Abstract
Metazoan genomes encode an abundant collection of mRNA-like, long noncoding (lnc)RNAs. Although lncRNAs greatly expand the transcriptional repertoire, we have a limited understanding of how these RNAs contribute to developmental regulation. Here, we investigate the function of the Drosophila lncRNA called yellow-achaete intergenic RNA (yar). Comparative sequence analyses show that the yar gene is conserved in Drosophila species representing 40–60 million years of evolution, with one of the conserved sequence motifs encompassing the yar promoter. Further, the timing of yar expression in Drosophila virilis parallels that in D. melanogaster, suggesting that transcriptional regulation of yar is conserved. The function of yar was defined by generating null alleles. Flies lacking yar RNAs are viable and show no overt morphological defects, consistent with maintained transcriptional regulation of the adjacent yellow (y) and achaete (ac) genes. The location of yar within a neural gene cluster led to the investigation of effects of yar in behavioral assays. These studies demonstrated that loss of yar alters sleep regulation in the context of a normal circadian rhythm. Nighttime sleep was reduced and fragmented, with yar mutants displaying diminished sleep rebound following sleep deprivation. Importantly, these defects were rescued by a yar transgene. These data provide the first example of a lncRNA gene involved in Drosophila sleep regulation. We find that yar is a cytoplasmic lncRNA, suggesting that yar may regulate sleep by affecting stabilization or translational regulation of mRNAs. Such functions of lncRNAs may extend to vertebrates, as lncRNAs are abundant in neural tissues.
METAZOAN genomes encode an abundant collection of noncoding (nc) RNAs. These include housekeeping ncRNAs, such as transfer RNAs and ribosomal RNAs, and a growing number of regulatory ncRNAs. Regulatory ncRNAs have been categorized into two subclasses, on the basis of length (Prasanth and Spector 2007; Mercer et al. 2009). RNAs <200 nucleotides encompass the small ncRNAs class, which includes endogenous small interfering (endo si) RNAs, micro (mi) RNAs and piwi-interacting (pi) RNAs. RNAs >200 nucleotides encompass the long ncRNA (lncRNA) class. Many lncRNAs share properties with mRNAs, being transcribed by RNA polymerase II and processed by the splicing and polyadenylation machinery. Emerging evidence indicates that regulatory RNAs make multiple contributions to cellular functions (Mercer et al. 2009; Chen and Carmichael 2010; Taft et al. 2010; Clark and Mattick 2011). Small ncRNAs function primarily in the cytoplasm, working as guides for the recognition of regulated target RNAs by associated protein complexes. LncRNAs localize both to the nucleus and cytoplasm. Nuclear lncRNAs have many regulatory roles, including organization of nuclear architecture and control of transcription, splicing, and nuclear trafficking (Mercer et al. 2009; Chen and Carmichael 2010; Taft et al. 2010; Clark and Mattick 2011). Recently, cytoplasmic roles for lncRNAs have been uncovered, including regulation of mRNA decay and miRNA function (Panzitt et al. 2007; Matouk et al. 2009; Wang et al. 2010; Clark and Mattick 2011). These observations demonstrate that regulatory RNAs expand the functional repertoire of the transcriptome in developing organisms.
The Drosophila melanogaster genome has been estimated to encode >100 lncRNAs (Tupy et al. 2005; Willingham et al. 2006; Graveley et al. 2011). Many of these lncRNA genes are transcribed during embryogenesis and display spatially restricted expression, with predominant RNA accumulation in the developing central and peripheral nervous system (Inagaki et al. 2005; Li et al. 2009). While many Drosophila lncRNAs have been identified, mutations in only a small number of these genes are known and are limited to genes encoding nuclear lncRNAs. Two lncRNA genes that have been studied genetically encode the nuclear retained roX1 and roX2 RNAs, essential RNAs involved in dosage compensation (Meller and Rattner 2002; Deng and Meller 2006). Although the roX RNAs display limited sequence identity, these RNAs share a role in assembly and targeting of the dosage compensation complex to the male X chromosome (Ilik and Akhtar 2009; Koya and Meller 2011). A third genetically studied Drosophila lncRNA gene is hsr-ω gene, which encodes the heat inducible hsr-ω-n transcript (Jolly and Lakhotia 2006). This essential gene encodes a large, nuclear retained lncRNA, which forms nucleoplasmic omega speckles that accumulate heterogeneous nuclear RNA binding proteins (hnRNPs) (Prasanth et al. 2000). Recent evidence suggests that hsr-ω-n functions as a hub for coordination of transcriptional regulators and hnRNPs, impacting cellular responses such as apoptosis (Mallik and Lakhotia 2010). While our understanding of the in vivo functions of lncRNAs remains limited, the essential roles of these three nuclear-retained lncRNAs suggest that lncRNAs make multiple contributions to development and cell differentiation.
The Drosophila yellow-achaete (ac) intergenic RNA (yar) is a newly identified lncRNA gene. This gene encodes multiple alternatively spliced poly(A)+ RNAs that are highly expressed during midembryogenesis. As yar RNAs lack a predicted translation product >75 amino acids, yar has been classified as a lncRNA gene. Within the Drosophila genome, yar resides within a neural gene cluster (Soshnev et al. 2008). Upstream of yar is yellow (y), a gene that encodes a secreted protein required for cuticle coloration and male sexual behavior (Nash and Yarkin 1974; Biessmann 1985; Chia et al. 1986; Geyer et al. 1986; Geyer and Corces 1987; Drapeau et al. 2003). Downstream of yar is achaete (ac), a gene that encodes one of four related bHLH transcription factors of the achaete–scute complex (AS-C) required for proper development of the central and peripheral nervous systems (Modolell and Campuzano 1998; Gibert and Simpson 2003; Negre and Simpson 2009). The order and transcriptional orientation of genes in the AS-C complex is remarkably conserved among insect species, and this organization extends to the y gene in most species (Negre and Simpson 2009). This linkage cannot be explained by shared enhancers, as y, yar, and ac show distinct temporal patterns of embryonic gene expression (Campuzano et al. 1985; Chia et al. 1986; Soshnev et al. 2008). Interestingly, transcription of yar coincides with down-regulation of the ac gene, while transcription of y coincides with down-regulation of yar (Soshnev et al. 2008). These observations suggest that temporal regulation of y, yar, and ac might be linked, a possibility supported by previously identified regulatory contributions of other ncRNA genes (Ogawa and Lee 2002; Martens et al. 2004; Petruk et al. 2006; Martianov et al. 2007).
Here, we use genomic and genetic approaches to define the role of yar in the y–yar–ac region. Genomic analyses revealed the presence of large blocks of sequence identity within yar that have been conserved over 40–60 million years of evolution. This conservation does not extend to the putative open reading frames within yar RNAs, supporting that yar is a lncRNA gene. Interestingly, the second largest block of sequence identity encompasses the three yar promoters (Soshnev et al. 2008). This conservation is reflected in the parallel temporal pattern of embryonic yar expression in the distantly related D. melanogaster and D. virilis species. We show that the D. melanogaster yar gene is globally expressed during midembryogenesis, with yar RNA accumulating in the cytoplasm. Using homologous recombination, two null alleles were generated. Flies lacking yar RNAs are viable and appropriately regulate y and ac transcription, but show defects in sleep. We uncovered that yar mutants exhibit shortened sleep bouts within a normal circadian sleep–wake cycle and have diminished levels of sleep rebound following deprivation. Importantly, both phenotypes are rescued by a transgene encompassing the yar gene, demonstrating that yar is required for sleep regulation. As yar is a cytoplasmic RNA, its regulatory effects are likely to depend upon stabilization or translational regulation of target RNAs. Our findings represent the first example of a lncRNA gene involved in Drosophila sleep behavior.
Materials and Methods
Fly stocks and crosses
Flies were raised at 25°, 70% humidity on standard cornmeal/agar medium. Description of the alleles used can be found at www.flybase.org.
Analyses of the y–ac intergenic region
Genomic sequences of the y–ac intergenic region from eight species of Drosophila were compared with D. melanogaster, including species in the subgenus Sophophora estimated to represent 10 million years (MY) of evolution (D. yakuba and D. erecta), and 20–30 MY (D. ananassae, D. pseudoobscura, and D. willistoni), and species in the subgenus Drosophila estimated to represent 40–60 MY of evolution (D. virilis, D. mojavensis, and D. grimshawi) (Stark et al. 2007). Sequences were obtained through FlyBase using Release 4 (www.flybase.org). Sequence alignments based on percentage of identity with nucleotide-level alignments were generated with MultiPipMaker (Schwartz et al. 2000). In all species except D. mojavensis, the y and ac genes are oriented the same as D. melanogaster, so sequence alignments were obtained with the corresponding intergenic interval. In D. mojavensis, y is not adjacent to ac. In this case, the “intergenic” regions were defined as an ∼12-kb fragment either upstream of ac or downstream of y. Alignments in the genome of D. mojavensis revealed sequence conservation in the upstream region of ac that includes yar. For this reason, all reported analyses only include the intergenic interval upstream of ac.
Nucleotide alignments revealed overall sequence conservation among nine Drosophila species and provided a guide for subdividing the intergenic region of each species into two approximately equal segments. Sequence motifs within each segment that were conserved across the nine species were identified using MEME (Figure 1) (Bailey et al. 2006). High-scoring sequence motifs were identified as regions of homology shared among the maximum set of species (a large gap is present in the genome sequence of segment 1 for D. grimshawi). These regions are likely to be constrained due to an evolutionarily conserved function. Each was given a unique identifier x.y, with x representing segment 1 or 2 and y representing the ordinal score in each MEME analysis. Each conserved MEME motif was mapped onto the sequence alignment of the D. melanogaster Release 5 (Figure 1, supporting information, Figure S1).
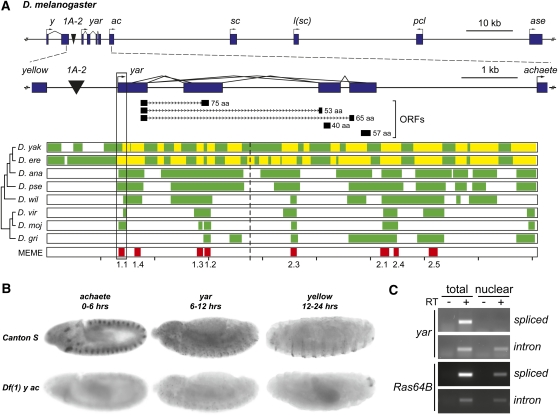
Figure 1.
Conservation and tissue localization of yar RNA in D. melanogaster. (A) Multiple DNA motifs are conserved in the y–ac intergenic region. (A, top) Structure of the D. melanogaster genomic region that includes y, yar, the four AS-C genes, and pepsinogen-like (pcl). Genes are shown as rectangles, with promoters shown as bent arrows. The 1A-2 regulatory element is indicated by black inverted triangle. (A, middle) An expanded view of the 9-kb region separating y and ac, including yar, with a representation of its splicing pattern. The tracks located under the yar gene structure indicate the location of the potential yar ORFs, with the size of the peptide noted. (A, bottom) Aligned with the 9-kb region are the percentage of identity plots obtained from MultiPipMaker analyses of the corresponding regions from nine drosophilid species. Regions of no alignment are indicated in white, regions with significant BLASTZ alignment to D. melanogaster are indicated in green, and regions of nongapped alignments of >100 nucleotides with >70% identity are indicated in yellow. Conserved motifs identified by MEME are indicated on the bottom track. Motif 1.1 identifies the yar promoter. A gap in the genome sequence of D. grimshawi spans the region corresponding to motifs 1.1, 1.4, and 1.3. The dashed line indicates where the intergenic regions were split in two fragments for MEME analyses. (B) Whole mount RNA in situ hybridization of aged D. melanogaster embryos. ac mRNA is detected in the neuroectoderm clusters in the early embryogenesis, yar is globally expressed in midembryogenesis, and y is expressed in late embryogenesis in denticle belts. Df(1) y ac embryos serve as a negative control. (C) Analyses of cellular localization of yar transcripts. Total RNA isolated from equal amounts of unfractionated embryos and nuclear fraction was reverse transcribed and analyzed by semiquantitative PCR. The (−) RT lanes control for genomic DNA contamination. Spliced products were detected with primer pairs flanking the intron; intronic sequences were detected with primer pairs located within the intron. The housekeeping gene Ras64B serves as a positive control.
Analyses of y–ac intergenic transcription in D. virilis
Embryogenesis in D. virilis is prolonged relative to D. melanogaster, lasting 32 vs. 22 hr, respectively, after egg laying (Markow et al. 2009). RNA was isolated from aged D. virilis embryos as described previously (Parnell et al. 2006). This RNA was converted into cDNA using the HighCapacity cDNA kit (Applied Biosystems) with either random hexamers or oligo-dT primers for first strand synthesis. Primer pairs for PCR amplification of this cDNA were anchored within the MEME-identified conserved motifs or within the Genscan-predicted exons of the intergenic transcript (Burge and Karlin 1997) (Figure 2, Figure S2, Table S1). As a control, all primer pairs were tested for amplification of D. virilis genomic DNA. In all cases, genomic DNA fragments of the appropriate size were obtained. Three independent RNA isolations were analyzed by PCR.
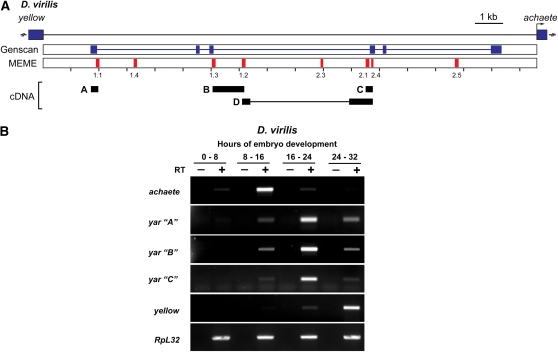
Figure 2.
yar is conserved in D. virilis. (A) Structure of the D. virilis 17-kb y–ac intergenic region. The positions of the y and ac genes are shown by rectangles. The Genscan-predicted gene (blue) and MEME-identified motifs (red) are indicated below. Four cDNAs (A–D) obtained from PCR analyses are shown; the thin line in cDNA D indicates an intron. (B) Semiquantitative PCR analyses of RNAs isolated during the indicated times of D. virilis embryogenesis. Primer pairs corresponding to ac, three of the yar cDNAs, and y were analyzed. RpL32 represents a constitutively expressed RNA and serves as a control. The (−) RT lanes control for genomic DNA contamination. RNAs were isolated from aged embryos, as indicated by hours of development.
Whole mount in situ hybridization
Whole mount in situ hybridization on Drosophila embryos was performed as described previously (Tautz and Pfeifle 1989), with the overnight hybridization at 48°. Digoxigenin-labeled probes were generated from cDNAs encompassing the second exon of y (EcoRI–BglII fragment), first, third, and fourth exon of yar (GenBank accession no. GQ329854), and the complete ac transcription unit. Images were collected using an Olympus BX-51 bright field microscope and processed with ImageJ and Adobe Photoshop. The homozygous deficiency line Df(1) y ac was used as a negative control, as this line carries a deletion of the genomic region encompassing the y, yar, and ac genes. Three biological replicates were performed.
Nuclear RNA isolation and analyses
Aged 6- to 12-hr embryos were collected on orange juice/agar plates, dechorionated with 50% hypochlorite solution, and frozen in liquid nitrogen. Frozen embryos were ground on dry ice, resuspended in buffer A (60 mM KCl, 15 mM NaCl, 15 mM Tris-HCl pH 7.4, 1 mM EDTA, 0.1 mM EGTA, 0.15 mM spermine, 0.45 mM spermidine, 0.45 mM DTT), and filtered through Miracloth (Calbiochem). The filtered material was split into two equivalent volumes (“total” and “nuclear”). The nuclear fraction was processed using nuclei isolation protocol as described previously (Parnell et al. 2003). RNA was isolated from both nuclear and total volumes using TRIzol extraction. Equal volumes were DNase I treated and reverse transcribed using High Capacity cDNA kit with random hexamer primers and analyzed by semiquantitative PCR at 25 cycles for spliced transcripts and 30 cycles for intronic sequences. Amplified fragments were resolved on 1% agarose gel with ethidium bromide.
Northern analyses
Northern analyses were performed as described previously (Soshnev et al. 2008). Briefly, embryos were collected from D. melanogaster on orange juice and aged for 6–12 hr in a 25° incubator, while embryos were collected from D. virilis on grape juice plates and aged for 16–24 hr in a 20° incubator. RNA was isolated using TRIzol. Poly(A)+ RNA was selected using Qiagen mRNA Midi kit, according to the manufacturer’s instructions. Three to 10 μg of poly(A)+ selected RNA were resolved on a formaldehyde–agarose gel, transferred to Nytran N membrane (Whatman), and hybridized to 32P-dATP labeled DNA probes generated from D. melanogaster yar clone (GenBank accession no. GQ329854) or D. virilis yar cDNA. Membranes were exposed to X-ray film, stripped, and probed with a 32P-dATP labeled DNA from the constitutively expressed RpL32 gene as a loading control.
Enhancer-blocking assays
Insulator activity of the region encompassing the yar promoter was tested using two independent enhancer-blocking reporter P-element transposons. For these studies, a 150-bp fragment was PCR amplified from y1 w67c23 genomic DNA, denoted yarP. This fragment includes MEME motif 1.1 (Figure 1) and contains two of the three yar transcription start sites. yarP was cloned between direct repeats of loxP sites and inserted either between the wing and body enhancers and promoter of the y gene to generate P[yarP-yellow enhancer blocking (YEB)] or between the eye enhancer and promoter of the white (w) gene to generate P[yarP-white enhancer blocking (WEB)] (Figure S3). For the y reporter, yarP was cloned in both orientations relative to the y promoter. As no differences in phenotypes were observed between transgenic lines carrying these distinct transposons, we represent these independent transposons together. P-element vectors were injected into the host y1 w67c23 strain, and resulting progeny were screened for phenotypic changes indicative of carrying a second marker gene included on the transposon (Genetic Services, Cambridge, MA). Transgenic lines with single transposon insertions were established and analyzed. Phenotypes were determined by crossing transgenic males to y1 w67c23 virgin females. Pigmentation of the wing and body cuticle in the resulting P[yarP-YEB] flies was determined in 3- to 4-day-old females, using a scale of 1–5, where 1 represents the null phenotype and 5 represents the wild-type state. Eye pigmentation in P[yarP-WEB] flies was determined in 3-day-old males and females, using a score of 1–5, where 1 represents white eyes and 5 represents red eyes. At least three independent crosses were set up for each genotype, and two people scored at least 20 flies from each cross. Lines that had low yellow pigmentation scores were analyzed further. In these cases, crosses were made with flies expressing Cre recombinase to catalyze excision of the yarP, as described previously (Chen et al. 2002). Resulting progeny were used to establish stocks. Confirmation of the deletion of the yarP was achieved by PCR analysis.
Ends-out gene targeting
Two transposons were constructed from the pW25 targeting vector (Gong and Golic 2003, 2004), kindly provided by Kent Golic. pW25 contains a multicloning site on either side of the whs gene flanked by loxP sites. The P[ΔHR2 target] (XGL440) transposon was generated to establish a 0.5-kb deletion encompassing yarP, which includes all three yar transcription start sites (Soshnev et al. 2008), whereas the P[ΔHR1 target] (MDW47) transposon was generated to establish a 0.2-kb deletion, which included yarP and twox of the three yar start sites. Both transposons were made in a two-step procedure. First, PCR primers containing NotI sites were used to isolate a 3.3-kb fragment (+6031 to +9318, relative to the y TSS, ΔHR1) or a 3-kb fragment [+6334 to +9318 relative to the y TSS, ΔHR2] of the y–ac intergenic region. These PCR fragments were sequenced to confirm appropriate amplification. Second, the PCR fragments were cloned into XGL235, a derivative of pW25 that carries a 6.6-kb y fragment (−1842 to +4796 relative to the y TSS), which includes the y transcription unit and the body enhancer, but lacks the wing enhancer, ultimately generating the targeting transposons. Transgenic lines of P[ΔHR1 target] and P[ΔHR2 target] were established by transformation of y1 w67c23 flies. Gene targeting was completed followed the procedure outlined in Gong and Golic (2004), screening for flies with darkly pigmented wings, to generate yarΔHR1w and yarΔHR2w. Next, the whs gene was removed by crossing red-eyed males carrying the targeted yar deletion to females carrying Cre recombinase, as described in Chen et al. (2002). The white-eyed flies were collected and used to establish homozygous stocks, called yarΔHR1 and yarΔHR2. A combination of Southern and PCR analyses identified correctly targeted events (Figure S4).
Real-time PCR analyses
RNA was isolated from embryos from three lines: Canton S, yarΔHR1 line MDW47 43-1, and yarΔHR2 line XGL440-114. RNA isolation and quantitative real-time PCR analyses were performed (Parnell et al. 2006). Primer sequences are shown in Table S1. Values obtained from technical replicates for each PCR amplification were averaged, with no greater than 0.5 cycle threshold (Ct) seen between replicates. Two to three experiments were performed for each primer set from at least two independent RNA samples. The expression level of each gene was determined using RpL32 as an internal control (ΔCt). The fold change in expression of each gene relative to the wild-type (Canton S) value was determined with the ΔΔCT method.
Reactive climbing assays
Climbing assays were performed as described previously (Pinto et al. 2008). Five males and five females for each genotype were collected 1 day after eclosion and housed in individual vials at 25°, 70% humidity, with a 12 hr day/night cycle. Five-day-old flies were placed in a 15-cm-long by 1.5-cm-wide graduated glass cylinder. The flies were equilibrated for several minutes, tapped to the bottom, and allowed to climb up the sides. The number of flies that crossed the 15-cm mark in a 30-sec time was recorded. This procedure was repeated five times with five replicates for each genotype (n = 25). The average of these replicates was plotted as the percentage of flies that climb 15 cm in 30 sec. The dMAN1Δ81 null mutant was used as a positive control (Pinto et al. 2008).
Sleep pattern analyses and yar rescue
Three- to 5-day-old virgin females were individually housed in a glass tube (5 [W] × 65 [L] mm) with regular fly food and subjected to 12-hr light and 12-hr dark cycles at 25°. Flies were acclimated to the experimental conditions for 1 day and then their locomotor activity was monitored using the Drosophila Activity Monitor system (Trikinetics). Locomotor activity data were collected at 1-min intervals for 3 days and analyzed with a Microsoft Excel-based script as described previously (Hendricks et al. 2003; Kume et al. 2005). Sleep was defined as ≥5 min of behavioral immobility in the DAM system.
To establish a genomic yar rescue construct P[yar w], a 6-kb yar genomic region was amplified (+4674 to +10696 relative to the y TSS), sequenced and cloned into the CaSpeR3 P-element vector carrying mini-w selectable marker (Figure 3). The rescue construct was injected directly into the yarΔHR2 background produced by homologous recombination (Genetic Services). Transgene insertions were identified by phenotypic rescue of the mutant eye color. Homozygous stocks were established by crossing yarΔHR2, P[yar] males and virgin females together to obtain homozygous P[yar] insertions in the same genetic background as the yar mutants. Southern analyses determined the structure and number of transgenes. Flies from two independent transformed lines were analyzed.
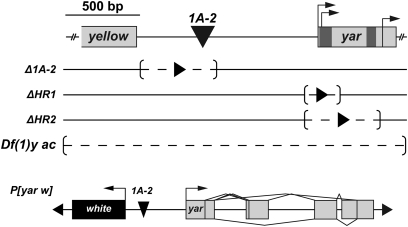
Figure 3.
Structure of the yar alleles used in the study. (Top) Schematic of the genomic region encompassing the 3′ end of y, the 1A-2 element (inverted triangle), and the first exon of yar. Previously identified alternative start sites are indicated by bent arrows (Soshnev et al. 2008). Motifs 1.1 and 1.4 are colored by darker shading in the yar gene. (Middle) Structure of the yar deletion alleles obtained by homologous recombination. Dashed line in brackets indicates deleted region; solid arrowhead represents the residual loxP site. The extant allele Df(1) y ac removes the region spanning the whole y–yar–ac locus. (Bottom) Structure of the P[yar w] transgene used in the rescue studies.
Results
Conservation of yar
The yar gene is located ∼1.2 kb downstream of y and ∼3.0 kb upstream of ac (Figure 1). To address whether yar is conserved, we compared genomic sequences of the y–ac intergenic region from D. melanogaster with eight Drosophila species. In these analyses, we included the region upstream of ac for D. mojavensis, the exceptional Drosophila species that does not have y linked to yar (Negre and Simpson 2009). Intergenic regions were analyzed using MultiPipMaker, a program that constructs multisequence alignments on the basis of percentage of identity plots (PIPs) from pairwise comparisons (Schwartz et al. 2000) and MEME, a sequence analysis program that identifies statistically supported conserved motifs, on the basis of length, similarity, and number of occurrences within and among the sequence set (Bailey et al. 2006). These analyses revealed multiple aligned regions that decreased in size with increasing evolutionary distance (Figure 1A). We identified eight motifs of identical order and orientation present in all species, with the exception of D. grimshawi, where fewer motifs were found due to a gap in the available genome sequence (Figure 1, Figure S1, Table S2). The sizes of the conserved motifs ranged from 40 (motif 2.4) to 111 bp (motif 2.1), showing extensive sequence identity. Motifs of conservation largely localize within or near the yar transcription unit. While four of the identified MEME motifs correspond to yar exons, none encompassed the short open reading frames of the potential polypeptides, providing further evidence that yar encodes lncRNAs. These findings extend previous analyses comparing D. melanogaster and D. virilis sequences that identified motifs 1.1 and 1.4, which lie upstream of the dorsocentral enhancer (Garcia-Garcia et al. 1999).
We noted that one of the largest most conserved motifs in the y–yar–ac region corresponded to motif 1.1, implying that transcription of yar might be conserved. To test this postulate, we determined whether yar was expressed in D. virilis, a species separated from D. melanogaster by 40–60 MY of evolution. RNA was isolated from D. virilis embryos, a developmental stage of maximal expression of D. melanogaster yar (Chia et al. 1986; Soshnev et al. 2008). Northern analysis of poly(A)+ RNA identified a major D. virilis transcript of ∼2.5 kb that accumulates in midembryogenesis, although at much lower levels than the major D. melanogaster yar transcript (Figure S2A). Semiquantitative reverse transcriptase polymerase chain reaction (RT-PCR) studies were undertaken to define the expression pattern for D. virilis RNAs generated in the y–ac intergenic region. Using primer pairs anchored in the conserved MEME motifs, the expression of four cDNAs was studied during three stages of embryonic development (Figure 2, Figure S2B). Importantly, the expression pattern of these cDNAs was coordinated. The temporal accumulation of yar in D. virilis embryos was reminiscent of D. melanogaster, with transcription of yar coinciding with ac down-regulation and transcription of y coinciding with yar down-regulation. Sequence analysis of these D. virilis cDNAs showed that the coding capacity of each was <75 amino acids, with no evidence of sequence conservation with the potential D. melanogaster polypeptides (data not shown). A novel D. virilis gene was predicted within the y–ac intergenic region using Genscan (Burge and Karlin 1997) (Figure S2A). The exons of the Genscan-predicted gene were close to the identified MEME motifs, with three motifs showing a partial overlap with the predicted Genscan exons. We completed RT-PCR analyses to test the accuracy of the Genscan representation (Figure S2B). Primer pairs anchored within the predicted exons failed to amplify D. virilis embryonic cDNAs. Further, tests using mixed primer pairs, including between one MEME motif primer and one Genscan exon primer, were only successful in one case (product A, Figure S2B). On the basis of these findings, we conclude that the Genscan prediction does not accurately reflect the D. virilis y–ac intergenic transcription unit. Taken together, our data provide compelling support that yar is a conserved lncRNA gene.
Several experiments tested the function of the conserved MEME motifs. First, we tested whether motif 1.1 had insulator activity. These experiments were predicated on previous studies showing that in certain genetic contexts, the enhancers of the y and ac genes were capable of directing inappropriate transcription (Campuzano et al. 1986; Parnell et al. 2003). Such observations imply that independent transcriptional regulation of the y and ac genes requires the presence of an intervening chromatin insulator. Many insulators have been associated with promoter activity (Kuhn and Geyer 2003; Chopra et al. 2009; Raab and Kamakaka 2010), consistent with the location of two out of three yar promoters within motif 1.1 (Soshnev et al. 2008). To this end, we tested the enhancer blocking capacity of motif 1.1 using two well-characterized reporter genes (Figure S3). Analysis of transgenic flies carrying these reporter genes showed that enhancer blocking did not occur, implying that motif 1.1 is not an insulator. Second, we investigated whether the conserved motifs, or other regions within the yar transcription unit, were processed into smaller RNAs, such as miRNAs. In these analyses, existing miRNA databases (Kozomara and Griffiths-Jones 2011), as well as structural predictions using Mfold were employed (Zuker 2003). These strategies provided no evidence for the generation of miRNAs from yar (data not shown).
Distribution of yar during embryogenesis
To gain insights into the function of yar, we examined the spatial distribution of this RNA during embryogenesis (Figure 1B). Whole mount in situ RNA hybridization was performed in aged embryos using probes corresponding to yar, y, and ac. The y and ac probes served as positive controls because the accumulation of these RNAs was previously defined (Romani et al. 1987; Walter et al. 1991). As a negative control, Df (1) y ac embryos were studied in parallel. These embryos carry a deletion of all three genes, thereby providing a null background. As expected, accumulation of ac RNA was restricted to neurogenic regions, while accumulation of y RNA was limited to stripes that underlie the ventral denticle belts. In contrast, yar RNA was found throughout the embryo (Figure 1B). In all cases, the level of hybridization for each probe was higher in wild-type embryos than in the Df(1) y ac controls. These data indicate that yar is globally expressed during embryogenesis.
The function of lncRNAs depends upon their subcellular location. As lncRNAs can localize to the nucleus and cytoplasm, we determined which subcellular compartment contains yar. In these experiments, RNA was isolated from unfractioned or nuclear fractions of homogenates made from 6- to 12-hr wild-type embryos (Figure 1C). These RNA samples were reverse transcribed and the level of yar was determined by PCR. Ras64B was chosen as a control because this gene encodes a globally expressed, spliced protein-coding mRNA. The PCR analyses involved two sets of primer pairs for analysis of yar and Ras64B RNAs (Table S1). For one pair, opposing primers were positioned at opposite exon junctions spanning a common intron to detect mature RNA. For the second pair, opposing primers were located within intronic sequences, which are expected to be nuclear restricted. We obtained a PCR product representing the mature yar RNA only from the sample of total RNA, whereas a PCR product representing yar intronic sequences was detected in both nuclear and total RNA (Figure 1C). These data imply that spliced yar RNAs are cytoplasmic, a conclusion that is supported by analyses of Ras64B RNA.
Investigation of yar contributions to the regulation of neighboring gene expression
Recent studies demonstrate that lncRNA genes regulate transcription both in cis and trans (Ogawa and Lee 2002; Martens et al. 2004; Petruk et al. 2006; Martianov et al. 2007; Barrandon et al. 2008; Brock et al. 2009; Mercer et al. 2009; Taft et al. 2010). The genomic location of yar, coupled with the conserved timing of embryonic expression, suggested that yar might regulate transcription of the adjacent y or ac genes. This postulate is supported by examples where transcription of an upstream noncoding RNA gene represses expression of the adjacent downstream gene by transcriptional interference (Martens et al. 2004; Petruk et al. 2006). Two observations supported that repression of ac would require yar transcription, and not yar RNA production. First, yar is a cytoplasmic RNA (Figure 1C), unlike nuclear lncRNAs that have a direct role in gene silencing. Second, the timing and level of ac and y expression are unchanged when levels of yar are reduced, as defined in studies of the hypomorphic yarΔ1A-2 mutant (Soshnev et al. 2008).
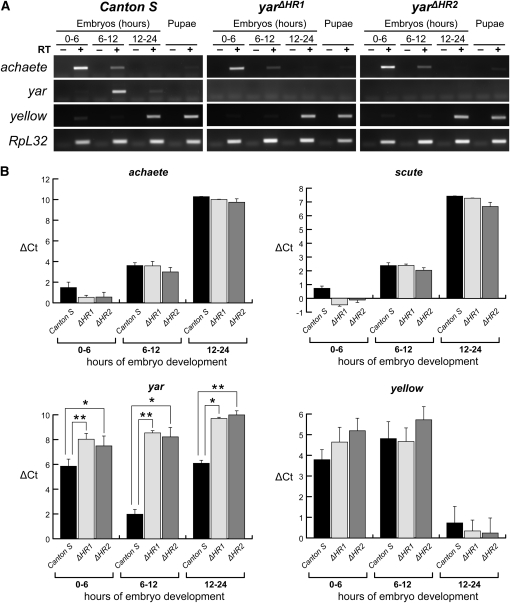
We generated two null alleles using ends-out gene targeting to test the role of yar transcription on neighboring gene expression. These experiments deleted either 200 bp (yarΔHR1) or 500 bp (yarΔHR2) of the yar promoter, removing two or three of the yar transcription start sites (Figure 3, Figure S4), respectively. Quantitative real-time PCR analyses demonstrated that yarΔHR1 and yarΔHR2 flies had 85-fold lower levels of yar RNA relative to Canton S, which were undetectable in agarose gel analyses (Figure 4). Measured levels of yar RNA in yarΔHR1 and yarΔHR2 flies was similar to those obtained from flies carrying a deletion of the yar locus (Df (1) y ac, data not shown), consistent with our prediction that the newly generated alleles remove the yar promoter. Using yarΔHR1 and yarΔHR2 mutants, we defined the timing and level of y, ac, and scute (sc) RNA using quantitative real-time PCR analyses. We found that embryonic expression of all genes was unchanged by loss of yar transcription (Figure 4). We conclude that yar does not contribute to the regulation of transcription of neighboring genes.
Figure 4.
Quantitative analyses of gene expression in yar null mutants. RNA was isolated from wild type (Canton S) and the yar null mutants [yarΔHR1 (ΔHR1) and yarΔHR2 (ΔHR2)]. (A) Semiquantitative PCR analyses of RNAs isolated from aged embryos and mixed stage pupae. RpL32 is a constitutively expressed gene and serves as a loading control. The (−) RT lanes control for genomic DNA contamination. (B) Quantitative RT-PCR of y, yar, ac, and sc, the gene downstream of ac. Cycle threshold (Ct) values were normalized to the constitutively expressed Ras64B gene to control for the amount of input cDNA (ΔCt). A higher ΔCt value indicates lower level of RNA accumulation. Error bars indicate standard deviation from two biological replicates. Asterisks indicate statistical significance by Student’s t-test, *P < 0.05, **P < 0.01.
Functional analysis of yar
Visual inspection of yar mutant flies revealed no overt morphological defects. The absence of changes in bristle number and cuticle pigmentation is consistent with normal transcription of the neighboring y and ac genes (Figure 4). A possible functional role for yar was suggested by consideration of the functions of the neighboring y, ac and sc genes. The ac and sc genes encode basic helix–loop–helix transcription factors required for formation of neural precursors (Modolell and Campuzano 1998), while the y gene encodes a secreted protein required for male sexual behavior (Nash and Yarkin 1974; Biessmann 1985; Chia et al. 1986; Geyer et al. 1986; Drapeau et al. 2003, 2006). These observations suggested that yar resides in a cluster of neural genes. As emerging evidence suggests that gene order within eukaryotic chromosomes is nonrandom (Lee and Sonnhammer 2003; Hurst et al. 2004; Yi et al. 2007; de Wit and van Steensel 2009), we predicted that yar may have a neural function.
One of the ultimate manifestations of neural function is behavior. To address possible roles of yar in fly behavior, two assays were used. First, we evaluated the general locomotor and geotactic ability in yar mutants. Second, we examined sleep, a fundamental biological process conserved among evolutionarily diverse animal species (Sehgal et al. 2007; Cirelli and Bushey 2008; Cirelli 2009). In both sets of experiments, flies corresponding to multiple independently generated yar mutant alleles were tested. We used homozygous and heteroallelic mutant combinations to assay behavior in multiple distinct genetic backgrounds. We tested four yar alleles, including one hypomorphic (yarΔ1A-2) and three null (yarΔHR1, yarΔHR2, and Df(1) y ac) alleles (Figure 3). We reasoned that if consistent behavioral changes were observed, then these data would support a neural function for yar.
In reactive climbing assays, three groups of 10 flies for each yar genotype were analyzed. These flies were placed in a graduated cylinder, tapped to the bottom, and the number of flies climbing to 15 cm in 30 sec was recorded. These studies showed that flies with decreased or eliminated yar RNA had normal climbing activity (data not shown). These experiments suggest that general locomotion and the tendency of flies to move against gravity are not perturbed.
The Drosophila Activity Monitoring (DAM) system was used to evaluate sleep behavior in yar mutants. In these studies, individual flies were loaded into an activity monitor tube and a computer recorded each time a fly crossed an infrared beam that bisects the tube. Previous studies have defined sleep as a period of quiescence lasting ≥5 min (Hendricks et al. 2000; Shaw et al. 2000). Parameters of sleep depend upon sex and age of the fly (Cirelli 2006, 2009; Koh et al. 2006; Sehgal et al. 2007). Here, we studied sleep behavior in 3- to 5-day-old females. We tested females carrying different homozygous and heterozygous combinations of yar alleles representing different genetic backgrounds. Each of these yar mutant backgrounds carried a deletion of the w gene (the w67c23 allele), a mutation that has significance to behavior assays. The w gene encodes an adenosine triphosphate (ATP)-binding cassette (ABC) transmembrane transporter protein (Mount 1987; Pepling and Mount 1990; Anaka et al. 2008), and affects fly behavior (Zhang and Odenwald 1995; Cirelli et al. 2005; Anaka et al. 2008). For this reason, our reference line carried a wild-type yar gene and the w deletion allele.
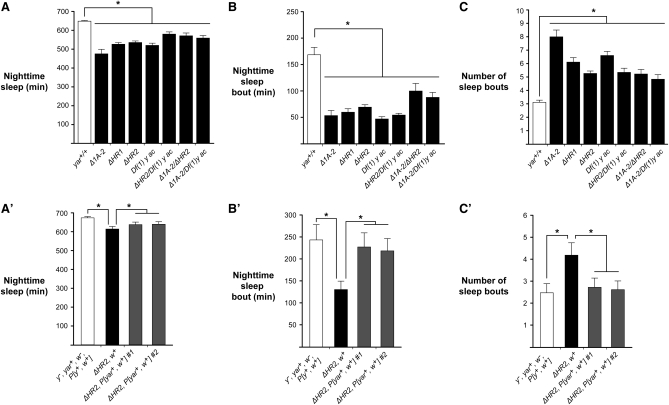
Females of each genotype were placed in activity monitor tubes and acclimated for 1 day. Locomotor activity was assessed at 1-min intervals over a 3-day period of consecutive 12-hr light (day) and dark (night) cycles. From these data, we determined the average time spent in nighttime sleep. We found that all yar mutant females showed a significant decrease in the level of total nighttime sleep, irrespective of mutant genotype (Figure 5A). Even though nighttime sleep in yar mutants was reduced, sleep remained higher in the night than day, indicating sleep reduction occurred in the context of a normal circadian rhythm. Further, yar mutants retained a circadian rhythm even when placed in total darkness for 10 days (Figure S5A). We assessed whether decreased sleep resulted from increased activity during nighttime periods of wakefulness. To this end, we divided the total activity counts by the total length of waking time. These calculations showed that the nighttime activity of yar mutants was not elevated compared to the reference strain (Figure S5B), indicating that reduced sleep is not due to a general increase in locomotion activity. To characterize the architecture of nighttime sleep, we analyzed the duration and number of sleep bouts in yar mutants relative to the reference strain. Notably, we observed that the length of nighttime sleep bouts was shortened in yar females, coinciding with an increase in the number of sleep episodes (Figure 5, B and C). These data indicate that yar mutants have reduced and fragmented sleep patterns. Similar findings were observed for males (data not shown).
Figure 5.
Loss of yar affects sleep behavior in Drosophila. (A) Baseline nighttime sleep in the parental yar+/+ (y1 yar+ w67c23, open bar) line and yar mutants (y1 yarmutant w67c23, solid bars). Average amounts of sleep for 3- to 5-day-old virgin females are shown (n ≥ 32). (B and C) Average duration of nighttime sleep bout and number of sleep episodes. (A′) Effect of rescue by P[yar w] on nighttime sleep duration. Shown are data obtained from the parental line (y1 yar+ w67c23, P[ΔHR2 target], open bar), the yar mutant (ΔHR2, solid bar). and the yar mutant carrying the P[yar w] rescue construct inserted at two independent genomic locations (ΔHR2 R1 and ΔHR2 R2, shaded bars). (B′ and C′) Effects of the rescue P[yar w] transposon on sleep bout duration and number of sleep bouts in yar mutants. Kruskal–Wallis one-way ANOVA, *P < 0.05. Error bars represent SEM.
A complementation assay was used to determine whether the altered sleep behaviors were caused by a loss of yar. In these studies, yarΔHR2 w67c23 flies were directly transformed with a P[yar w] transposon, to ensure that the mutant and P[yar w] transgenic flies had isogenic backgrounds. The P[yar w] transposon contains a genomic fragment encompassing the yar transcription unit, with ∼1.2 kb of upstream and ∼0.15 kb of downstream flanking DNA linked to the mini-w reporter gene (Figure 3). To assess effects of reintroduction of yar, we changed our reference line to reflect that the P[yar w] transposon carries a mini-w gene. The reference line, called yar+ w+, carried the w67c23 mutation and a P transposon with the mini-w transgene. We measured sleep parameters in females from two independent yarΔHR2, P[yar w] transgenic lines, in which the rescue transposon was located at a different genomic locations [rescue 1 (R1) and rescue 2 (R2)]. We found that reintroduction of the yar genomic fragment restored the total amount of nighttime sleep (Figure 5A′). Significantly, sleep bout length was increased in both independent yarΔHR2, P[yar w] lines, while the number of sleep episodes was decreased (Figure 5, B′ and C′). These data indicate that reintroducing yar restored sleep parameters, providing strong evidence that yar plays a role in sleep regulation.
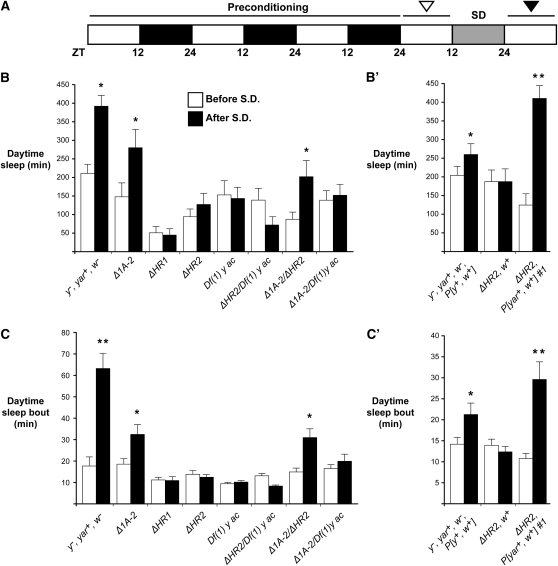
Sleep has a homeostatic component, in which individuals regain lost sleep after deprivation (Ho and Sehgal 2005; Cirelli 2009). We tested whether loss of yar affected homeostasis. In these studies, reference females and females corresponding to multiple yar genotypes were placed in activity monitor tubes, preconditioned for 3 days of 12-hr light and dark cycles, followed by 12 hr of mechanical sleep deprivation during the fourth night (Figure 6A). The amount of daytime sleep was assessed prior to (third and fourth days) and after (fifth day) sleep disruption. We found that daytime sleep bout length did not change between days 3 and 4 for either the reference or yar mutant females (data not shown), indicating that time spent in the activity tubes did not influence sleep. However, following sleep deprivation, reference females increased total daytime sleep and sleep bout length. In contrast, no increase in these parameters was observed for yar mutants, with one exception (Figure 6, B and C). The exceptional females were yarΔ1A-2, where daytime sleep was increased, but to a lower level than the reference line. The ability of yarΔ1A-2 females to respond to sleep deprivation may reflect the presence of low levels of yar RNA (Soshnev et al. 2008), a proposal supported by findings that sleep rebound was lowered or lost in females that were heterozygous for yarΔ1A-2 and a yar null allele (Figure 6, B and C). Importantly, defects in sleep homeostasis were rescued by introduction of P[yar w]. The rescued yarΔHR2, P[yar w] females showed increased daytime sleep due to lengthened sleep bouts following sleep deprivation compared to the isogenic yarΔHR2 females (Figure 6, B′ and C′). Together, these data suggest that yar mutants display an altered homeostatic sleep response.
Figure 6.
Loss of yar causes defect in homeostatic regulation of sleep. (A) Diagram of experimental strategy for determining effects of sleep deprivation. Flies were preconditioned in the DAM system for 3 days of 12-hr day and night cycles, and baseline daytime sleep bout duration was established on day 4 (open arrowhead). Flies were sleep deprived for one night (SD) and sleep parameters were measured the following morning (solid arrowhead). (B) Total daytime sleep before (open bars) and after (solid bars) sleep deprivation is shown. (C) The average length of sleep bouts before and after sleep deprivation. (B′ and C′) Response to sleep deprivation in the w+ reference and yar mutants carrying the P[yar w] rescue transgene. Kruskal–Wallis one-way ANOVA, *P < 0.05, **P < 0.001. Error bars represent SEM.
Discussion
yar is a conserved lncRNA gene
The yar gene resides within a cluster of neural genes, separating the well-characterized y and ac genes. The architecture of this region has been cited as a paradigmatic example of gene organization and function, because the order and orientation of these genes remains unchanged within genomes of distantly related insect species (Garcia-Bellido and De Celis 2009; Negre and Simpson 2009). However, these previous analyses did not recognize the presence of yar, likely because yar generates transcripts that lack a large ORF (Soshnev et al. 2008). In our studies, we obtained several lines of evidence demonstrating that yar is conserved within Drosophila species. First, genomic comparisons of the y–ac intergenic region identified eight conserved motifs that largely map to the yar transcription unit (Figure 1). Second, one of the most highly conserved motifs, motif 1.1, encompasses the yar promoters and regulates yar transcription. Importantly, motif 1.1 is not a regulatory element for either y or ac, as deletion of this region eliminates yar expression, without affecting transcription of neighboring genes (Figure 4). Third, the temporal pattern of embryonic transcription of yar is conserved in D. virilis (Figure 2). These data suggest that yar is conserved in drosophilids.
The yar gene was classified as an lncRNA gene on the basis of the absence of an ORF >75 amino acids. While the requirement for a large ORF is commonly used to distinguish ncRNAs, emerging evidence suggests that caution is needed when this is the only parameter used for lncRNA designation (Galindo et al. 2007; Kondo et al. 2007; Hanyu-Nakamura et al. 2008; Hashimoto et al. 2008; Timinszky et al. 2008). For example, the tarsal-less/polished rice (tal/pri) gene was originally identified as a putative mRNA-like lncRNA (Inagaki et al. 2005; Tupy et al. 2005). Subsequent analyses showed that tal/pri transcripts encode short peptides of 11 amino acids that contain full biological function (Galindo et al. 2007; Kondo et al. 2007). In cases where small peptides have been identified, evolutionary comparisons have demonstrated conservation of the small ORFs. To this end, we examined whether the eight conserved motifs provide evidence for a conserved coding capacity in yar transcripts. Of the eight, four motifs reside within yar exons but do not overlap with the putative short ORFs (Figure 1, Figure S1). Of the four motifs located within yar exons, only motif 1.3 contains an ATG; translation at this codon would generate a peptide of three amino acids. Further, our analysis of D. virilis yar transcripts failed to identify conservation with any of the potential D. melanogaster polypeptides. Together, these findings provide strong evidence that yar is a lncRNA gene.
yar is a cytoplasmic RNA
LncRNAs have been identified that localize to specific subcellular compartments. A large and growing list of lncRNAs are retained in the nucleus, where they contribute to nuclear organization and gene expression (Mercer et al. 2009; Chen and Carmichael 2010; Taft et al. 2010; Clark and Mattick 2011). A smaller number of lncRNAs have been characterized that function in the cytoplasm, with yar falling into this second class (Panzitt et al. 2007; Matouk et al. 2009; Wang et al. 2010; Clark and Mattick 2011). One of these cytoplasmic lncRNAs is the highly up-regulated in liver cancer (HULC) RNA, discovered in expression array studies that identified genes misregulated in hepatocellular carcinoma (Panzitt et al. 2007). Functional analyses of HULC found that this lncRNA contributes to a regulatory circuit that modulates miRNA activities, acting as a sponge to down-regulate a series of miRNAs (Wang et al. 2010). Similarly, RNAs generated from pseudogenes have been found to act as decoys for miRNAs by modulating interactions between miRNAs and target coding mRNAs (Poliseno et al. 2010). Prompted by these observations, we investigated a possible link between yar and Drosophila miRNAs. To this end, sequences encompassing the yar exons (2 kb) were submitted to the Web-based tool MicroInspector (Rusinov et al. 2005). We identified the presence of miRNA seed matches with a high free energy cutoff value of −25 kcal/mol at the temperature of 25°, using the Release 17 of Sanger Institute miRBase that includes both computationally predicted and experimentally confirmed miRs. These analyses uncovered 33 miRNA seed matches within yar exons corresponding to 19 confirmed miRNAs (Figure S6, Table S3). Of the exonic seeds for miRNAs, six map within the conserved motifs 1.1, 1.2, and 1.4, with one miRNA (dme-miR-4970-5p) having three seed matches within the yar exons. These data support a possible connection between yar and miRNA regulation. As a control, we submitted sequences corresponding to the yar intron (2.8 kb). These analyses identified 36 miRNA seed matches that correspond to 25 confirmed miRNAs (Figure S6, Table S3). Of the confirmed intronic miRNAs, one has two seed matches. These observations indicate that the yar exons are not enriched for miRNA sequences relative to control, a finding consistent with the small size of the miRNA seeds. Further studies are necessary to discern the functional significance between yar and the miRNAs, experiments that require an understanding of the targets of the yar exonic miRNAs, which are largely unknown at this time. Even so, we note that yar and many Drosophila lncRNAs are expressed during early embryogenesis following the developmental period of active changes in mRNA stability. As recent studies suggest that miRNAs promote turnover of maternal and zygotic RNAs (Bushati et al. 2008; Thomsen et al. 2010), these observations raise the possibility that yar and other cytoplasmic lncRNAs may function as sponges that titrate miRNAs during embryogenesis, permitting fine-tuning of the miRNA-dependent degradation pathway.
Loss of yar disrupts sleep regulation
Phenotypic analyses of yar mutants demonstrate that yar is required for both sleep maintenance and homeostasis. We find that nighttime sleep is decreased in loss-of-function yar mutants, correlating with reduced sleep bout length. Further, sleep homeostasis is affected by yar loss, as these mutants do not increase daytime sleep following sleep deprivation. Both defects are restored by introduction of the yar gene, providing compelling evidence that yar is required for sleep regulation. The yar mutant phenotypes are reminiscent of those described for mutations in the serotonin receptor 1A gene (Yuan et al. 2006), which affects sleep due to defects in the adult mushroom bodies. It is unclear whether the requirement for yar is developmental or due to a physiological role in sleep regulation. While yar expression is highest during early embryogenesis, recent deep sequencing studies have uncovered yar RNAs in poly(A)+ RNA isolated from male and female heads (Graveley et al. 2011). These data suggest that yar might directly regulate processes in the brain that impact sleep behavior. Of note, one of the miRNA seed matches within the yar exon corresponds to miRNAs from the miR-310 cluster (Figure S6), a match not found in similar analyses of yar intronic sequences or exonic sequences corresponding to three other genes (y, ac, and GAPDH2; data not shown). Loss of miRNAs 310 to 313 alters synaptic transmission at the larval neuromuscular junction, with no effect on viability or fertility (Tsurudome et al. 2010). These findings are consistent with the possibility that yar might participate in a regulatory circuit that influences levels of miRs within the brain, which may have the capacity to contribute to synaptic homeostasis. Further studies are needed to elucidate the temporal and tissue-specific requirements for yar, which will provide insights into how yar contributes to sleep regulation.
Many Drosophila lncRNA genes display spatially restricted embryonic expression that corresponds to RNA accumulation in the developing central and peripheral nervous system (Inagaki et al. 2005; Li et al. 2009). These observations suggest that ncRNAs might commonly contribute to neuronal function during Drosophila development. Further, large numbers of ncRNAs have been identified in mouse brain (Mercer et al. 2008), suggesting that such functions might extend to vertebrates.
Acknowledgments
We thank the members of Geyer and Kitamoto laboratories for critically reading this manuscript. We thank Kate Appleton, Kate Jandacek, Will Barry, and Yasir Ahmed for technical assistance. We thank Yikang Rong for providing pW25 and Kent Golic for providing the 70FLP and 70I-SceI flies. This work was supported by National Institutes of Health Grant GM42539 to P.K.G.
Literature Cited
- Anaka M., MacDonald C. D., Barkova E., Simon K., Rostom R., et al. , 2008. The white gene of Drosophila melanogaster encodes a protein with a role in courtship behavior. J. Neurogenet. 22: 243–276 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Williams N., Misleh C., Li W. W., 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34: W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrandon C., Spiluttini B., Bensaude O., 2008. Non-coding RNAs regulating the transcriptional machinery. Biol. Cell 100: 83–95 [DOI] [PubMed] [Google Scholar]
- Biessmann H., 1985. Molecular analysis of the yellow gene (y) region of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 82: 7369–7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock H. W., Hodgson J. W., Petruk S., Mazo A., 2009. Regulatory noncoding RNAs at Hox loci. Biochem. Cell Biol. 87: 27–34 [DOI] [PubMed] [Google Scholar]
- Burge C., Karlin S., 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Bushati N., Stark A., Brennecke J., Cohen S. M., 2008. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18: 501–506 [DOI] [PubMed] [Google Scholar]
- Campuzano S., Carramolino L., Cabrera C. V., Ruiz-Gomez M., Villares R., et al. , 1985. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell 40: 327–338 [DOI] [PubMed] [Google Scholar]
- Campuzano S., Balcells L., Villares R., Carramolino L., Garcia-Alonso L., et al. , 1986. Excess function hairy-wing mutations caused by gypsy and copia insertions within structural genes of the achaete-scute locus of Drosophila. Cell 44: 303–312 [DOI] [PubMed] [Google Scholar]
- Chen J. L., Huisinga K. L., Viering M. M., Ou S. A., Wu C. T., et al. , 2002. Enhancer action in trans is permitted throughout the Drosophila genome. Proc. Natl. Acad. Sci. USA 99: 3723–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. L., Carmichael G. G., 2010. Decoding the function of nuclear long non-coding RNAs. Curr. Opin. Cell Biol. 22: 357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia W., Howes G., Martin M., Meng Y. B., Moses K., et al. , 1986. Molecular analysis of the yellow locus of Drosophila. EMBO J. 5: 3597–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra V. S., Cande J., Hong J. W., Levine M., 2009. Stalled Hox promoters as chromosomal boundaries. Genes Dev. 23: 1505–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., 2006. Sleep disruption, oxidative stress, and aging: new insights from fruit flies. Proc. Natl. Acad. Sci. USA 103: 13901–13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., 2009. The genetic and molecular regulation of sleep: from fruit flies to humans. Nat. Rev. Neurosci. 10: 549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., Bushey D., 2008. Sleep and wakefulness in Drosophila melanogaster. Ann. N. Y. Acad. Sci. 1129: 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C., Bushey D., Hill S., Huber R., Kreber R., et al. , 2005. Reduced sleep in Drosophila Shaker mutants. Nature 434: 1087–1092 [DOI] [PubMed] [Google Scholar]
- Clark M. B., Mattick J. S., 2011. Long noncoding RNAs in cell biology. Semin. Cell Dev. Biol. (in press) [DOI] [PubMed] [Google Scholar]
- de Wit E., van Steensel B., 2009. Chromatin domains in higher eukaryotes: insights from genome-wide mapping studies. Chromosoma 118: 25–36 [DOI] [PubMed] [Google Scholar]
- Deng X., Meller V. H., 2006. Non-coding RNA in fly dosage compensation. Trends Biochem. Sci. 31: 526–532 [DOI] [PubMed] [Google Scholar]
- Drapeau M. D., Radovic A., Wittkopp P. J., Long A. D., 2003. A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. J. Neurobiol. 55: 53–72 [DOI] [PubMed] [Google Scholar]
- Drapeau M. D., Cyran S. A., Viering M. M., Geyer P. K., Long A. D., 2006. A cis-regulatory sequence within the yellow locus of Drosophila melanogaster required for normal male mating success. Genetics 172: 1009–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo M. I., Pueyo J. I., Fouix S., Bishop S. A., Couso J. P., 2007. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 5: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A., de Celis J. F., 2009. The complex tale of the achaete-scute complex: a paradigmatic case in the analysis of gene organization and function during development. Genetics 182: 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia M. J., Ramain P., Simpson P., Modolell J., 1999. Different contributions of pannier and wingless to the patterning of the dorsal mesothorax of Drosophila. Development 126: 3523–3532 [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Corces V. G., 1987. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1: 996–1004 [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Spana C., Corces V. G., 1986. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J. 5: 2657–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert J. M., Simpson P., 2003. Evolution of cis-regulation of the proneural genes. Int. J. Dev. Biol. 47: 643–651 [PubMed] [Google Scholar]
- Gong W. J., Golic K. G., 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100: 2556–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W. J., Golic K. G., 2004. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics 168: 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanyu-Nakamura K., Sonobe-Nojima H., Tanigawa A., Lasko P., Nakamura A., 2008. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature 451: 730–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Kondo T., Kageyama Y., 2008. Lilliputians get into the limelight: novel class of small peptide genes in morphogenesis. Dev. Growth Differ. 50(Suppl 1): S269–S276 [DOI] [PubMed] [Google Scholar]
- Hendricks J. C., Finn S. M., Panckeri K. A., Chavkin J., Williams J. A., et al. , 2000. Rest in Drosophila is a sleep-like state. Neuron 25: 129–138 [DOI] [PubMed] [Google Scholar]
- Hendricks J. C., Lu S., Kume K., Yin J. C., Yang Z., et al. , 2003. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J. Biol. Rhythms 18: 12–25 [DOI] [PubMed] [Google Scholar]
- Ho K. S., Sehgal A., 2005. Drosophila melanogaster: an insect model for fundamental studies of sleep. Methods Enzymol. 393: 772–793 [DOI] [PubMed] [Google Scholar]
- Hurst L. D., Pal C., Lercher M. J., 2004. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5: 299–310 [DOI] [PubMed] [Google Scholar]
- Ilik I., Akhtar A., 2009. roX RNAs: non-coding regulators of the male X chromosome in flies. RNA Biol. 6: 113–121 [DOI] [PubMed] [Google Scholar]
- Inagaki S., Numata K., Kondo T., Tomita M., Yasuda K., et al. , 2005. Identification and expression analysis of putative mRNA-like non-coding RNA in Drosophila. Genes Cells 10: 1163–1173 [DOI] [PubMed] [Google Scholar]
- Jolly C., Lakhotia S. C., 2006. Human sat III and Drosophila hsr omega transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res. 34: 5508–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K., Evans J. M., Hendricks J. C., Sehgal A., 2006. A Drosophila model for age-associated changes in sleep:wake cycles. Proc. Natl. Acad. Sci. USA 103: 13843–13847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Hashimoto Y., Kato K., Inagaki S., Hayashi S., et al. , 2007. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat. Cell Biol. 9: 660–665 [DOI] [PubMed] [Google Scholar]
- Koya S. K., Meller V. H., 2011. roX RNAs and genome regulation in Drosophila melanogaster. Prog. Mol. Subcell. Biol. 51: 147–160 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S., 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn E. J., Geyer P. K., 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15: 259–265 [DOI] [PubMed] [Google Scholar]
- Kume K., Kume S., Park S. K., Hirsh J., Jackson F. R., 2005. Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 25: 7377–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M., Sonnhammer E. L., 2003. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 13: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu M., Zhang L., Zhang W., Gao G., et al. , 2009. Detection of intergenic non-coding RNAs expressed in the main developmental stages in Drosophila melanogaster. Nucleic Acids Res. 37: 4308–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallik M., Lakhotia S. C., 2010. Improved activities of CREB binding protein, heterogeneous nuclear ribonucleoproteins and proteasome following downregulation of noncoding hsromega transcripts help suppress poly(Q) pathogenesis in fly models. Genetics 184: 927–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow T. A., Beall S., Matzkin L. M., 2009. Egg size, embryonic development time and ovoviviparity in Drosophila species. J. Evol. Biol. 22: 430–434 [DOI] [PubMed] [Google Scholar]
- Martens J. A., Laprade L., Winston F., 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A., 2007. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445: 666–670 [DOI] [PubMed] [Google Scholar]
- Matouk I. J., Abbasi I., Hochberg A., Galun E., Dweik H., et al. , 2009. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur. J. Gastroenterol. Hepatol. 21: 688–692 [DOI] [PubMed] [Google Scholar]
- Meller V. H., Rattner B. P., 2002. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 21: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Mariani J., Kosik K. S., Mehler M. F., et al. , 2008. Noncoding RNAs in long-term memory formation. Neuroscientist 14: 434–445 [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Mattick J. S., 2009. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Modolell J., Campuzano S., 1998. The achaete-scute complex as an integrating device. Int. J. Dev. Biol. 42: 275–282 [PubMed] [Google Scholar]
- Mount S. M., 1987. Sequence similarity. Nature 325: 487. [DOI] [PubMed] [Google Scholar]
- Nash W. G., Yarkin R. J., 1974. Genetic regulation and pattern formation: a study of the yellow locus in Drosophila melanogaster. Genet. Res. 24: 19–26 [DOI] [PubMed] [Google Scholar]
- Negre B., Simpson P., 2009. Evolution of the achaete-scute complex in insects: convergent duplication of proneural genes. Trends Genet. 25: 147–152 [DOI] [PubMed] [Google Scholar]
- Ogawa Y., Lee J. T., 2002. Antisense regulation in X inactivation and autosomal imprinting. Cytogenet. Genome Res. 99: 59–65 [DOI] [PubMed] [Google Scholar]
- Panzitt K., Tschernatsch M. M., Guelly C., Moustafa T., Stradner M., et al. , 2007. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132: 330–342 [DOI] [PubMed] [Google Scholar]
- Parnell T. J., Viering M. M., Skjesol A., Helou C., Kuhn E. J., et al. , 2003. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. USA 100: 13436–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell T. J., Kuhn E. J., Gilmore B. L., Helou C., Wold M. S., et al. , 2006. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol. Cell. Biol. 26: 5983–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepling M., Mount S. M., 1990. Sequence of a cDNA from the Drosophila melanogaster white gene. Nucleic Acids Res. 18: 1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S., Sedkov Y., Riley K. M., Hodgson J., Schweisguth F., et al. , 2006. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 127: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto B. S., Wilmington S. R., Hornick E. E., Wallrath L. L., Geyer P. K., 2008. Tissue-specific defects are caused by loss of the drosophila MAN1 LEM domain protein. Genetics 180: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L., Salmena L., Zhang J., Carver B., Haveman W. J., et al. , 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K. V., Spector D. L., 2007. Eukaryotic regulatory RNAs: an answer to the 'genome complexity' conundrum. Genes Dev. 21: 11–42 [DOI] [PubMed] [Google Scholar]
- Prasanth K. V., Rajendra T. K., Lal A. K., Lakhotia S. C., 2000. Omega speckles: a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J. Cell Sci. 113(Pt 19): 3485–3497 [DOI] [PubMed] [Google Scholar]
- Raab J. R., Kamakaka R. T., 2010. Insulators and promoters: closer than we think. Nat. Rev. Genet. 11: 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani S., Campuzano S., Modolell J., 1987. The achaete-scute complex is expressed in neurogenic regions of Drosophila embryos. EMBO J. 6: 2085–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinov V., Baev V., Minkov I. N., Tabler M., 2005. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 33: W696–W700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Zhang Z., Frazer K. A., Smit A., Riemer C., et al. , 2000. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 10: 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A., Joiner W., Crocker A., Koh K., Sathyanarayanan S., et al. , 2007. Molecular analysis of sleep: wake cycles in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 72: 557–564 [DOI] [PubMed] [Google Scholar]
- Shaw P. J., Cirelli C., Greenspan R. J., Tononi G., 2000. Correlates of sleep and waking in Drosophila melanogaster. Science 287: 1834–1837 [DOI] [PubMed] [Google Scholar]
- Soshnev A. A., Li X., Wehling M. D., Geyer P. K., 2008. Context differences reveal insulator and activator functions of a Su(Hw) binding region. PLoS Genet. 4: e1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Lin M. F., Kheradpour P., Pedersen J. S., Parts L., et al. , 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft R. J., Pang K. C., Mercer T. R., Dinger M., Mattick J. S., 2010. Non-coding RNAs: regulators of disease. J. Pathol. 220: 126–139 [DOI] [PubMed] [Google Scholar]
- Tautz D., Pfeifle C., 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98: 81–85 [DOI] [PubMed] [Google Scholar]
- Thomsen S., Anders S., Janga S. C., Huber W., Alonso C. R., 2010. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 11: R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky G., Bortfeld M., Ladurner A. G., 2008. Repression of RNA polymerase II transcription by a Drosophila oligopeptide. PLoS ONE 3: e2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurudome K., Tsang K., Liao E. H., Ball R., Penney J., et al. , 2010. The Drosophila miR-310 cluster negatively regulates synaptic strength at the neuromuscular junction. Neuron 68: 879–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupy J. L., Bailey A. M., Dailey G., Evans-Holm M., Siebel C. W., et al. , 2005. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 102: 5495–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M. F., Black B. C., Afshar G., Kermabon A. Y., Wright T. R., et al. , 1991. Temporal and spatial expression of the yellow gene in correlation with cuticle formation and dopa decarboxylase activity in Drosophila development. Dev. Biol. 147: 32–45 [DOI] [PubMed] [Google Scholar]
- Wang J., Liu X., Wu H., Ni P., Gu Z., et al. , 2010. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 38: 5366–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham A. T., Dike S., Cheng J., Manak J. R., Bell I., et al. , 2006. Transcriptional landscape of the human and fly genomes: nonlinear and multifunctional modular model of transcriptomes. Cold Spring Harb. Symp. Quant. Biol. 71: 101–110 [DOI] [PubMed] [Google Scholar]
- Yi G., Sze S. H., Thon M. R., 2007. Identifying clusters of functionally related genes in genomes. Bioinformatics 23: 1053–1060 [DOI] [PubMed] [Google Scholar]
- Yuan Q., Joiner W. J., Sehgal A., 2006. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr. Biol. 16: 1051–1062 [DOI] [PubMed] [Google Scholar]
- Zhang S. D., Odenwald W. F., 1995. Misexpression of the white (w) gene triggers male-male courtship in Drosophila. Proc. Natl. Acad. Sci. USA 92: 5525–5529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]