Abstract
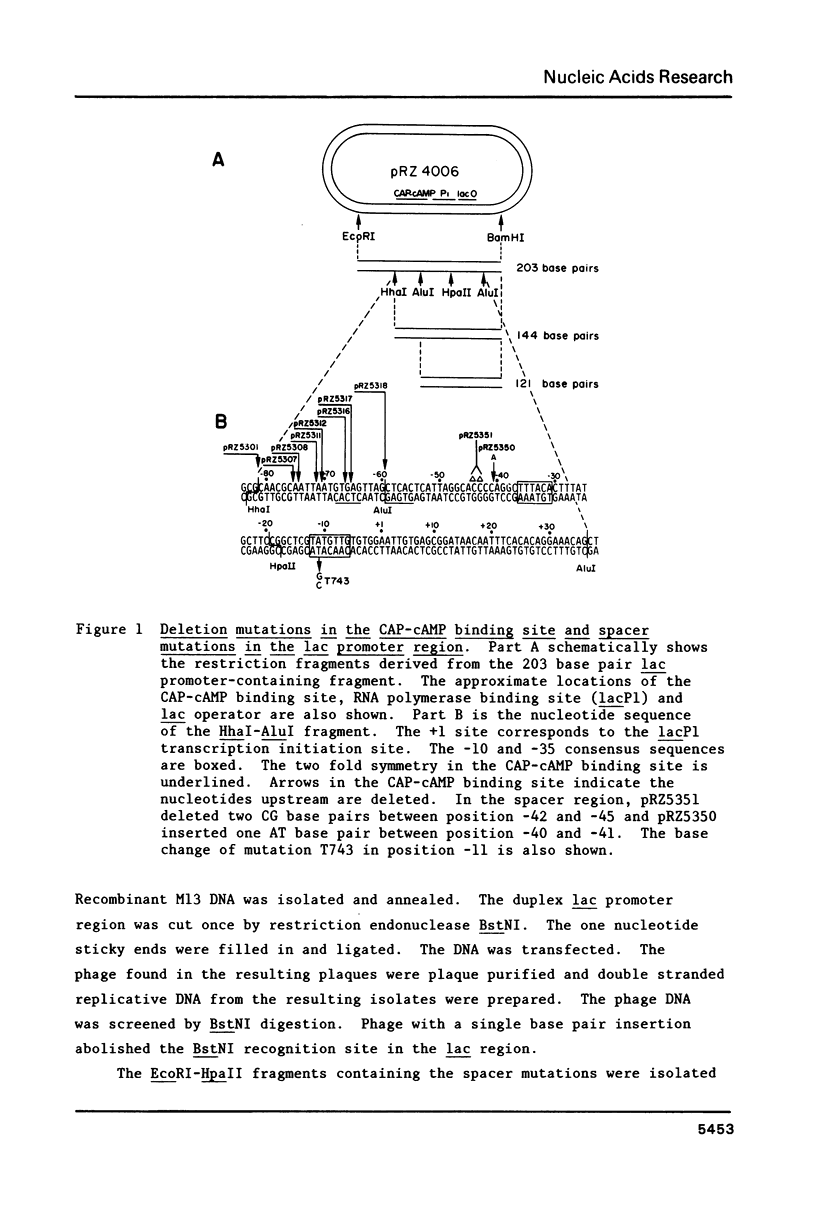
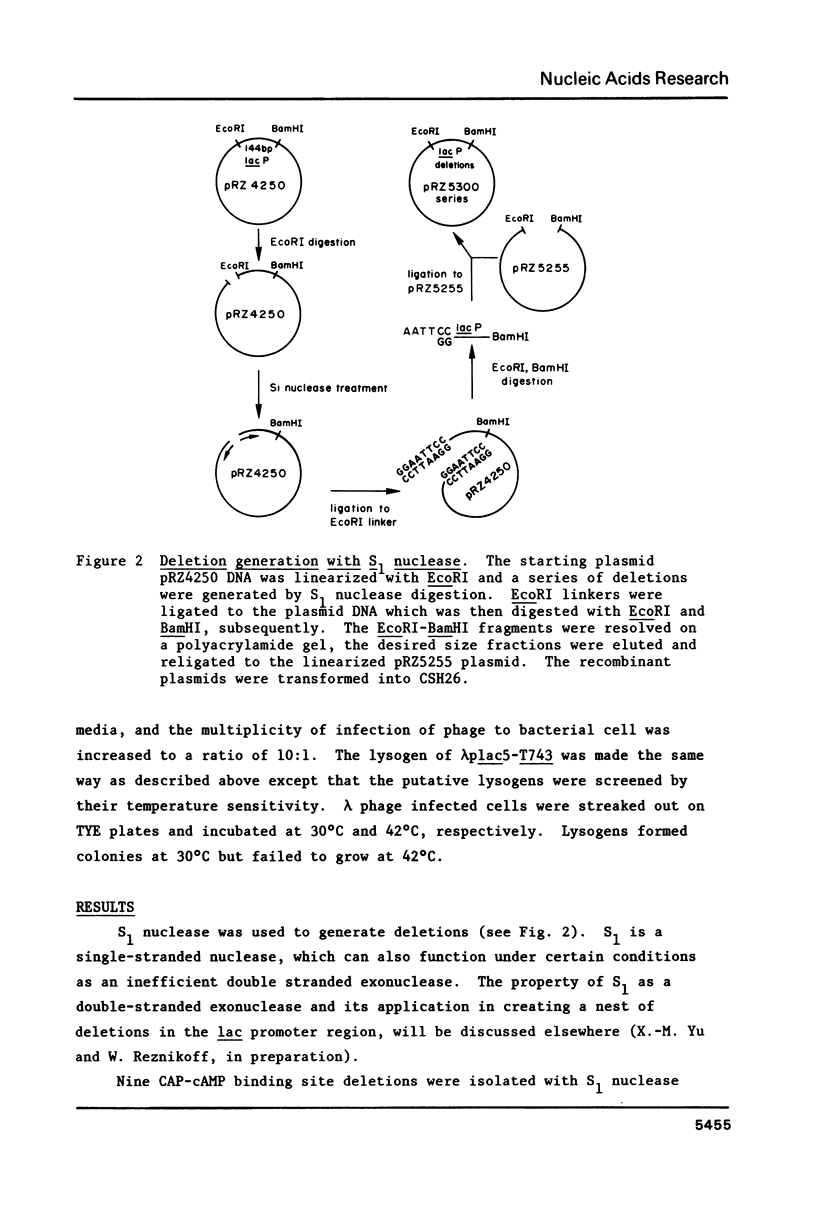
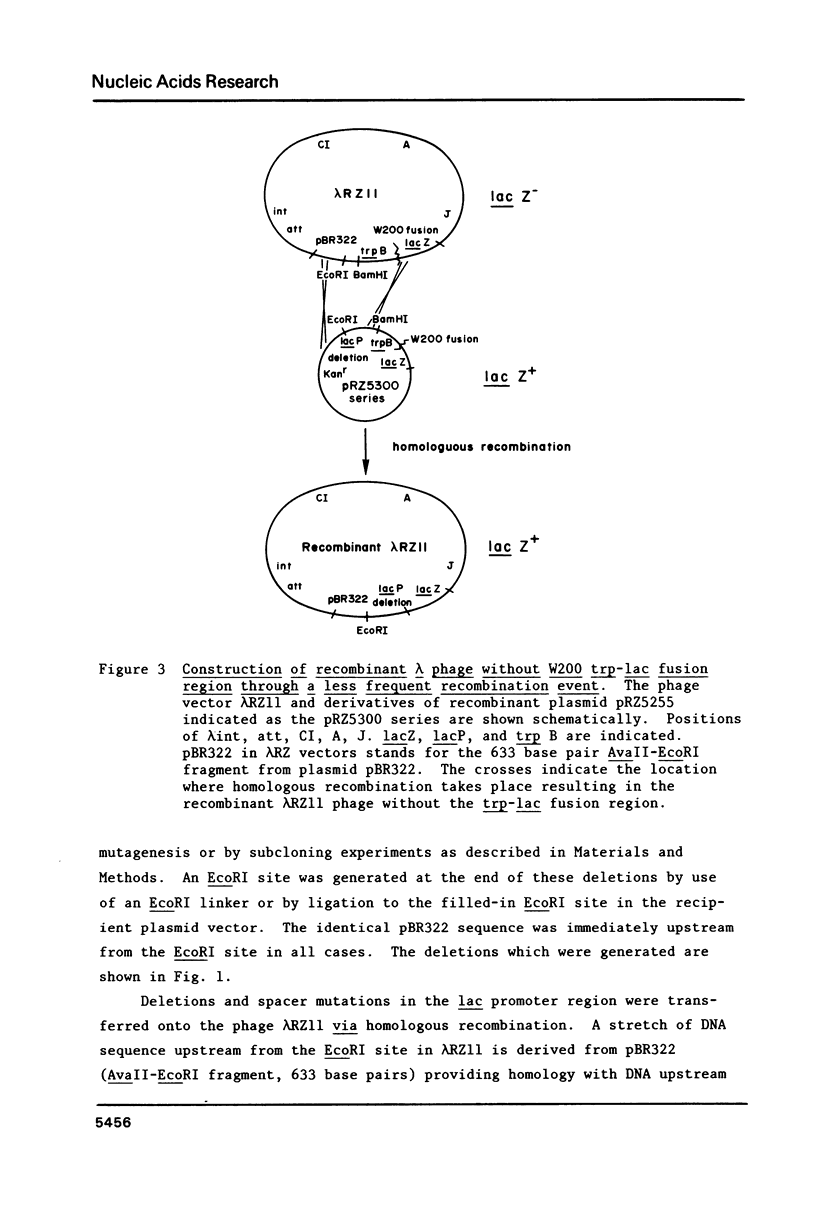
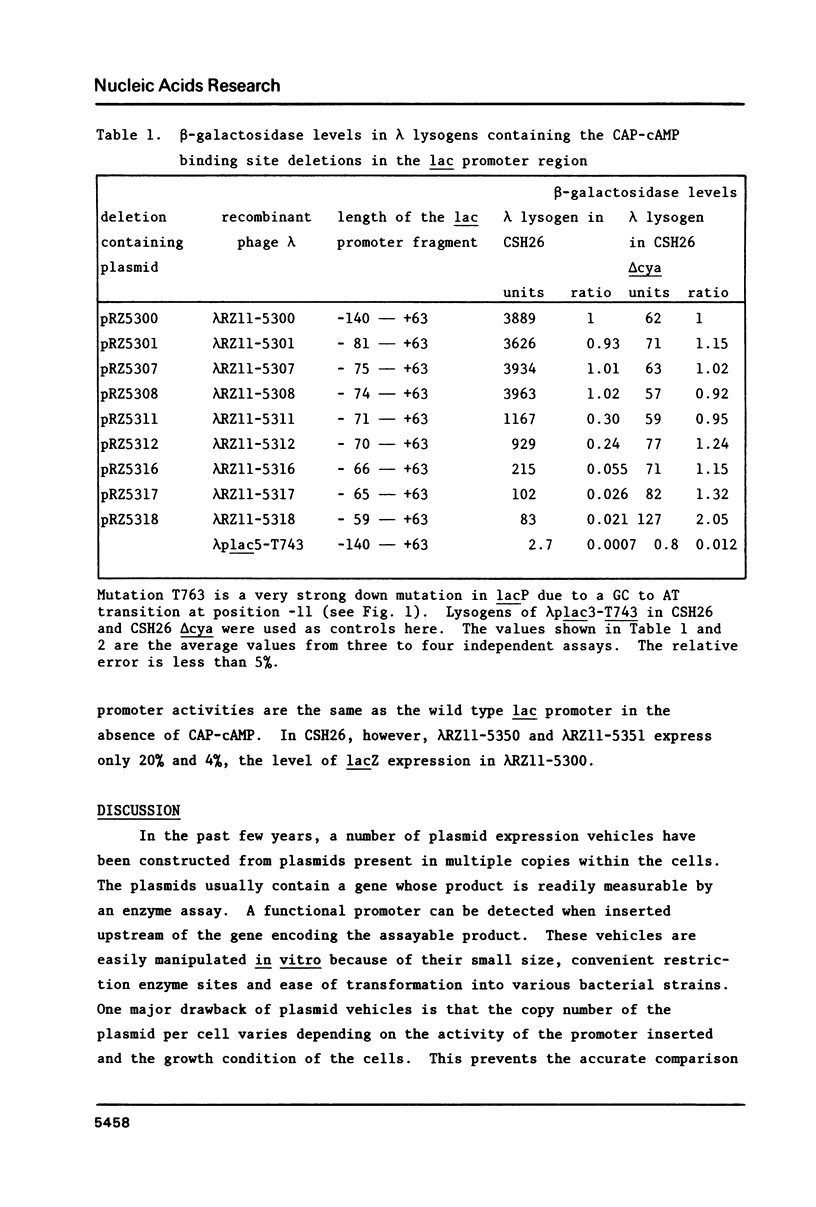
S1 nuclease was used to generate a series of deletions which extend into the CAP-cAMP binding site from upstream of the Escherichia coli lactose operon promoter (lacP). Deletion and insertion mutations were also created which changed the spacing region between the CAP-cAMP binding site and the lacP -35 region. The promoter activities of these mutations were compared by measuring the levels of beta-galactosidase gene expression in vivo. The results show that sequence information prior to 74 base pairs (-74) upstream from the transcription start site (designated as +1) is not necessary for the full activation of the lac promoter by the CAP-cAMP complex. However, the deletion which extends to the -71 position retains only one third of the promoter activity in the presence of the CAP-cAMP complex. Removal of one symmetrical element from the two fold symmetry in the CAP-cAMP binding site abolished the CAP-cAMP stimulation of the lac promoter. Spacer mutations which increase by one base pair or decrease by two base pairs the length of the spacing region between the CAP-cAMP binding site and the lacP -35 region drastically reduced the CAP-cAMP stimulation of the lac promoter. This suggests that the distance between the lac promoter transcription start site and CAP-cAMP binding site is crucial for the function of the lac promoter, despite the fact that this distance varies in other E. coli promoters positively regulated by CAP-cAMP. A deletion which extends to the -59 position results in a two fold enhanced expression of lac in the absence of CAP-cAMP. This is consistent with the existance of a competitive RNA polymerase binding site in this region which would normally act to inhibit RNA polymerase binding.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Caruthers M. H., Beaucage S. L., Efcavitch J. W., Fisher E. F., Goldman R. A., deHaseth P. L., Mandecki W., Matteucci M. D., Rosendahl M. S., Stabinsky Y. Chemical synthesis and biological studies on mutated gene-control regions. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):411–418. doi: 10.1101/sqb.1983.047.01.048. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Abelson J., Barnes W. M., Reznikoff W. S. Genetic regulation: the Lac control region. Science. 1975 Jan 10;187(4171):27–35. doi: 10.1126/science.1088926. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Abelson J., Johnson P. Nucleotide sequence changes produced by mutations in the lac promoter of Escherichia coli. J Mol Biol. 1977 Mar 25;111(1):65–75. doi: 10.1016/s0022-2836(77)80132-0. [DOI] [PubMed] [Google Scholar]
- Manley J. L. Synthesis and degradation of termination and premature-termination fragments of beta-galactosidase in vitro and in vivo. J Mol Biol. 1978 Nov 15;125(4):407–432. doi: 10.1016/0022-2836(78)90308-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Schmitz A. Cyclic AMP receptor proteins interacts with lactose operator DNA. Nucleic Acids Res. 1981 Jan 24;9(2):277–292. doi: 10.1093/nar/9.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. B. Interaction of the cAMP receptor protein with the lac promoter. Nucleic Acids Res. 1980 Feb 25;8(4):759–766. [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Wu R., Ruben G., Siegel B., Jay E., Spielman P., Tu C. P. Synchronous digestion of SV40 DNA by exonuclease III. Biochemistry. 1976 Feb 24;15(4):734–740. doi: 10.1021/bi00649a003. [DOI] [PubMed] [Google Scholar]
- Yu X. M., Munson L. M., Reznikoff W. S. Molecular cloning and sequence analysis of trp-lac fusion deletions. J Mol Biol. 1984 Jan 25;172(3):355–362. doi: 10.1016/s0022-2836(84)80032-7. [DOI] [PubMed] [Google Scholar]


