Abstract
To realize the full potential of human embryonic stem cells (hESCs), it is important to develop culture conditions that maintain hESCs in a pluripotent, undifferentiated state. A low O2 atmosphere (∼4% O2), for example, prevents spontaneous differentiation and supports self-renewal of hESCs. To identify genes whose expression is sensitive to O2 conditions, microarray analysis was performed on RNA from hESCs that had been maintained under either 4% or 20% O2. Of 149 genes differentially expressed, 42 were up-regulated and 107 down-regulated under 20% O2. Several of the down-regulated genes are most likely under the control of hypoxia-inducing factors and include genes encoding enzymes involved in carbohydrate catabolism and cellular redox state. Although genes associated with pluripotency, including OCT4, SOX2, and NANOG were generally unaffected, some genes controlled by these transcription factors, including LEFTY2, showed lowered expression under 20% O2, while a few genes implicated in lineage specification were up-regulated. Although the differences between O2 conditions were generally subtle, they were observed in two different hESC lines and at different passage numbers. The data are consistent with the hypothesis that 4% O2 favors the molecular mechanisms required for the maintenance of pluripotency.
Introduction
Human embryonic stem cells (hESCs) are generally derived from the inner cell mass (ICM) of preimplantation embryos and retain the potential of their embryonic founder cells to differentiate into cell types representing all three germ layers, ectoderm, mesoderm, and endoderm [1,2]. The pluripotent nature of hESCs and the study of their directed differentiation are beginning to provide insights into the process of lineage specification and the accompanying events that lead to functional differentiation [1,3]. In this regard, hESCs provide a unique model for the study of basic human developmental biology. In addition, hESCs have tremendous potential as a source of cells for tissue replacement and repair [1,4]. However, a serious hindrance to the use of hESCs for either of these purposes is the tendency of these cells to undergo spontaneous differentiation in culture [4,5]. As a result, hESC research has focused as much on the ability to propagate hESCs in an undifferentiated state as to the development of techniques that direct differentiation along specific cell lineages. It is clear that the full potential of hESCs will not be realized until the delicate balance between the processes of self-renewal and early differentiation are elucidated.
Since the derivation of hESCs was first reported in 1998 [6], it has been routine practice to culture these cells under atmospheric oxygen (20% O2). The efficacy of this practice is questionable as a preimplantation human conceptus would most likely be exposed to O2 tensions well below this concentration in utero [7,8]. One consequence of this culture practice is that overt differentiation within hESC colonies generally becomes visible after about 1 week if the cultures are maintained under 20% O2. Either undifferentiated cells must be separated from those already differentiated at the time the cells are passaged or else passage must be performed before differentiation is observable. In either case, pluripotent stem cells are likely to be contaminated with cells that are already cryptically differentiated and possibly committed to one lineage or another. We recently demonstrated that maintaining hESC cultures under low (physiological) O2 (2–5% O2) inhibits spontaneous differentiation and supports pluripotency [9]. Moreover, hESCs continuously maintained under 4% O2 for more than a single passage prior to being placed under 20% O2 conditions exhibit an even more prolonged delay before onset of differentiation [9]. These studies provided evidence that molecular processes that mediate pluripotency and suppress differentiation are supported under 4% O2 conditions and that the cells retain a “memory” of their prior environment.
Hypoxia-inducible factors-1α and −2α (HIF1A and −2A) exhibit reduced stability as O2 concentrations rise. They regulate transcriptional responses as heterodimers with a constitutively expressed protein, HIF1β (ARNT) [10] through binding to cis-acting hypoxia-response elements (HREs) on their target genes when O2 tensions are low [10–12] but may also have important roles in regulating pluripotent stem cell self-renewal and differentiation [13–15]. In most mammalian species examined, physiological O2 improves in vitro embryo development [16–18] and increases cell number, particularly of the ICM [13,19,20], suggesting that O2–regulated gene expression supports the maintenance of pluripotent cells. Interestingly, bovine blastocysts express HIF2A rather than HIF1A in their ICM [13]. Presumably, the former is responsible for the transcriptional responses in these pluripotent cells to physiological oxygen concentrations. Recently, HIF1A and HIF2A have been implicated in the maintenance of hESC pluripotency [21,22]. HIF2A, for example, regulates OCT4 expression [21], one of the core transcription factors long known to be essential for maintaining ESCs in an undifferentiated state [23]. A not unreasonable assumption, therefore, is that the HIF transcription factors play a role in suppressing the differentiation of hESCs under physiological oxygen concentrations.
The main objective of the experiments described in this paper was to examine the transcriptional profiles of two hESC lines, H1 and H9, which had been maintained under either 4% or 20% O2 conditions. As part of our experimental design, hESC RNA was collected prior to the time when obvious morphological differentiation first became apparent in the hESC colonies cultured under 20% O2. We addressed four main questions: (1) Are there consistent differences in gene expression that accompany culture in 20% O2? (2) Are the transcriptional profiles more consistent in cells cultured under 4% O2 as compared to cells cultured under 20% O2? (3) Does transcriptome profiling reveal any indications of differentiation occurring under 20% O2? (4) Is there evidence for the involvement of the HIF transcription factors in maintaining the pluripotency of hESCs cultured under 4% O2?
Materials and Methods
Culture of human ESCs
Human ESCs H1 (WA01) at passage 23 and H9 (WA09) at passage 19 were purchased from WiCell Research Institute (Madison, WI, USA) and cultured in six-well tissue culture plates (Nunc, Sigma-Aldrich, St. Louis, MO, USA) on a monolayer of γ-irradiated (8,000 cGy) mouse embryonic fibroblast (MEF) feeder cells until passage number 33 and 30, respectively, as previously described [9]. Thereafter, cells were cultured on a substratum of 1:30 dilutions of Matrigel™ (BD Biosciences, San Diego, CA, USA) in medium conditioned by MEF [9,24].
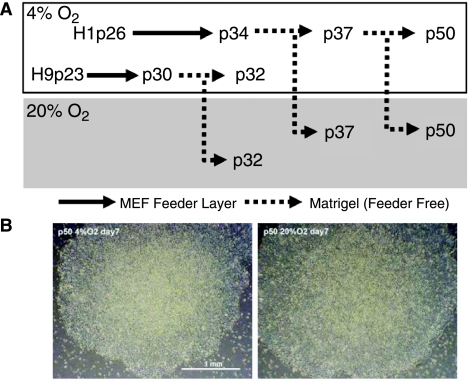
To identify differences in global gene expression between hESCs cultured under 20% and 4% O2, hESCs were passaged into six-well culture dishes and placed in either a standard humidified tissue culture CO2 incubator, (NuAire, IR Autoflow) set at 5% CO2 in air, or a tri-gas incubator (Heraeus HERAcell 150; Kendro Laboratory Products GmbH, Langenselbold, Germany) at a 4% O2, 5% CO2 setting. Oxygen levels, measured by using a Bacharach CO2/O2 Analyzer (Bacharach, Model 2830, New Kensington, PA, USA), were ∼19% in the standard incubator and 3.5–4.5% in the low O2 incubator. To minimize differences unrelated to O2 tension, experimental groups within replicates were cultured in the same medium and passaged at approximately the same time and as swiftly as possible. Cells maintained under 4% O2 were never exposed to air for more than 30 min, which is more than enough time to passage a six-well plate. We have measured how rapidly the level of dissolved oxygen in the culture media changes following changes in the oxygen tension in the air above the cells using an oxygen polarographic electrode (Pinpoint II Oxygen Meter, Aquatic Eco-Systems Inc, Apopka, FL, USA). When culture medium is shifted from a 20% environment to a 4% environment, it takes about ∼3 h for the DOC to decrease back to 3.5 ppm, with a half-maximal change taking about 40 min. The reverse change, that is, medium transferred from a 4% environment to a 20% environment, takes about ∼100 min for equilibrium, with a half time of 28 min. The H1 and H9 hESC lines had been maintained in a physiological O2 (3–5% O2) atmosphere since passage 26 and 23, respectively (Fig. 1A). RNA was isolated from H1 cells after 7 days of culture at two independent passage numbers (p37 and p50) and had been maintained on Matrigel™ for 3 and 16 passages, respectively. RNA from H9 cells was isolated at passage 32 and had been grown on Matrigel™ for 3 passages (Fig. 1A).
FIG. 1.
(A) Schematic diagram depicting conditions under which human embryonic stem cells (hESCs) were maintained and passaged prior to RNA extraction. H1 and H9 ESCs were maintained under 4% O2 condition from p26 and p23, respectively. The H1 and H9 cells were propagated under 4% O2 (represented by upper open box) for seven passages on MEF feeder layer (represented by solid arrow lines); thereafter the both cells were switched to Matrigel coated culture wells for at least three passages (represented by broken arrow lines) prior to RNA collection. RNA extraction was performed within 3 min following removal of the culture plates from respective O2 conditions and well before dissolved O2 in the culture medium showed any detectable changes. Total six RNA samples were collected from H1 cells at two different passage numbers (p37 and p50) and H9 cells at passage 32 (p32) under 4% and 20% O2 (represented by lower gray box) conditions, respectively. The cells maintained under 4% O2 were split into both 4% and 20% O2 conditions and cultured for 7 days before of RNA collection. (B) Phase contrast images of H1p50 hESC colonies under 4% (left) and 20% (right) O2 as they appeared prior to RNA extraction. RNA was isolated at 7 days postpassage, prior to the time at which morphological differentiation is apparent in H1 hESCs maintained under 20% O2. Bar 1 mm.
RNA extraction and preparation for microarray analysis
RNA was isolated from hESCs by using RNA STAT-60 reagent (Tel-Test Inc, Friendswood, TX, USA). RNA (5 μg) was used to prepare the biotin-labeled antisense RNA (aRNA) target by using the GeneChip® one-cycle target labeling and control reagents (#900493, Affymetrix, Santa Clara, CA, USA). The biotin-labeled aRNA (10 μg) was then hybridized to the Affymetrix Human U133 Plus 2.0 gene chip (Affymetrix) [25]. Each sample was hybridized to an individual chip, for a total of six chips. After hybridization, the chips were washed and stained with R-phycoerythrin-streptavidin on an Affymetrix fluidics station 450 by the Fluidics Protocol EukGE-WS2v5. The image data were acquired on an Affymetrix Genechip Scanner 3000.
Microarray data analysis
The study comprised three independent experiments (two samples of H1 and one H9 sample) cultured under two different O2 conditions, 4% and 20% O2. An absolute analysis of the resulting output was performed with Affymetrix Data Mining Tool, Version 3.0 software. The relative abundance of the transcripts based on signal and detection (present, absent or marginal) was assessed and a preliminary assessment of changes in relative transcript concentrations was performed. In addition, Affymetrix data were imported into Genespring 7.2 software (Silicon Genetics; Redwood City, CA, USA) for default normalization, that is, setting signal values to below 0.01 to 1.0, total chip normalization to the 50th percentile, and normalization of each gene to the median based on its measured expressed values. Those probe sets that did not meet the following criteria: (1) a signal value of at least 100 in either 20% or 4% O2, and (2) a consistent response to O2 conditions, that is, an increase or decrease in expression under 20% O2 in all three paired comparisons were removed from further analysis.
A separate statistical assessment was performed by first computing the logarithm of the intensity as a variance stabilizing transformation. The transformed intensity was then analyzed in a two stage mixed linear model. In the first stage, the across-array fixed effect treatment as well as random model effect array were modeled. The residuals were then modeled by probe by using a fixed effect model for treatment. A t-test for between-treatment comparisons within each probe was calculated. The Wilcoxon rank sum test was also calculated for each probe to add confidence for significant results.
Quantitative real-time PCR
Two-step real-time PCR was carried out to analyze and confirm the expression of select genes. RNA (1.2 μg) from different hESC treatment groups was reverse transcribed into cDNA in a reaction primed by oligo deoxynucleotide T (dT) 12–15 primer by using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitative TaqMan real-time PCR analyses were conducted on an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) with the following PCR conditions: (50°C for 2 min; 95°C for 10 min; 45 cycles at 95°C for 15 s; 60°C for 1 min). Each 50 μl real-time PCR reaction contained 20 ng of reverse-transcribed cDNA and 25 μl of 2× TaqMan Universal Master Mix (4304437, Applied Biosystems). The final concentration of gene-specific forward and reverse primers (Sigma-Genosys, Sigma-Aldrich) and gene-specific Taqman FAM-MGB probe (Applied Biosystems) was 20 nM and 100 nM, respectively. Each target transcript was analyzed in triplicate and independently normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (NM_002046) and ribosomal protein L19 (NM_000981). Supplemental Table 1 (supplementary table 1 is available online at http://www.liebertpub.com/scd) lists the sequences of the gene-specific primers and probes that were used. Fold changes were calculated according to the formula, Fold Change = 2ΔΔCt where ΔΔCt = ΔCt L–ΔCt A and ΔCt = Ct target gene−Ct reference (GAPDH or RPL19).
Immunocytochemistry
H1 ESCs (p33) were cultured for 10 days under 20% and 4% O2 conditions, at which time spontaneous differentiation in colonies under 20% O2 conditions can be easily observed [9]. Cell colonies were fixed by immersing the coverslips in 2% paraformaldehyde/PBS and permeabilized in 1.0% Triton X-100/PBS. Nonspecific immunoglobulin binding sites were blocked by incubation in 5% goat serum and 5% BSA prior to the addition of primary antibodies. The serological reagents used were a rabbit anti-HIF1A polyclonal antibody (NB100-134, Novus Biologicals. Littleton, CO, USA), a mouse anti-HIF2A monoclonal antibody (NB100-132, Novus Biologicals), mouse antistage-specific embryonic antigen (anti-SSEA)-4 (MC813-70, Developmental Studies Hybridoma Bank, IA, USA) and anti-Oct4 [9]. Secondary antibody staining was performed with Alexa Fluor 568 and 488 goat antirabbit, goat antimouse and/or goat antirat antibodies (Molecular Probes, Invitrogen). Nuclei were labeled with TO-PRO-3 (Molecular Probes, Invitrogen) or Hoechst 33258 (Sigma-Aldrich). Images were captured on a Zeiss META NLO two photon confocal system (Carl Zeiss, Obercohen, Germany) coupled with a Zeiss Axiovert 200M microscope.
Results
General features of gene expression in H1 and H9 ESCs under 20% and 4% O2 conditions
We compared the transcriptional profiles of hESCs after 1 week of culture under 20% and 4% O2 conditions after initial maintenance of the cell lines for several passages on a Matrigel substratum under 4% O2 conditions (Fig. 1A). Despite the differences in O2 atmosphere, the hESC colonies were indistinguishable morphologically and size (658,000 ± 108,764 μm2 and 636,614 ± 220,157 μm2 in 20% and 4% O2, respectively), with no signs of differentiation evident under 20% O2 (Fig. 1B). A total of 21,891 and 22,534 (out of 54,675) probe-sets were assessed as “present” in all three samples by Genespring software, under 20% and 4% O2, respectively.
The overall gene expression profiles of the H1 and H9 ESCs used for these experiments are similar to those previously reported for other hESCs, including the H1, BG01, BG02 and BG03 lines [26–31]. Several genes highly expressed in these cell lines [26,28,31,32], for example, most ribosomal proteins, MTCO1 (cytochrome c oxidase), ACTB (beta-actin), and EEF1A1L14 (eukaryotic translation elongation factor alpha 1), were also found to be amongst the top 50 genes expressed in the H1 and H9 samples analyzed here (Supplemental Table 2; supplementary table 2 is available online at http://www.liebertpub.com/scd) and showed no significant changes in expression in the shift from 4% to 20% O2 conditions. Other genes reported to be highly expressed in hESCs [28,29,32] that were not amongst the top 50 genes in our analysis, for example, HMG1, FLJ10713, USP9X, CDC2, and HSPCA (Supplemental Table 2)were also not regulated by the increase in O2. These data tend to confirm the general similarity of the H1 and H9 cells to other undifferentiated hESC lines, irrespective of they are cultured under 20% or 4% O2 conditions.
The transcriptional profiles of the three samples of RNA collected from cultures maintained continuously under 4% O2 conditions as assessed as a Pearson correlation coefficient (PCC) over 54,675 probe-sets were quite similar to each other (PCC = 0.87) (Supplemental Fig. 1; supplementary figure 1 is available online at http://www.liebertpub.com/scd). By contrast, the hESC samples cultured under 20% O2 exhibited a greater variance in their gene expression profiles (PCC = 0.67) (Supplemental Fig. 1). In other words, H1 and H9 ESC samples cultured under 4% O2 clustered more tightly with each other than with their respective partner samples under 20% O2.
A total of 149 transcripts (42 with increased expression, 107 with decreased expression under 20% O2 conditions) demonstrated consistent differences in all three samples sets (Supplemental Table 3; supplementary table 3 is available online at http://www.liebertpub.com/scd). Tables 1 and 2 list the top 25 genes within each class that displayed the greatest changes in expression. Over 400 regulated genes were identified (Supplementary Table 4; supplementary table 4 is available online at http://www.liebertpub.com/scd) if a less stringent selection criterion was employed, namely a 1.5-fold or greater average change in expression but not necessarily in all three sample comparisons. Of these genes, 123 increased expression while 301 transcripts were down-regulated under 20% O2 culture conditions.
Table 1.
List of Top 25 Genes Up and Down-Regulated in 20% O2
| Probe Set ID | Accession | Unigene | Symbol | Descriptions | Fold |
|---|---|---|---|---|---|
| Up-regulated in 20% O2 | |||||
| 209160_at | NM_003739 | Hs.78183 | AKR1C3 | Aldo-keto reductase family 1, member C3 | 2.2 |
| 225809_at | AI659927 | Hs.105460 | — | DKFZP564O0823 protein | 2.1 |
| 207057_at | NM_004731 | Hs.439643 | SLC16A7 | Solute carrier family 16 (monocarboxylic acid transporters), member 7 | 2.0 |
| 226278_at | AI150224 | Hs.349096 | — | Hypothetical protein DKFZp313A2432 | 2.0 |
| 226751_at | AW193693 | Hs.212885 | C2orf32 | Chromosome 2 open reading frame 32 | 1.8 |
| 230960_at | AI740721 | Hs.128292 | — | Transcribed locus | 1.7 |
| 211075_s_at | Z25521 | Hs.446414 | CD47 | CD47 antigen (Rh-related antigen, integrin-associated signal transducer)* | 1.7 |
| 235371_at | AI452595 | Hs.431092 | PPP4R2 | Protein phosphatase 4, regulatory subunit 2 | 1.7 |
| 230130_at | AI692523 | Hs.29802 | — | Transcribed locus | 1.7 |
| 209459_s_at | AF237813 | Hs.336768 | ABAT | 4-aminobutyrate aminotransferase | 1.6 |
| 205328_at | NM_006984 | Hs.26126 | CLDN10 | Claudin 10 | 1.6 |
| 201467_s_at | NM_000903 | Hs.406515 | NQO1 | NAD(P)H dehydrogenase, quinone 1* | 1.6 |
| 204493_at | NM_001196 | Hs.172894 | BID | BH3 interacting domain death agonist | 1.6 |
| 48808_at | NM_000791 | Hs.83765 | DHFR | Dihydrofolate reductase* | 1.6 |
| 223122_s_at | AF311912 | Hs.481022 | SFRP2 | Secreted frizzled-related protein 2 | 1.5 |
| 211737_x_at | BC005916 | Hs.371249 | PTN | Pleiotrophin (heparin binding growth factor 8)* | 1.5 |
| 225688_s_at | AK025444 | Hs.477114 | PHLDB2 | Pleckstrin homology-like domain, family B, member 2 | 1.5 |
| 209773_s_at | BC001886 | Hs.75319 | RRM2 | Ribonucleotide reductase M2 polypeptide | 1.4 |
| 232037_at | AW204060 | Hs.567396 | PUNC | Putative neuronal cell adhesion molecule | 1.4 |
| 225418_at | AI520949 | Hs.8372 | UQCR | Ubiquinol-cytochrome c reductase (6.4 kD) subunit | 1.4 |
| 1568678_s_at | BC037785 | Hs.487175 | FGFR1OP | FGFR1 oncogene partner | 1.4 |
| 201117_s_at | NM_001873 | Hs.75360 | CPE | Carboxypeptidase E | 1.3 |
| 215646_s_at | R94644 | Hs.443681 | CSPG2 | Chondroitin sulfate proteoglycan 2 (versican)* | 1.3 |
| 201202_at | NM_002592 | Hs.147433 | PCNA | Proliferating cell nuclear antigen | 1.3 |
| 202345_s_at | NM_001444 | Hs.408061 | FABP5 | Fatty acid binding protein 5 (psoriasis-associated) | 1.3 |
| Down-regulated in 20% O2 | |||||
| 1569287_at | BC017942 | Hs.621322 | — | Similar to otoconin 90* | −6.7 |
| 201848_s_at | NM_004052 | Hs.144873 | BNIP3 | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 | −4.6 |
| 201010_s_at | NM_006472 | Hs.533977 | TXNIP | Thioredoxin interacting protein* | −4.1 |
| 236180_at | W57613 | Hs.419240 | SLC2A3 | Solute carrier family 2 (facilitated glucose transporter), member 3 | −3.2 |
| 202718_at | NM_000597 | Hs.162 | IGFBP2 | Insulin-like growth factor binding protein 2 | −3.1 |
| 202887_s_at | NM_019058 | Hs.523012 | DDIT4 | DNA-damage-inducible transcript 4 | −3.0 |
| 1569453_a_at | BG772667 | Hs.560022 | — | Hypothetical locus LOC692247 | −2.9 |
| 206701_x_at | NM_003991 | Hs.82002 | EDNRB | Endothelin receptor type B* | −2.8 |
| 225283_at | AV70117 | Hs.6093 | ARRDC4 | Arrestin domain containing 4 | −2.6 |
| 1557636_a_at | BC031107 | Hs.258357 | — | Hypothetical protein LOC136288 | −2.5 |
| 1562529_s_at | BC040965 | Hs.569497 | RORA | RAR-related orphan receptor A | −2.4 |
| 206012_at | NM_003240 | Hs.25195 | LEFTY2 | Left-right determination factor 2 | −2.3 |
| 223734_at | AF329088 | Hs.84549 | OSAP | Corneal endothelium specific protein 1 | −2.2 |
| 207543_s_at | NM_000917 | Hs.500047 | P4HA1 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase, alpha polypeptide I | −2.2 |
| 218717_s_at | NM_018192 | Hs.374191 | LEPREL1 | Leprecan-like 1 | −2.2 |
| 214624_at | NM_007000 | Hs.150309 | UPK1A | Uroplakin 1A | −2.2 |
| 223710_at | NM_006072 | Hs.131342 | CCL26 | Chemokine (C-C motif) ligand 26 | −2.1 |
| 201968_s_at | NM_002633 | Hs.1869 | PGM1 | Phosphoglucomutase 1* | −2.1 |
| 201105_at | NM_002305 | Hs.227751 | LGALS1 | Lectin, galactoside-binding, soluble, 1 (galectin 1) | −2.1 |
| 200737_at | NM_000291 | Hs.7877 | PGK1 | Phosphoglycerate kinase 1* | −2.0 |
| 227271_at | AU151265 | Hs.380704 | FGF11 | Fibroblast growth factor 11 | −2.0 |
| 206424_at | NM_000783 | Hs.150595 | CYP26A1 | Cytochrome P450, subfamily XXVIA, polypeptide 1 | −2.0 |
| 202022_at | NM_005165 | Hs.155247 | ALDOC | Aldolase C, fructose-bisphosphate | −2.0 |
| 227404_s_at | AI459194 | Hs.326035 | EGR1 | Early growth response 1 | −2.0 |
| 214823_at | AF033199 | Hs.8198 | ZNF204 | Zinc finger protein 204 | −1.9 |
Genes exhibiting an increase (Up-regulated) or decrease (Down-regulated) in all three samples (H1p37, H1p50, and H9p32) are represented. Fold changes shown are the average of the three paired comparisons. Positive or negative values indicate fold-increases or fold-decreases in gene expression, respectively, in human embryonic stem cells cultured under 20% O2 compared to cells under 4% O2. The complete list of genes which are consistently up- or down-regulated in 20% O2 is provided in Supplemental Table 3.
Genes represented by more than one probe-set, (redundant unigene entries were removed from list).
Table 2.
Expression of Pluripotent Markers in Human Embryonic Stem Cells under 20% and 4% O2 Conditions
| |
20% O2 |
4% O2 |
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Probe Set ID | Accession | Unigene | Symbol | Description | Signal | STDEV | Signal | STDEV | Change call | Average fold change |
| 220668_s_at | NM_006892 | Hs.251673 | DNMT3B | DNA (cytosine-5-)-methyltransferase 3 beta | 28,878 | 2,796 | 26,378 | 2,086 | NC (I) | |
| 206012_at | NM_003240 | Hs.25195 | LEFTY2 | Left-right determination factor 2 | 1,606 | 150 | 3,866 | 1,450 | D | −2.4 ± 1.0 |
| 204271_s_at | NM_000115 | Hs.82002 | EDNRB | Endothelin receptor type B | 6,282 | 3,308 | 13,223 | 841 | D | −2.1 ± 1.4 |
| 206783 _at | NM_002007 | Hs.1755 | FGF4 | Fibroblast growth factor 4 | 476 | 77 | 602 | 271 | NC | |
| 220053_at | NM_020634 | Hs.86232 | GDF3 | Growth differentiation factor 3 | 519 | 76 | 542 | 187 | NC | |
| 214022_s_at | NM_003641 | Hs.458414 | IFITM1 | Interferon induced transmembrane protein 1 | 14,449 | 1,316 | 16,809 | 631 | D | −1.2 ± 0.1 |
| 219823_at | NM_024674 | Hs.86154 | LIN28 | Lin-28 homolog (C. elegans) | 24,203 | 2,986 | 23,495 | 3,453 | NC | |
| 220184_at | NM_024865 | Hs.504647 | NANOG | Nanog Homeobox | 11,469 | 1,626 | 11,871 | 2,177 | NC | |
| 230916_at | NM_018055 | Hs.65853 | NODAL | Nodal Homolog (mouse) | 686 | 225 | 575 | 408 | NC (MI) | |
| 201578_at | NM_005397 | Hs.16426 | PODXL | Podocalyxin-like | 26,319 | 3,328 | 27,443 | 3,908 | NC | |
| 208286_x_at | NM_002701 | Hs.2860 | POU5F1 | POU domain, class 5, transcription factor 1, Oct4 | 11,180 | 3,548 | 11,283 | 3,417 | NC | |
| 213721_at | L07335 | Hs.518438 | SOX2 | SRY (sex determining region 1) -box 2 | 243 | 89 | 332 | 133 | NC (D) | |
| 207199_at | NM_003219 | Hs.492203 | TERT | Telomerase reverse transcriptase | 243 | 89 | 332 | 133 | NC | |
| 208275_x_at | NM_003577 | Hs.158307 | UTF1 | Undifferentiated embryonic cell transcription factor 1 | 859 | 720 | 944 | 642 | NC (D) | |
| 1554777_at | AF450454 | Hs.335787 | ZFP42 (Rex1) | Zinc Finger Protein 42 | 1,935 | 1,362 | 2,184 | 1,277 | NC (D) | |
Change calls (NC = no change, D = decrease, MI = mild increase) and a brief description of the observed change are listed.
Changes in expression of genes associated with pluripotency
Several genes associated with pluripotency [27,33–35], namely OCT4 [36], NANOG [37], LIN28 [38], SOX2 [39], ZFP42/REX1 [40], TERT [41], and PODXL [27] exhibited expression levels that did not vary greatly according to the O2 conditions under which the cells were maintained (Table 2).In particular, OCT4, NANOG, and SOX2 are believed to function as core transcription factors in maintaining pluripotency of ESCs [33,34,42]. Although the expression of SOX2 varied somewhat between paired comparisons, that of OCT4 and NANOG remained relatively constant (Table 2).
On the other hand, several genes considered to be under the transcriptional control of the OCT4/NANOG/SOX2 triad [33,34,43] displayed markedly lower expression in cells cultured under 20% O2 as compared to parallel cultures maintained under 4% O2 conditions. These down-regulated genes included LEFTY2 [33] and endothelin receptor type B (ENDRB) (Table 2).In addition, genes known to be regulated by OCT4, such as SALL1 [34,43], TRIM2 [43], ZIC2 [43], and FGFR2 [33,34,43], also exhibited decreased expression in hESCs cultured under 20% O2 (Table 3).These data suggest that although transcriptional profiles of the core pluripotency genes may not be greatly affected by O2 conditions, the expression of their downstream targets might well be.
Table 3.
OCT4-Regulated Genes Exhibit Decreased Expression in 20% O2
| Probe Set ID | Accession | Unigene | Symbol | Descriptions | Fold |
|---|---|---|---|---|---|
| 202887_s_at | NM_019058 | Hs.523012 | DDIT4 | DNA-damage-inducible transcript 4 | −3.0 |
| 206012_at | NM_003240 | Hs.25195 | LEFTY2 | Left-right determination factor | −2.3 |
| 209220_at | NM_004484 | Hs.567276 | GPC3 | Glypican 3 | −1.7 |
| 206893_at | NM_002968 | Hs.135787 | SALL1 | Sal-like 1 (Drosophila) | −1.4 |
| 221605_s_at | AF136970 | Hs.462585 | PIPOX | Pipecolic acid oxidase | −1.4 |
| 223642_at | NM_007129 | Hs.591205 | ZIC2 | Zic family member 2 (odd-paired homolog, Drosophila) | −1.4 |
| 208789_at | BC004295 | Hs.437191 | PTRF | Polymerase I and transcript release factor | −1.3 |
| 202342_s_at | NM_015271 | Hs.435711 | TRIM2 | Tripartite motif protein | −1.3 |
| 203638_s_at | NM_022969 | Hs.533683 | FGFR2 | Fibroblast growth factor receptor 2 | −1.3 |
| 225992_at | AL562031 | Hs.30385 | MLLT10 | Myeloid/lymphoid or mixed-lineage leukemia translocated to, 10 | −1.3 |
| 228356_at | AV711366 | Hs.335003 | ANKRD11 | Ankyrin repeat domain 11 | −1.2 |
Fold decreases shown are the average of three paired comparisons.
Transforming growth factor beta (TGFβ) signaling pathway regulated genes
The TGFβ/ACTIVIN/NODAL signaling pathways play a critical role in maintaining hESC pluripotency [44–46]. Expression of a member of the TGFβ family of genes, LEFTY2 was reduced under 20% O2 (Table 2).The TGFβ-inducible protein, SERPINE1 (PAI-1) [47] and transcription factor KLF11 [48] also exhibited reduced expression (1.76- and 1.68-fold, respectively) in cells cultured under 20% O2.
Changes in expression of genes associated with differentiation
Several genes previously associated with hESC differentiation [26,27,49,50] were up-regulated in at least one of the three replicates under 20% O2 (Supplemental Table 4).For example, HAND1 exhibited a 2-fold and 2.5-fold increase in the H1p37 and H9 samples, respectively. EOMES and H19 (imprinted maternally expressed untranslated mRNA), GATA6 and MSX2 were increased in the H9 ESC comparison. On the other hand, expression of the TDGF1 (CRIPTO) gene, which has been shown to be down-regulated in association with the initial steps of hESC differentiation [27,49], was not altered under 20% O2. Nor did we note any changes in expression of the gene for leukemia inhibitory factor (LIF) and its receptor, both of which have been reported to be increased during initial hESC differentiation [51]. In fact, the hybridization signal for LIF was sufficiently low that it was considered to be absent under both 4% and 20% O2 conditions. The expression of several genes associated with more advanced stages of differentiation and early embryoid body formation, including GATA4, SOX1, FGF5, and T (Brachyury) [27,50] was either very low or assessed as “absent” under both 4% and 20% O2 conditions.
Genes associated with an oxidative stress response
A number of genes associated with oxidative stress responses [52] were increased under 20% O2 conditions (Supplemental Table 5; supplementary table 5 is available online at http://www.liebertpub.com/scd). NRF2 (NFE2L2), a transcription factor responsible for the regulation of cytoprotective and antioxidant proteins [53,54] and at least two of its downstream targets, NQO1 (NAD(P)H quinine oxidoreductase) [53] and AKR1C3 (aldo-keto reductase family 1, member C3) [53], were among those exhibiting increased expression under 20% O2 (Supplemental Table 5).In addition, expression levels of the mRNAs for VCAN (versican, CSPG2), a proteoglycan isoform that protects cells from oxidative stress-induced apoptosis [55], and CTNS (cystinosin), a gene encoding a lysosomal membrane transporter for cystine [56] and whose deficiency can lead to increased ratio of oxidized to reduced glutathione [57], were increased under 20% O2 (Supplemental Table 5).
Functional classes of regulated genes
The major physiological pathway down-regulated in 20% O2 was glycolysis (Table 4).Regulated genes included the rate limiting enzymes HK2 and HK1 (hexokinase 2, −1), and PKM2 (pyruvate kinase). A few of these genes, for example, those encoding PGM1 (phosphoglucomutase 1), ALDOA (aldolase A), and GPI (glucose phosphate isomerase), are also linked to the pentose phosphate pathway (Table 4).These regulatory switches relating to O2 conditions were not unexpected as most genes encoding enzymes of the glycolytic pathway are considered to be under the control of HIF1A [12].
Table 4.
Consistently Decreased Transcripts for Enzymes of the Glycolytic and Pentose Phosphate Pathways Under 20% O2
| Probe Set ID | Accession | Unigene | Symbol | Descriptions | Fold |
|---|---|---|---|---|---|
| Glycolysis | |||||
| 201968_s_at | NM_002633 | Hs.1869 | PGM1 | Phosphoglucomutase 1 | −2.1 |
| 202022_at | NM_005165 | Hs.155247 | ALDOC | Aldolase C, fructose-bisphosphate | −2.0 |
| 200737_at | NM_000291 | Hs.78771 | PGK1 | Phosphoglycerate kinase 1 | −2.0 |
| 238996_x_at | NM_000034 | Hs.513490 | ALDOA | Aldolase A, fructose-bisphosphate | −1.8 |
| 200650_s_at | NM_005566 | Hs.2795 | LDHA | Lactate dehydrogenase A | −1.7 |
| 208308_s_at | NM_000175 | Hs.466471 | GPI | Glucose phosphate isomerase | −1.5 |
| 201251_at | NM_002654 | Hs.534770 | PKM2 | Pyruvate kinase, muscle | −1.5 |
| 200822_x_at | NM_000365 | Hs.524219 | TPI1 | Triosephosphate isomerase 1 | −1.4 |
| 202934_at | NM_000189 | Hs.198427 | HK2 | Hexokinase 2 | −1.4 |
| 217294_s_at | U88968 | Hs.517145 | EN01 | Enolase 1, (alpha) | −1.3 |
| 200697_at | NM_000188 | Hs.118625 | HK1 | Hexokinase 1 | −1.2 |
| Pentose phosphate pathway | |||||
| 201968_s_at | NM_002633 | Hs.1869 | PGM1 | Phosphoglucomutase 1 | −2.1 |
| 238996_x_at | NM_000034 | Hs.513490 | ALDOA | Aldolase A, fructose-bisphosphate | −1.8 |
| 208308_s_at | NM_000175 | Hs.466471 | GPI | Glucose phosphate isomerase | −1.5 |
Fold increases shown are the average of three paired comparisons.
Several genes believed to be under the control of HIF1A [11,22,58] and that contribute to apoptosis, cellular redox regulation, and proliferation were down-regulated under 20% O2, for example, BNIP3 (−4.6-fold) [59, 60], TXNIP (thioredoxin interacting protein; −4.5-fold), DDIT4 (DNA-damage-inducible transcript 4, −3.0-fold) [61], IGFBP2 (−3.1-fold), LGALS1 (−2.1-fold) and VEGF (−1.6-fold) (Table 5).
Table 5.
HIF1A-Regulated Genes Showing Differential Expression between 20% and 4% O2 Conditions
| Probe Set ID | Accession | Unigene | Symbol | Descriptions | Fold |
|---|---|---|---|---|---|
| 201848_s_at | NM_004052 | Hs.144873 | BNIP3 | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 | −4.6 |
| 201010_s_at | NM_006472 | Hs.533977 | TXNIP | Thioredoxin interacting protein | −4.1 |
| 236180_at | W57613 | Hs.419240 | SLC2A3 | Solute carrier family 2 (facilitated glucose transporter), member 3 | −3.2 |
| 202718_at | NM_000597 | Hs.162 | IGFBP2 | Insulin-like growth factor binding protein 2 | −3.1 |
| 202887_s_at | NM_019058 | Hs.523012 | DDIT4 | DNA-damage-inducible transcript 4 | −3.0 |
| 207543_s_at | NM_000917 | Hs.500047 | P4HA1 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase, alpha polypeptide I | −2.2 |
| 201105_at | NM_002305 | Hs.227751 | LGALS1 | Lectin, galactoside-binding, soluble, 1 | −2.1 |
| 1562529_s_at | BC040965 | Hs.569497 | RORA | RAR-related orphan receptor A | −2.1 |
| 202022_at | NM_005165 | Hs.155247 | ALDOC | Aldolase C, fructose-bisphosphate | −2.0 |
| 200737_at | NM_000291 | Hs.78771 | PGK1 | Phosphoglycerate kinase 1 | −2.0 |
| 238996_x_at | NM_000034 | Hs.513490 | ALDOA | Aldolase A, fructose-bisphosphate | −1.8 |
| 200650_s_at | NM_005566 | Hs.2795 | LDHA | Lactate dehydrogenase A | −1.7 |
| 203234_at | NM_003364 | Hs.488240 | UPP1 | Uridine phosphorylase 1 | −1.6 |
| 210512_s_at | NM_003376 | Hs.73793 | VEGF | Vascular endothelial growth factor | −1.6 |
| 202627_s_at | NM_000602 | Hs.414795 | SERPINE1 (PAI-1) | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | −1.5 |
| 208308_s_at | NM_000175 | Hs.466471 | GPI | Glucose phosphate isomerase | −1.5 |
| 201251_at | NM_002654 | Hs.534770 | PKM2 | Pyruvate kinase, muscle | −1.5 |
| 200822_x_at | NM_000365 | Hs.524219 | TPI1 | Triosephosphate isomerase 1 | −1.4 |
| 202934_at | NM_000189 | Hs.198427 | HK2 | Hexokinase 2 | −1.4 |
| 217294_s_at | U88968 | Hs.517145 | ENO1 | Enolase 1, (alpha) | −1.3 |
| 209773_s_at | BC001886 | Hs.75319 | RRM2 | Ribonucleotide reductase M2 polypeptide | 1.4 |
Genes in the top panel are positively-regulated by HIF1A and exhibit decreased expression under 20% O2. The gene in the bottom panel, RRM2, has been shown to be repressed HIF1A and are up-regulated in human embryonic stem cells cultured under 20% O2. Fold changes shown are the average of three paired comparisons.
Hypoxia-inducible factors and related genes
HIF1A mRNA (NM_001530) was present in relatively high amounts in H1 and H9 cells, and the transcript concentrations did not change in response to placing the cells under 20% O2 conditions (12,998 ± 4,589 and 11,924 ± 3,863 in 20% and 4% O2, respectively). This lack of change in HIFA message in response to higher O2 was further supported by quantitative real-time PCR analysis (Supplemental Table 6; supplementary table 6 is available online at http://www.liebertpub.com/scd) and is not unexpected as HIF1A is primarily regulated at the level of protein turnover [14]. The expression of HIF2A was much lower than that of HIF1A (586 ± 409 and 276 ± 68 in 20% and 4% O2, respectively), but its transcript concentration was increased at 20 % O2. Transcripts for ARNT, like those for HIF2A, were also present in low concentrations (463 ± 51 and 550 ± 21 in 20% and 4% O2, respectively) and were unaffected by O2. Finally, expression of the von Hippel-Lindau (VHL) tumor suppressor gene, the recognition component of an E3 ubiquitin-protein ligase complex that targets both HIF1A and −2A for proteasomal degradation [10], was present in both H1 and H9 cells in similar concentrations and showed no differential expression between the two conditions (6,833 ± 461 and 6,266 ± 1134, in 20% and 4% O2, respectively).
Validation by real-time, quantitative PCR (RT-QPCR)
To validate the results from the microarray analyses, RT-QPCR was performed on the same RNA samples used in the array hybridization experiment. Selected genes included, AKR1C3, FGF2, OCT4, and LEFTY2 (Supplemental Table 6).Data in RT-QPCRs were independently normalized to transcripts for either ribosomal protein L19 (RPL19) or GAPDH. The latter, despite being a component of the glycolytic pathway and regulated by oxygen in endothelial cells [62], showed no evidence for regulation by O2 in the microarray analyses and was therefore a suitable standard to employ. Although minor discrepancies were observed, the overall changes in expression observed with real-time PCR were consistent with the microarray data (Supplemental Table 6).
Expression patterns of HIF1A and −2A in undifferentiated and differentiated hESCs
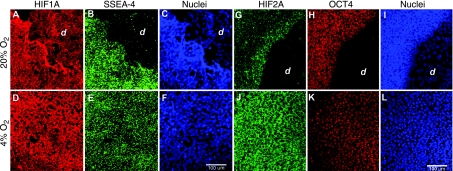
Since HIF gene products are largely regulated at the protein level, we examined the expression pattern of HIF1A and HIF2A by immunocytochemistry in H1 cell colonies cultured under 20% and 4% O2 conditions for 10 days, a time when signs of overt differentiation are visible in the 20% O2 but not the 4% O2 cultures (Fig. 2). In contrast to SSEA-4, a marker of pluripotent, undifferentiated hESCs, HIF1A was expressed in both the undifferentiated and differentiated areas of H1 cells cultured under 20% O2 (Fig. 2). Under both 20% and 4% O2 conditions, the protein was primarily localized to the cytoplasm. In contrast, the HIF2A protein, like OCT4, was expressed only in undifferentiated cells, but, like HIF1A, was concentrated in the cytoplasm (Fig. 2).
FIG. 2.
HIFA and SSEA-4 (A–F) and HIF2A and OCT4 (G–L) expression in human embryonic stem cells (hESCs) cultured under either 20% (A–C and G–I) or 4% (D–F and J–L) O2. H1 cells (p33) were cultured for 10 days under either 20% or 4% O2 conditions. Immunofluorescent detection of HIF1A (red) and SSEA-4 (green) expression in colonies cultured under 20% (A and B) or 4% (D and E) O2. Spontaneous differentiation of H1 cells (d) under 20% O2 can be seen in the upper right (A–C) or bottom right corner (G–I) of the figure. Expression of HIF2A (green) and OCT4 (red) as observed by immunofluorescence in hESC colonies under 20% (G and H) or 4% (J and K) O2. Nuclei are stained blue using TO-PRO-3 (C, F, I, and L). All images were captured by confocal microscopy using a 20× objective. Bar, 0.1 mm.
Discussion
Despite a decade of research, optimal conditions required for maintaining pluripotency and avoiding spontaneous differentiation of hESCs have not been established, although it is clear that reduced O2 conditions are helpful in this regard. For example, hESCs under low O2 exhibit a lowered tendency to differentiate spontaneously [9] and enhanced clonal recovery and genomic stability [63]. Physiological O2 also prevents differentiation of a range of adult stem cells [64,65] and enhances the derivation of ESC lines from mouse blastocysts [66]. Clearly there are benefits to culturing stem cells under physiological O2, but the molecular mechanisms favoring pluripotency under these conditions are unknown.
The experiments described in this paper were undertaken to address four main questions relating to the effects of O2 on hESC pluripotency. The first question we asked was whether there are consistent differences in gene expression associated with culture under 20% versus 4% O2. As the appearance of differentiated cell populations in hESC cultures under 20% O2 does not occur until about day 10 if the cells were cultured under physiological O2 in previous passage, we expected that any changes in gene expression between cells cultured under the two conditions at 7 days post passage would likely be subtle and not of major magnitude. This hypothesis was born out. Only 149 genes consistently exhibited altered expression at high versus low O2 conditions, and the changes observed were generally quite small. Importantly, several genes invariably associated with a pluripotent phenotype, including OCT4, NANOG, SOX2, did not display changes in expression. Such an outcome was not unexpected, as these stem cell lines have been maintained under atmospheric O2 for at least 10 years without loss of pluripotency, emphasizing the robustness of the transcriptional network that maintains stemness even under conditions that are nonphysiological. OCT4 concentrations, in particular, must be tightly controlled, as partial down-regulation of its expression can lead to the emergence of trophectoderm and its up-regulation to endoderm [67,68]. In addition, OCT4, NANOG, and SOX2 appear to regulate each other and to function together to repress genes involved in lineage specification [33,34].
The second question we addressed was whether transcriptional profiles were more consistent in cells under 4% than 20% O2. As expected, they were. Whereas profiles observed across cell lines and at different passage number were remarkably alike under low O2, the greater heterogeneity under 20% O2 suggested that cryptic differentiation might have already begun despite the lack of morphological changes in the colonies, thereby allowing us to address our third question relating to whether signs of lineage specification were evident in day 7 cultures under 20% O2. Indeed, some genes regulated by the OCT4, NANOG, and SOX2 triad of transcription factors showed significantly reduced expression at the higher O2 concentration (Table 2), while other genes, including ones associated with lineage-specific differentiation, for example, HAND1, EOMES, MEIS2, GATA3 & −6, MSX2, TFAP2A [69], and H19 [27] exhibited increased expression in at least one of the samples cultured under 20% O2 (Supplemental Table 4).As the latter cells were indistinguishable from cells grown under 4% O2 and harvested prior to any visible signs of differentiation (Fig. 1B), our results suggest that morphological cues are poor guides when selecting hESCs for passage. Thus, culture under 20% O2 is likely to provide a mixed group of cells, including subpopulations already programmed to differentiate.
Perhaps the most interesting regulated gene was LEFTY2, which displayed a 2- to 3-fold higher expression under 4% as compared to 20% O2. LEFTY2, the human ortholog of murine Lefty A, is regulated by SOX2 and OCT4, and is able to prevent differentiation initiated by Nodal signaling by blocking the formation of an active Nodal/Activin receptor complex [70–72]. Conceivably, LEFTY2 is a key gene in preventing the spontaneous differentiation of hESCs along the mesoderm and endoderm lineages. Its down-regulation as O2 concentrations rise may render the pluripotent gene network less stable and more responsive to signals that drive differentiation, a hypothesis that can be readily tested by knockdown of LEFTY2 expression.
The final question we attempted to address was whether there is involvement of the HIF transcription factors in maintaining the pluripotency of hESCs, particularly when the cells are cultured under physiological O2. Mammalian cells rely on HIFs to ensure accommodation to low O2 levels, primarily through the action of HIF1A and HIF2A and their common binding partner, ARNT [12,73,74] and an extensive list of HIF target genes have been identified. The higher expression of genes encoding glycolytic enzymes and other known HIF targets such as BNIP3, DDIT4, IGFBP2, and VEGF in hESCs cultured under 4% O2 (Table 5)is entirely consistent with the view that the HIFAs are most active under low O2 environments where they have a major role in maintaining cellular homeostasis. The gene list in Table 5 provides few clues as to any potential involvement in controlling pluripotency. While some workers have argued that HIF1A and HIF2A display redundancy regarding HIF targets [75], others have indicated differences in gene preference [73,76], especially under acute hypoxia [77]. Although no exclusive HIF2A target gene has been identified [78], HIF2A may regulate OCT4 expression specifically [21] and has been shown to activates SERPINE1 (PAI-1) [79]. Such a link to OCT4 expression would place HIF2A as a central player in the network of genes maintaining the pluripotent phenotype. Our observation that HIF2A was confined to the undifferentiated cells of day 10 colonies cultured under 20% O2, while HIF1A was located throughout the colonies, including the overtly differentiated areas (Fig. 2), is consistent with this notion, as is the report that HIF2A is localized to the pluripotent ICM of bovine blastocysts, while HIF1A is absent from these cells but present in trophectoderm [13].
In conclusion, this report emphasizes the importance of employing physiological concentrations of O2 when culturing hESCs. The transcript profiles of cultures under 20 % O2 suggest that the cells are more poised to differentiate than when they are under the lower 4% O2 conditions and that the down-regulation of LEFTY2 under 20% O2 may destabilize the network of genes maintaining ESC pluripotency. Finally, the association of HIF2A with undifferentiated but not differentiating cells is consistent with a particular role for that transcription factor in control of pluripotency.
Supplementary Material
Acknowledgments
We gratefully acknowledge Christine Schramm and Dr. Richard Tsika for providing insightful suggestions and assistance with the real time PCR and Norma McCormack for preparing the manuscript and figures for submission. This work was supported by NIH grant 1R01 HD042201 (R.M.R).
Disclaimers: The authors indicate no potential conflicts of interest.
References
- 1.Odorico JS. Kaufman DS. Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 2.Stojkovic M. Lako M. Strachan T. Murdoch A. Derivation, growth and applications of human embryonic stem cells. Reproduction. 2004;128:259–267. doi: 10.1530/rep.1.00243. [DOI] [PubMed] [Google Scholar]
- 3.Menendez P. Bueno C. Wang L. Human embryonic stem cells: a journey beyond cell replacement therapies. Cytotherapy. 2006;8:530–541. doi: 10.1080/14653240601026654. [DOI] [PubMed] [Google Scholar]
- 4.Jones JM. Thomson JA. Human embryonic stem cell technology. Semin Reprod Med. 2000;18:219–223. doi: 10.1055/s-2000-12560. [DOI] [PubMed] [Google Scholar]
- 5.Amit M. Itskovitz-Eldor J. Derivation and spontaneous differentiation of human embryonic stem cells. J Anat. 2002;200:225–232. doi: 10.1046/j.1469-7580.2002.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 7.Fischer B. Bavister BD. Oxygen tension in the oviduct and uterus of rhesus monkeys, hamsters and rabbits. J Reprod Fertil. 1993;99:673–679. doi: 10.1530/jrf.0.0990673. [DOI] [PubMed] [Google Scholar]
- 8.Ottosen LD. Hindkaer J. Husth M. Petersen DE. Kirk J. Ingerslev HJ. Observations on intrauterine oxygen tension measured by fibre-optic microsensors. Reprod Biomed Online. 2006;13:380–385. doi: 10.1016/s1472-6483(10)61443-5. [DOI] [PubMed] [Google Scholar]
- 9.Ezashi T. Das P. Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci USA. 2005;102:4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wenger RH. Gassmann M. Oxygen(es) and the hypoxia-inducible factor-1. Biol Chem. 1997;378:609–616. [PubMed] [Google Scholar]
- 11.Greijer AE. van der Groep P. Kemming D. Shvarts A. Semenza GL. Meijer GA. van de Wiel MA. Belien JA. van Diest PJ. van der Wall E. Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1) J Pathol. 2005;206:291–304. doi: 10.1002/path.1778. [DOI] [PubMed] [Google Scholar]
- 12.Semenza GL. Roth PH. Fang HM. Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- 13.Harvey AJ. Kind KL. Pantaleon M. Armstrong DT. Thompson JG. Oxygen-regulated gene expression in bovine blastocysts. Biol Reprod. 2004;71:1108–1119. doi: 10.1095/biolreprod.104.028639. [DOI] [PubMed] [Google Scholar]
- 14.Iyer NV. Kotch LE. Agani F. Leung SW. Laughner E. Wenger RH. Gassmann M. Gearhart JD. Lawler AM. Yu AY. Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keith B. Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giles JR. Foote RH. Effects of gas atmosphere, platelet-derived growth factor and leukemia inhibitory factor on cell numbers of rabbit embryos cultured in a protein-free medium. Reprod Nutr Dev. 1997;37:97–104. doi: 10.1051/rnd:19970110. [DOI] [PubMed] [Google Scholar]
- 17.Petersen A. Mikkelsen AL. Lindenberg S. The impact of oxygen tension on developmental competence of post-thaw human embryos. Acta Obstet Gynecol Scand. 2005;84:1181–1184. doi: 10.1111/j.0001-6349.2005.00630.x. [DOI] [PubMed] [Google Scholar]
- 18.Thompson JG. Simpson AC. Pugh PA. Donnelly PE. Tervit HR. Effect of oxygen concentration on in-vitro development of preimplantation sheep and cattle embryos. J Reprod Fertil. 1990;89:573–578. doi: 10.1530/jrf.0.0890573. [DOI] [PubMed] [Google Scholar]
- 19.Booth PJ. Holm P. Callesen H. The effect of oxygen tension on porcine embryonic development is dependent on embryo type. Theriogenology. 2005;63:2040–2052. doi: 10.1016/j.theriogenology.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Rinaudo PF. Giritharan G. Talbi S. Dobson AT. Schultz RM. Effects of oxygen tension on gene expression in preimplantation mouse embryos. Fertil Steril 86 Suppl. 2006;4:1252–1265. doi: 10.1016/j.fertnstert.2006.05.017. e1251–1236. [DOI] [PubMed] [Google Scholar]
- 21.Covello KL. Kehler J. Yu H. Gordan JD. Arsham AM. Hu CJ. Labosky PA. Simon MC. Keith B. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu CJ. Iyer S. Sataur A. Covello KL. Chodosh LA. Simon MC. Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Mol Cell Biol. 2006;26:3514–3526. doi: 10.1128/MCB.26.9.3514-3526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan GJ. Chang ZY. Scholer HR. Pei D. Stem cell pluripotency and transcription factor Oct4. Cell Res. 2002;12:321–329. doi: 10.1038/sj.cr.7290134. [DOI] [PubMed] [Google Scholar]
- 24.Xu C. Inokuma MS. Denham J. Golds K. Kundu P. Gold JD. Carpenter MK. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 25.Harbig J. Sprinkle R. Enkemann SA. A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array. Nucleic Acids Res. 2005;33:e31. doi: 10.1093/nar/gni027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharya B. Cai J. Luo Y. Miura T. Mejido J. Brimble SN. Zeng X. Schulz TC. Rao MS. Puri RK. Comparison of the gene expression profile of undifferentiated human embryonic stem cell lines and differentiating embryoid bodies. BMC Dev Biol. 2005;5:22. doi: 10.1186/1471-213X-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai J. Chen J. Liu Y. Miura T. Luo Y. Loring JF. Freed WJ. Rao MS. Zeng X. Assessing self-renewal and differentiation in human embryonic stem cell lines. Stem Cells. 2006;24:516–530. doi: 10.1634/stemcells.2005-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim CG. Lee JJ. Jung DY. Jeon J. Heo HS. Kang HC. Shin JH. Cho YS. Cha KJ. Kim CG. Do BR. Kim KS. Kim HS. Profiling of differentially expressed genes in human stem cells by cDNA microarray. Mol Cells. 2006;21:343–355. [PubMed] [Google Scholar]
- 29.Rao RR. Calhoun JD. Qin X. Rekaya R. Clark JK. Stice SL. Comparative transcriptional profiling of two human embryonic stem cell lines. Biotechnol Bioeng. 2004;88:273–286. doi: 10.1002/bit.20245. [DOI] [PubMed] [Google Scholar]
- 30.Rao RR. Stice SL. Gene expression profiling of embryonic stem cells leads to greater understanding of pluripotency and early developmental events. Biol Reprod. 2004;71:1772–1778. doi: 10.1095/biolreprod.104.030395. [DOI] [PubMed] [Google Scholar]
- 31.Zeng X. Miura T. Luo Y. Bhattacharya B. Condie B. Chen J. Ginis I. Lyons I. Mejido J. Puri RK. Rao MS. Freed WJ. Properties of pluripotent human embryonic stem cells BG01 and BG02. Stem Cells. 2004;22:292–312. doi: 10.1634/stemcells.22-3-292. [DOI] [PubMed] [Google Scholar]
- 32.Brimble SN. Zeng X. Weiler DA. Luo Y. Liu Y. Lyons IG. Freed WJ. Robins AJ. Rao MS. Schulz TC. Karyotypic stability, genotyping, differentiation, feeder-free maintenance, and gene expression sampling in three human embryonic stem cell lines derived prior to August 9, 2001. Stem Cells Dev. 2004;13:585–597. doi: 10.1089/scd.2004.13.585. [DOI] [PubMed] [Google Scholar]
- 33.Boyer LA. Lee TI. Cole MF. Johnstone SE. Levine SS. Zucker JP. Guenther MG. Kumar RM. Murray HL. Jenner RG. Gifford DK. Melton DA. Jaenisch R. Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loh YH. Wu Q. Chew JL. Vega VB. Zhang W. Chen X. Bourque G. George J. Leong B. Liu J. Wong KY. Sung KW. Lee CW. Zhao XD. Chiu KP. Lipovich L. Kuznetsov VA. Robson P. Stanton LW. Wei CL. Ruan Y. Lim B. Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 35.Wang J. Rao S. Chu J. Shen X. Levasseur DN. Theunissen TW. Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 36.Nichols J. Zevnik B. Anastassiadis K. Niwa H. Klewe-Nebenius D. Chambers I. Scholer H. Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 37.Mitsui K. Tokuzawa Y. Itoh H. Segawa K. Murakami M. Takahashi K. Maruyama M. Maeda M. Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 38.Richards M. Tan SP. Tan JH. Chan WK. Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- 39.Avilion AA. Nicolis SK. Pevny LH. Perez L. Vivian N. Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben-Shushan E. Thompson JR. Gudas LJ. Bergman Y. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol Cell Biol. 1998;18:1866–1878. doi: 10.1128/mcb.18.4.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armstrong L. Saretzki G. Peters H. Wappler I. Evans J. Hole N. von Zglinicki T. Lako M. Overexpression of telomerase confers growth advantage, stress resistance, and enhanced differentiation of ESCs toward the hematopoietic lineage. Stem Cells. 2005;23:516–529. doi: 10.1634/stemcells.2004-0269. [DOI] [PubMed] [Google Scholar]
- 42.Boyer LA. Mathur D. Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Babaie Y. Herwig R. Greber B. Brink TC. Wruck W. Groth D. Lehrach H. Burdon T. Adjaye J. Analysis of OCT4 dependent transcriptional networks regulating self renewal and pluripotency in human embryonic stem cells. Stem Cells. 2007;25:500–510. doi: 10.1634/stemcells.2006-0426. [DOI] [PubMed] [Google Scholar]
- 44.Besser D. Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. J Biol Chem. 2004;279:45076–45084. doi: 10.1074/jbc.M404979200. [DOI] [PubMed] [Google Scholar]
- 45.James D. Levine AJ. Besser D. Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 46.Vallier L. Alexander M. Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- 47.Kutz SM. Higgins CE. Samarakoon R. Higgins SP. Allen RR. Qi L. Higgins PJ. TGF-beta 1-induced PAI-1 expression is E box/USF-dependent and requires EGFR signaling. Exp Cell Res. 2006;312:1093–1105. doi: 10.1016/j.yexcr.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 48.Buck A. Buchholz M. Wagner M. Adler G. Gress T. Ellenrieder V. The tumor suppressor KLF11 mediates a novel mechanism in transforming growth factor beta-induced growth inhibition that is inactivated in pancreatic cancer. Mol Cancer Res. 2006;4:861–872. doi: 10.1158/1541-7786.MCR-06-0081. [DOI] [PubMed] [Google Scholar]
- 49.Calhoun JD. Rao RR. Warrenfeltz S. Rekaya R. Dalton S. McDonald J. Stice SL. Transcriptional profiling of initial differentiation events in human embryonic stem cells. Biochem Biophys Res Commun. 2004;323:453–464. doi: 10.1016/j.bbrc.2004.08.117. [DOI] [PubMed] [Google Scholar]
- 50.Dvash T. Mayshar Y. Darr H. McElhaney M. Barker D. Yanuka O. Kotkow KJ. Rubin LL. Benvenisty N. Eiges R. Temporal gene expression during differentiation of human embryonic stem cells and embryoid bodies. Hum Reprod. 2004;19:2875–2883. doi: 10.1093/humrep/deh529. [DOI] [PubMed] [Google Scholar]
- 51.Aghajanova L. Skottman H. Stromberg AM. Inzunza J. Lahesmaa R. Hovatta O. Expression of leukemia inhibitory factor and its receptors is increased during differentiation of human embryonic stem cells. Fertil Steril. 2006;86(Suppl 4):1193–1209. doi: 10.1016/j.fertnstert.2005.12.081. [DOI] [PubMed] [Google Scholar]
- 52.Mathers J. Fraser JA. McMahon M. Saunders RD. Hayes JD. McLellan LI. Antioxidant and cytoprotective responses to redox stress. Biochem Soc Symp. 2004:157–176. doi: 10.1042/bss0710157. [DOI] [PubMed] [Google Scholar]
- 53.Chen XL. Kunsch C. Induction of cytoprotective genes through Nrf2/antioxidant response element pathway: a new therapeutic approach for the treatment of inflammatory diseases. Curr Pharm Des. 2004;10:879–891. doi: 10.2174/1381612043452901. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi M. Yamamoto M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid Redox Signal. 2005;7:385–394. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- 55.Wu Y. Wu J. Lee DY. Yee A. Cao L. Zhang Y. Kiani C. Yang BB. Versican protects cells from oxidative stress-induced apoptosis. Matrix Biol. 2005;24:3–13. doi: 10.1016/j.matbio.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Winchester BG. Lysosomal membrane proteins. Eur J Paediatr Neurol. 2001;5(Suppl A):11–19. doi: 10.1053/ejpn.2000.0428. [DOI] [PubMed] [Google Scholar]
- 57.Levtchenko E. de Graaf-Hess A. Wilmer M. van den Heuvel L. Monnens L. Blom H. Altered status of glutathione and its metabolites in cystinotic cells. Nephrol Dial Transplant. 2005;20:1828–1832. doi: 10.1093/ndt/gfh932. [DOI] [PubMed] [Google Scholar]
- 58.Manalo DJ. Rowan A. Lavoie T. Natarajan L. Kelly BD. Ye SQ. Garcia JG. Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 59.Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci USA. 2000;97:9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kothari S. Cizeau J. McMillan-Ward E. Israels SJ. Bailes M. Ens K. Kirshenbaum LA. Gibson SB. BNIP3 plays a role in hypoxic cell death in human epithelial cells that is inhibited by growth factors EGF and IGF. Oncogene. 2003;22:4734–4744. doi: 10.1038/sj.onc.1206666. [DOI] [PubMed] [Google Scholar]
- 61.Shoshani T. Faerman A. Mett I. Zelin E. Tenne T. Gorodin S. Moshel Y. Elbaz S. Budanov A. Chajut A. Kalinski H. Kamer I. Rozen A. Mor O. Keshet E. Leshkowitz D. Einat P. Skaliter R. Feinstein E. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol. 2002;22:2283–2293. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graven KK. Troxler RF. Kornfeld H. Panchenko MV. Farber HW. Regulation of endothelial cell glyceraldehyde-3-phosphate dehydrogenase expression by hypoxia. J Biol Chem. 1994;269:24446–24453. [PubMed] [Google Scholar]
- 63.Forsyth NR. Musio A. Vezzoni P. Simpson AH. Noble BS. McWhir J. Physiologic oxygen enhances human embryonic stem cell clonal recovery and reduces chromosomal abnormalities. Cloning Stem Cells. 2006;8:16–23. doi: 10.1089/clo.2006.8.16. [DOI] [PubMed] [Google Scholar]
- 64.Fehrer C. Brunauer R. Laschober G. Unterluggauer H. Reitinger S. Kloss F. Gully C. Gassner R. Lepperdinger G. Reduced oxygen tension attenuates differentiation capacity of human mesenchymal stem cells and prolongs their lifespan. Aging Cell. 2007;6:745–757. doi: 10.1111/j.1474-9726.2007.00336.x. [DOI] [PubMed] [Google Scholar]
- 65.D'Ippolito G. Diabira S. Howard GA. Roos BA. Schiller PC. Low oxygen tension inhibits osteogenic differentiation and enhances stemness of human MIAMI cells. Bone. 2006;39:513–522. doi: 10.1016/j.bone.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 66.Wang F. Thirumangalathu S. Loeken MR. Establishment of new mouse embryonic stem cell lines is improved by physiological glucose and oxygen. Cloning Stem Cells. 2006;8:108–116. doi: 10.1089/clo.2006.8.108. [DOI] [PubMed] [Google Scholar]
- 67.Niwa H. Miyazaki J. Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 68.Shimozaki K. Nakashima K. Niwa H. Taga T. Involvement of Oct3/4 in the enhancement of neuronal differentiation of ES cells in neurogenesis-inducing cultures. Development. 2003;130:2505–2512. doi: 10.1242/dev.00476. [DOI] [PubMed] [Google Scholar]
- 69.Hay DC. Sutherland L. Clark J. Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004;22:225–235. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- 70.Schier AF. Shen MM. Nodal signalling in vertebrate development. Nature. 2000;403:385–389. doi: 10.1038/35000126. [DOI] [PubMed] [Google Scholar]
- 71.Sakuma R. Ohnishi Yi Y. Meno C. Fujii H. Juan H. Takeuchi J. Ogura T. Li E. Miyazono K. Hamada H. Inhibition of Nodal signalling by Lefty mediated through interaction with common receptors and efficient diffusion. Genes Cells. 2002;7:401–412. doi: 10.1046/j.1365-2443.2002.00528.x. [DOI] [PubMed] [Google Scholar]
- 72.Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- 73.Hu CJ. Wang LY. Chodosh LA. Keith B. Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 75.Sowter HM. Raval RR. Moore JW. Ratcliffe PJ. Harris AL. Predominant role of hypoxia-inducible transcription factor (Hif)-1alpha versus Hif-2alpha in regulation of the transcriptional response to hypoxia. Cancer Res. 2003;63:6130–6134. [PubMed] [Google Scholar]
- 76.Wang V. Davis DA. Haque M. Huang LE. Yarchoan R. Differential gene up-regulation by hypoxia-inducible factor-1α and hypoxia-inducible factor-2α in HEK293T Cells. Cancer Res. 2005;65:3299–3306. doi: 10.1158/0008-5472.CAN-04-4130. [DOI] [PubMed] [Google Scholar]
- 77.Holmquist-Mengelbier L. Fredlund E. Lofstedt T. Noguera R. Navarro S. Nilsson H. Pietras A. Vallon-Christersson J. Borg A. Gradin K. Poellinger L. Pahlman S. Recruitment of HIF-1[alpha] and HIF-2[alpha] to common target genes is differentially regulated in neuroblastoma: HIF-2[alpha] promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 78.Keith B. Simon MC. Hypoxia-Inducible Factors, Stem Cells, and Cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sato M. Tanaka T. Maemura K. Uchiyama T. Sato H. Maeno T. Suga T. Iso T. Ohyama Y. Arai M. Tamura J. Sakamoto H. Nagai R. Kurabayashi M. The PAI-1 gene as a direct target of endothelial PAS domain protein-1 in adenocarcinoma A549 cells. Am J Respir Cell Mol Biol. 2004;31:209–215. doi: 10.1165/rcmb.2003-0296OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.