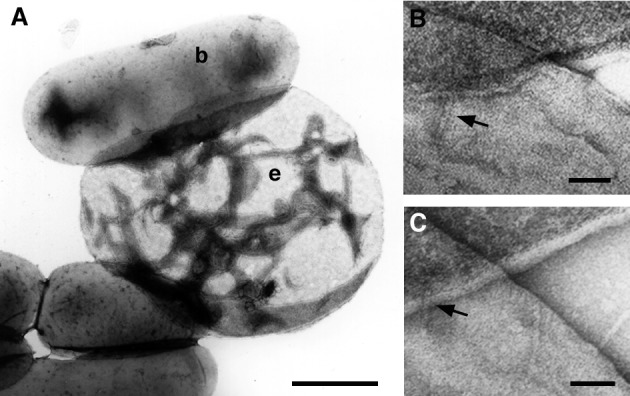
Structural investigation of the bacterium–erythrocyte interaction. (A) After centrifugation the contact between the bacteria (b) and the RBCs (e) was strong enough to withstand water-induced hemolysis. (B and C) Closer inspection showed that the needles of the bacteria pierced the erythrocyte membranes. In both pictures the erythrocyte ghost and the bacterium are in the upper and lower half of the picture, respectively. (The bar is 1 μm for A and 50 nm for B and C.)
To clearly demonstrate that the YscF needles had pierced the RBC membranes, we examined the RBC ghosts after the bacteria had been removed from the RBC surface by suspension in cold PBS buffer. Under these conditions some of the needles were still found to be penetrating the membranes (Fig. 4 A and B). This association was confirmed by immunoblotting (Fig. 4C). Lysed RBC membranes before (lane 1) and after (lane 2) contact with bacteria were analyzed by SDS/PAGE. The separated proteins were immunoblotted with antibodies against the needle protein and various Yop proteins such as YopB, D, E, and H. The antibody against YscF detected a weak band with an apparent molecular mass identical to that of YscF (Fig. 4C, lane 3), which was not detected before contact with the bacteria. No bands that were reactive with the various Yop antibodies were detected (data not shown).
Figure 4.
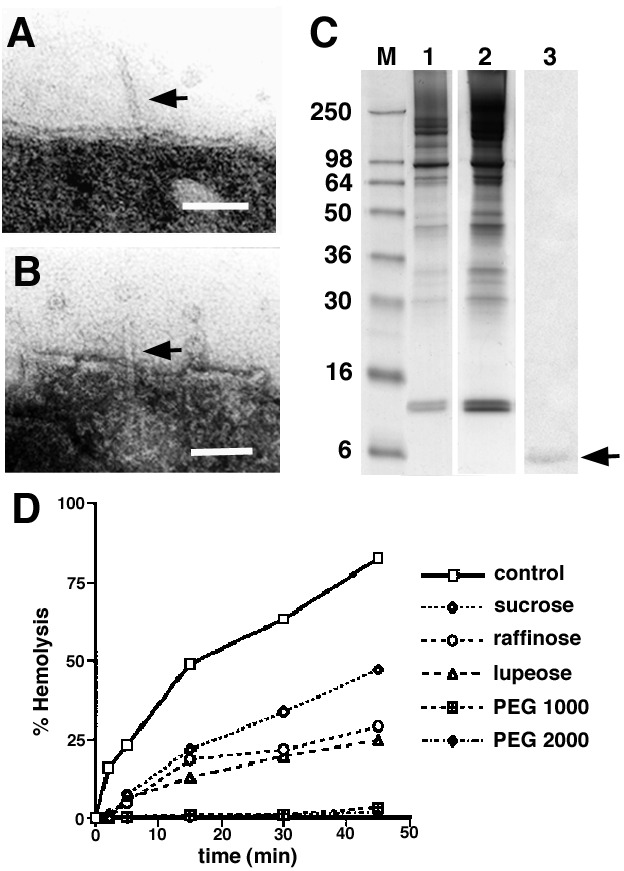
Examination of the interaction between YscF needles and erythrocyte membranes. (A and B) After separation of bacteria and RBCs, needles are still sometimes found inserted in the RBC membranes. (Bars are 50 nm.) (C) Analysis of the protein pattern of the RBC membrane found before (lane 1) and after (lane 2) contact hemolysis. Bacteria and RBCs were immediately resuspended without incubation at 37°C. Lane 3, immunoblot of lane 2, with an antibody against YscF. (D) Osmoprotection assays were used to determine the size of the bacterially induced lesions. Sugar with a molecular radius of ≥1.8 nm (polyethylene glycol 1000, 2000) blocked the release of hemoglobin and offered osmoprotection.
Finally, we used osmoprotection experiments to determine whether the size of the induced RBC lesions could be explained by the insertion of the YscF needles into the membrane (15). For Y. enterocolitica it was found that lupeose (molecular radius ≈ 1.2 nm) showed a significant protection against hemolysis, whereas the addition of polyethylene glycols with a molecular radius larger than 1.8 nm completely abolished hemolysis (Fig. 4D). Based on these experiments, the inner radius of the pore was estimated to be ≤1.8 nm, a value that correlated well with the electron microscopically estimated inner diameter of the YscF needle. Together these data suggest that hemolysis was caused by the insertion of the needle into the RBC membrane.
The YscF Needle Is Hydrophobic.
The ultrastructural interaction of the YscF needle with the RBC membrane after centrifugation prompted us to examine the needle's hydrophobicity. Although the amino acid sequence of YscF suggested a mostly hydrophilic protein, several assays clearly indicated that the needles had hydrophobic properties (30). These properties were documented by the needles' exclusive partition into the hexadecane phase (Fig. 5A) and their tendency to aggregate in the presence of salt (Fig. 5B). Interestingly, this aggregation involved the ends of the needles, the part of the structure that is inserted into the RBC membrane. Furthermore, the needles readily attached to hydrophobic surfaces such as latex beads (Fig. 5C). Together, these data suggest that the needles are hydrophobic enough to insert into membranes.
Figure 5.
The hydrophobic properties of YscF needles. (A) YscF was quantitatively extracted by n-hexadecane (lane 2, arrow) from a solution of needles (lane 1). (B) Upon the addition of salt the needles dimerize via their end (arrows), which contacts the host cell membrane. (The scale bar is 50 nm.) (C) Needles attached to a hydrophobic 0.3-μm latex bead.
The Role of Needles in the Cytotoxic Activity of Y. enterocolitica.
When Y. enterocolitica was added to HeLa cells, the bacteria started to adhere to the eukaryotic cells. This cell contact is thought to trigger the translocation of Yop proteins, a process that eventually leads to the death of the HeLa cells (1). To study the role of the needles in this cellular response, we used bacteria grown overnight without needles. We added them to HeLa cells and incubated them for 4 h at 37°C in different media that induced the formation of various quantities of needles. Electron microscopy was used to check the quantity of needles while the cellular response of the HeLa cells was observed with a light microscope. The data in Fig. 6 show that the cytotoxic effect on HeLa cells (detectable as rounded cells) was proportional to the number of needles formed. Under conditions where no needles were induced, there was no cytotoxic damage (Fig. 6A). With increasing formation of needles there was increasing cytotoxic damage to the HeLa cells (Fig. 6 B–D). Most importantly, when bacteria with preformed needles were incubated with HeLa cells under conditions where no new needles were induced, there was no cytotoxic damage to the HeLa cells (data not shown). Hence damage can only be inflicted during the formation of needles, not by preexisting needles.
To strengthen the correlation between needle formation and cell damage, HeLa cells were grown on electron microscope grids and incubated with bacteria under needle-inducing conditions (Fig. 7A). Within 30 min the bacteria had formed needles, which pierced the HeLa cell membranes (Fig. 7 B–D). If bacteria and HeLa cells were separated by a 10-μm-thick filter, no damage of HeLa cells could be observed, even though secreted Yops should readily cross the filter (data not shown and ref. 1).
Figure 7.

Structural investigation of the bacteria-HeLa cell interaction. (A) At low magnification the bacteria–HeLa cell contacts after cultivation on a grid are similar to the contacts observed by light microscopy (see Fig. 6). (B–D) Higher magnification reveals that the bacteria form needles during incubation at 37°C that pierce the HeLa cell membrane (arrows). This piercing and the secretion of Yops lead to a rapid rounding up of the HeLa cell. B, bacterium; H, HeLa cell; N, cell nucleus. (The scale bar in A is 1 μm and all other bars are 50 nm.)
Discussion
In this paper we demonstrate that Y. enterocolitica, like S. typhimurium and S. flexneri, uses short needle-like surface structures for protein translocation (14, 15). These needles are hollow tubes with a length of 60–80 nm, an outer diameter of 6–7 nm, and an inner diameter of about 2 nm. Biochemical analyses show that all of these needles are formed by small homologous proteins (8) and that in Y. enterocolitica each needle was made up of about 200–300 copies of YscF. No proteins other than YscF were detected when the needles were purified by calcium-induced aggregation. Nevertheless, we cannot rule out the possibility that the needles contain proteins that are present in only a few copies per needle and therefore may have escaped detection. Moreover, additional needle proteins may have been lost during purification of the needles.
As YscF homologue proteins are present in all animal pathogenic bacteria but absent from plant pathogenic bacteria (8), we studied the functional significance of the needle for protein translocation. With the use of HeLa cells, we showed that preexisting needles could not inflict damage on the cells. Only when bacteria assembled new needles during contact with the HeLa cells was cytotoxic damage observed. These results indicated that the assembly of the needles was crucial for the translocation of proteins into the host cell. Thereby the needles formed a continuous conduit from the bacterial surface to the host cell cytoplasm for Yop secretion. Experiments with erythrocytes suggested that the physical force needed to puncture a membrane could also be generated by centrifuging needle-bearing bacteria onto erythrocytes. Thus our data support a model in which the needles in Y. enterocolitica and other pathogenic bacteria puncture the host cell membrane (Fig. 8). According to this model, the bacteria attach tightly to the host cell surface with adhesion proteins such as YadA (8). This host cell contact or chemical signals of the medium trigger the formation of new needles, which reach and puncture the host cell membrane. The needles possess all of the structural and biochemical features necessary for this function. First, the needles are hollow structures that allow the passage of small or partially unfolded proteins (Yops). This idea is supported by the observation that a N-terminal truncated YscF specifically abolishes the translocation of larger Yops such as YopB (42 kDa) and YopD (33 kDa) but does not impair translocation of smaller Yops such as the 23-kDa YopE (29). As the inner diameter of the needles is certainly smaller in the truncated YscF mutant, the needles could mechanically impair the passage of larger proteins. Second, the needles are long enough to bridge the gap between bacterium and eukaryotic cell, which in thin sections is usually less than 40 nm (unpublished data). Third, the hydrophobic surface properties of the needles could explain the interaction of the structure with membranes. This conclusion is supported by the observation that YscF partially localizes to the RBC membranes after centrifugation and that the interaction of the needle with RBC and HeLa cell membranes can be visualized with an electron microscope. Finally, polymerization of YscF into needles could generate the force needed to puncture the membrane. Although we have not measured the force of needle protrusion in Y. enterocolitica, studies of pili in Neisseria gonorrhoeae suggest that a comparable molecular machine can generate substantial force and even mediate bacterial motility (31). The similarity between these two systems is based on the conservation of certain key components. Both machines use homologous outer membrane proteins as channels for their pilus or needle, respectively (refs. 32 and 33; YscC in Fig. 8). In addition, both systems seem to be energized by related ATPases, which are localized in the inner membrane and the cytoplasm (refs. 8 and 34; YscN in Fig. 8). These ATPases provide the energy necessary for needle and pilus extension by a constant incorporation of new monomers into the growing structure. If this extension generates the force necessary for membrane penetration, it would explain why only animal pathogenic bacteria have needles. A structure such as the needle could hardly penetrate the much thicker plant cell walls. However, it has been suggested that a much longer pilus-like structure might play a role in protein translocation by plant pathogenic bacteria (35).
Figure 8.
Schematic illustration of the role of YscF needle polymerization in Yop secretion and translocation. In a first step, the bacterium forms the necessary inner and outer membrane components of the translocation machinery and attaches to the host cell surface with adhesion proteins such as YadA. This host cell contact or chemical signals of the medium trigger the formation of the needle by polymerization of YscF, which serves as a driving force to puncture the host cell membrane. Thereby a continuous conduit is formed from the bacterial surface to the host cell cytoplasm for Yop translocation. The dotted lines indicate that the outer and inner membrane components might interact in the absence of the needle. YscC, outer membrane protein channel; YscN, putative ATPase.
In sum, our data provide evidence that the needle-like part of the type III secretion systems of animal pathogenic bacteria functions as a molecular syringe. Therefore, the bacteria need no proteins other than YscF to form a conduit for protein translocation into the host cell. Surprisingly, the basic principle of such an injection mechanism is widespread among life forms and is found in organisms as diverse as insects, snails, and fish.
Acknowledgments
We thank Jürgen Heesemann and Andreas Roggenkamp of the Max von Pettenkofer Institut for the gift of the Yersinia yopD mutant and various Yop antisera, Joseph Fernandez of the Protein/DNA Technology Center at The Rockefeller University for protein sequencing, and Erica Johnson and Yuh-Min Chook for critical reading of the manuscript. This work was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft.
Abbreviations
- Yops
Yersinia outer proteins
- BHI
Brain Heart Infusion
- TSB
Tryptic Soy Broth
References
- 1.Cornelis G R. J Bacteriol. 1998;180:5495–5504. doi: 10.1128/jb.180.21.5495-5504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hueck C J. Microbiol Mol Biol. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Gijsegem F, Genin S, Boucher C. Trends Microbiol. 1993;1:175–180. doi: 10.1016/0966-842x(93)90087-8. [DOI] [PubMed] [Google Scholar]
- 4.Rosqvist R, Håkansson S, Forsberg A, Wolf-Watz H. EMBO J. 1995;14:4187–4195. doi: 10.1002/j.1460-2075.1995.tb00092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ham J H, Bauer D W, Fouts D E, Collmer A. Proc Natl Acad Sci USA. 1998;95:10206–10211. doi: 10.1073/pnas.95.17.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossier O, Wengelnik K, Hahn K, Bonas U. Proc Natl Acad Sci USA. 1999;96:9368–9373. doi: 10.1073/pnas.96.16.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis G R, Wolf-Watz H. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M P, Stainier I. Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsberg Å, Rosqvist R, Wolf-Watz H. Trends Microbiol. 1994;2:14–19. doi: 10.1016/0966-842x(94)90339-5. [DOI] [PubMed] [Google Scholar]
- 10.Rosqvist R, Magnusson K E, Wolf-Watz H. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persson C, Nordfelth R, Holmstrom A, Håkansson S, Rosqvist R, Wolf-Watz H. Mol Microbiol. 1995;18:135–150. doi: 10.1111/j.1365-2958.1995.mmi_18010135.x. [DOI] [PubMed] [Google Scholar]
- 12.Ginocchio C C, Olmsted S B, Wells C L, Galan J E. Cell. 1994;76:717–724. doi: 10.1016/0092-8674(94)90510-x. [DOI] [PubMed] [Google Scholar]
- 13.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan J E, Aizawa S I. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 15.Blocker A, Gounon P, Larquet E, Niebuhr K, Cabiaux V, Parsot C, Sansonetti P. J Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamano K, Aizawa S I, Katayama E, Nonaka T, Imajoh-Ohmi S, Kuwae A, Nagai S, Sasakawa C. EMBO J. 2000;19:3876–3887. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimbrough T G, Miller S I. Proc Natl Acad Sci USA. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. . (First Published September 12, 2000; 10.1073/pnas.200209497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubori T, Sukhan A, Aizawa S I, Galan J E. Proc Natl Acad Sci USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. . (First Published August 15, 2000; 10.1073/pnas.170128997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reed K A, Clark M A, Booth T A, Hueck C J, Miller S I, Hirst B H, Jepson M A. Infect Immun. 1998;66:2007–2017. doi: 10.1128/iai.66.5.2007-2017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Håkansson S, Schesser K, Persson C, Galyov E E, Rosqvist R, Homble F, Wolf-Watz H. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 21.Hartland E L, Bordun A M, Robins-Browne R M. Infect Immun. 1996;64:2308–2314. doi: 10.1128/iai.64.6.2308-2314.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neyt C, Cornelis G R. Mol Microbiol. 1999;33:971–981. doi: 10.1046/j.1365-2958.1999.01537.x. [DOI] [PubMed] [Google Scholar]
- 23.Tardy F, Homble F, Neyt C, Wattiez R, Cornelis G R, Ruysschaert J M, Cabiaux V. EMBO J. 1999;18:6793–6799. doi: 10.1093/emboj/18.23.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee V T, Schneewind O. Mol Microbiol. 1999;31:1619–1629. doi: 10.1046/j.1365-2958.1999.01270.x. [DOI] [PubMed] [Google Scholar]
- 25.Michiels T, Wattiau P, Brasseur R, Ruysschaert J M, Cornelis G. Infect Immun. 1990;58:2840–2849. doi: 10.1128/iai.58.9.2840-2849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsberg Å, Bolin I, Norlander L, Wolf-Watz H. Microb Pathog. 1987;2:123–137. doi: 10.1016/0882-4010(87)90104-5. [DOI] [PubMed] [Google Scholar]
- 27.Brinton C C., Jr Trans NY Acad Sci. 1965;27:1003–1054. doi: 10.1111/j.2164-0947.1965.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez J, Gharahdaghi F, Mische S M. Electrophoresis. 1998;19:1036–1045. doi: 10.1002/elps.1150190619. [DOI] [PubMed] [Google Scholar]
- 29.Allaoui A, Schulte R, Cornelis G R. Mol Microbiol. 1995;18:343–355. doi: 10.1111/j.1365-2958.1995.mmi_18020343.x. [DOI] [PubMed] [Google Scholar]
- 30.Rosenberg M, Doyle R J. In: Microbial Cell Surface Hydrophobicity. Doyle R J, Rosenberg M, editors. Washington, DC: Am. Soc. Microbiol.; 1990. pp. pp.1–30. [Google Scholar]
- 31.Merz A J, So M, Sheetz M P. Nature (London) 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 32.Wolfgang M, van Putten J P M, Hayes S F, Dorwald D, Koomey M. EMBO J. 2000;19:6408–6418. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koster M, Bitter W, de Cock H, Allaoui A, Cornelis G R, Tommassen J. Mol Microbiol. 1997;26:789–798. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 34.Wolfgang M, Lauer P, Park H P, Brossay L, Hebert J, Koomey M. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 35.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E L, Kalkkinen N, Romantschuk M, He S Y. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]