Abstract
Inactivation of the p53 tumour suppressor, either by mutation or by overexpression of its inhibitors Hdm2 and HdmX is the most frequent event in cancer. Reactivation of p53 by targeting Hdm2 and HdmX is therefore a promising strategy for therapy. However, Hdm2 inhibitors do not prevent inhibition of p53 by HdmX, which impedes p53-mediated apoptosis. Here, we show that p53 reactivation by the small molecule RITA leads to efficient HdmX degradation in tumour cell lines of different origin and in xenograft tumours in vivo. Notably, HdmX degradation occurs selectively in cancer cells, but not in non-transformed cells. We identified the inhibition of the wild-type p53-induced phosphatase 1 (Wip1) as the major mechanism important for full engagement of p53 activity accomplished by restoration of the ataxia telangiectasia mutated (ATM) kinase-signalling cascade, which leads to HdmX degradation. In contrast to previously reported transactivation of Wip1 by p53, we observed p53-dependent repression of Wip1 expression, which disrupts the negative feedback loop conferred by Wip1. Our study reveals that the depletion of both HdmX and Wip1 potentiates cell death due to sustained activation of p53. Thus, RITA is an example of a p53-reactivating drug that not only blocks Hdm2, but also inhibits two important negative regulators of p53 – HdmX and Wip1, leading to efficient elimination of tumour cells.
Keywords: p53, HdmX, RITA, Wip1, cancer
By regulating cell-cycle control, DNA repair and apoptosis, p53 serves as a major gatekeeper of tumour suppression. Upon oncogene activation, p53 is released from its negative regulators, Hdm2 and HdmX, becomes activated and induces transcription of its target genes.1 Hdm2 and HdmX are structurally alike and both inhibit the transcriptional activity of p53 by direct binding. In addition, Hdm2 possesses an intrinsic E3 ubiquitin ligase activity, which targets p53 for proteasomal degradation.1 Upon certain types of DNA damage Hdm2 autoubiquitinates itself and HdmX, leading to the degradation of both proteins,2, 3, 4, 5 allowing continuous p53 activation.
The function of p53 is impaired in the majority, if not all human cancers, either by mutation, or by functional deregulation of its inhibitors, Hdm2 and HdmX which are overexpressed in many tumours.6, 7 Major efforts aimed at p53 reactivation have been focused on targeting the Hdm2/p53 interaction,8, 9, 10 the enzymatic activity of Hdm2,11 or restoring mutant p53 function.12 Several lines of evidence highlight the importance of developing inhibitors towards HdmX, since downregulation of HdmX by means of siRNA in cancer cell lines induces p53, suppresses colony outgrowth13 and reduces tumour xenograft growth.14 Importantly, HdmX forms heterodimers with Hdm2, which enhances the E3 ligase activity of Hdm2, leading to decreased p53 levels.15 A recent study has demonstrated that the p53-reactivating compound nutlin3a has a 30-fold lower efficiency in disrupting the p53/HdmX complex compared with p53/Hdm2.16 Furthermore, activation of p53 after nutlin3a treatment is significantly enhanced upon downregulation of HdmX by siRNA in a number of human tumour cell lines,16 which is in line with the notion that downregulation of HdmX is necessary for complete p53 activation.17 There have been reports of short peptides inhibiting HdmX/p53;18 however, the clinical application of peptides is not optimal due to their proteolytic degradation and insufficient cellular uptake. Recently, the chemical inhibitor of HdmX/p53 interaction, SJ-172550 has been found, which displays additive effects in cancer cells when combined with an Hdm2 inhibitor.19
Wild-type p53-induced phosphatase 1 (Wip1), encoded by the PPM1D gene, is a serine/threonine phosphatase that exhibits oncogenic features. It is overexpressed in a vast number of tumours20 and is important for tumour stem cell survival.21 Wip1 is a direct transcriptional target of p5322 which inhibits p53, creating a negative feedback loop. Wip1 counteracts p53 activation following DNA damage on several levels, including reversion of the activating p53 phosphorylation at S1523 and stabilization of p53's negative regulators Hdm2 and HdmX by dephosphorylating sites that trigger their proteasomal degradation.24 Importantly, the two major DNA-damage sensors ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) are targeted by Wip1, blocking DNA-damage signalling.25 This interference with the DNA damage response confers resistance to standard treatments in tumours overexpressing Wip1 and therefore makes Wip1 an attractive target for anti-cancer therapy.26
Here, we report that p53 induced by RITA, a chemical activator of p53, efficiently counteracts the potent oncogenes Wip1 and HdmX by reducing their levels in cancer cells of different origin. This disables the p53/Wip1 negative feedback loop and prevents inhibition of p53 by HdmX, thus facilitating robust apoptosis induction.
Results
RITA-activated p53 induces depletion of HdmX in tumour cells
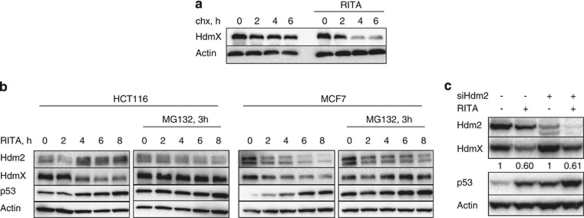
Comparison of the biological effects of two inhibitors of the p53/Hdm2 interaction, nutlin3a and RITA showed that in HCT116 and MCF7 cells, RITA induces apoptosis whereas nutlin3a triggers mainly growth arrest.27, 28 Several studies have demonstrated that HdmX impedes the induction of apoptosis upon inhibition of Hdm2 by nutlin3a in MCF7 cells. We found that HdmX is potently downregulated after RITA treatment, but not after nutlin3a treatment (Figures 1a and 5c). Kinetic analysis demonstrated that RITA treatment reduced HdmX protein levels in MCF7 and HCT116 cells in a time-dependent manner (Figure 1b). In addition, we observed oscillation of Hdm2 levels (Figures 1b and 3b), which is in line with our previous study.29 Several reports have demonstrated Hdm2 oscillation upon p53 activation by DNA damage due to opposing processes: transcriptional activation of its expression by p53 and enhanced proteasomal degradation.30
Figure 1.
HdmX protein levels are downregulated in a p53-dependent manner upon RITA treatment as assessed by western blotting. (a) HdmX protein levels were decreased in HCT116 and MCF7 cells 8 h after treatment with 1 μM RITA, but unchanged upon 10 μM nutlin3a or in control DMSO-treated cells. (b) The p53-positive cell lines HCT116 and MCF7 showed a continuous decline in HdmX protein levels over time, while HdmX was unaffected in the p53-null cells HCT116 TP53−/− after treatment with 1 μM RITA for the indicated time spans. (c) The p53-null cell lines H1299 and Saos-2 did not respond to RITA by HdmX downregulation. (d) Pretreatment with the p53-inhibitor pifithrin-α for 3 h prevented the decline of HdmX protein in HCT116 and MCF7 cells. (e) HCT116, HCT116 TP53−/− cells were injected into mice and the arising tumours were treated intra-tumourally with RITA (1 mg/kg). In line with cell culture results, HdmX protein was downregulated in the p53-positive, but not in the p53-null cells. (f) HdmX protein was not downregulated in the untransformed cell lines NHDF, MCF10A and 184A1 upon 8 h of 1 μM RITA treatment. (g) In a longer time-course HdmX protein levels were unchanged upon 1 μM RITA treatment in the untransformed NHDF cells
Next, we investigated whether the downregulation of HdmX was p53-dependent. We compared the effect of RITA in the wild-type p53-expressing HCT116 cells with that in the isogenic HCT116 p53-null derivative, HCT116 TP53−/−. In contrast to p53-positive cells, HdmX was not affected by RITA in p53-null cells (Figure 1b). Concordantly, HdmX was not depleted by RITA treatment in two other p53-null cell lines, H1299 and Saos-2 (Figure 1c), suggesting that the effect of RITA on HdmX is p53-dependent. This conclusion was further corroborated employing the previously described p53-inhibitor pifithrin-α (PFT-α),31 which abolished HdmX downregulation in HCT116 and MCF7 cells (Figure 1d). We extended our studies using a set of wild-type p53-expressing human tumour cell lines of different origin. The observed decline of HdmX levels upon RITA treatment was unambiguous in the majority of these cell lines (Figure 2a).
Figure 2.
Downregulation of HdmX contributes to RITA-induced apoptosis. (a) A panel of different cell lines was treated with RITA for 8 h, and HdmX as well as Wip1 was downregulated in all of them except RKO and SJSA, as detected by immunoblotting. (b) Assessment of the kinetics of HDMX levels by western blotting in RKO and SJSA cells did not detect downregulation of HdmX protein after treatment with 1 μM RITA. (c) Induction of apoptosis in different cell lines by 48 h of RITA treatment correlated with HdmX downregulation. The cells were stained with PI and the sub-G1 fraction representing apoptotic cells with fragmented DNA was quantified by FACS analysis (mean±S.E.M.; n=3). RKO and SJSA were the only cell lines that did not show a substantial increase of the sub-G1 fraction. (d) Depletion of HdmX by shRNA results in lower levels of HdmX upon RITA treatment. HCT116 and HCT116 shHdmX cells were treated with DMSO or RITA for the indicated time points and detected by immunoblotting. (e) Knockdown of HdmX by means of siRNA increased the efficiency of RITA to induce sub-G1 cells as assessed by FACS as in (c) in HCT116, HCT116 TP53−/− and HCT116 siHdmX. (f) HdmX depletion facilitates the killing of tumour cells by RITA, as detected using long-term viability assay. Cells were seeded at low density and exposed to 1 μM RITA for 24 h, the medium was changed and the cells were grown for 7 days. The colonies were then stained with crystal violet
To investigate the importance of our finding in a more physiological setting, we examined the level of HdmX upon RITA treatment in human xenografts grown in SCID mice. Notably, we detected a decline of HdmX in p53-positive HCT116, but not in HCT116 TP53−/− tumour xenografts (Figure 1e).
Since HdmX is important to keep p53 activity low in the absence of stress in untransformed cells, depletion of HdmX might lead to toxicity in normal tissues. Therefore, we tested whether the decline of HdmX upon RITA treatment occurs in untransformed cells. As previously reported, the p53 protein level is not elevated by RITA in untransformed cells,9, 27 and in line with that we did not detect significant changes in HdmX levels in untransformed cell lines, including normal human diploid fibroblasts (NHDF) and mammary epithelial cells, 184A1 and MCF10A (Figures 1f and g). Thus, HdmX downregulation requires p53 activation by RITA, which only occurs in oncogene-expressing cells.9, 27 Taken together, these results demonstrate that RITA induces p53-dependent HdmX downregulation exclusively in tumour cells.
Downregulation of HdmX correlates with RITA-induced apoptosis
HdmX levels after RITA treatment were examined in a panel of human tumour cell lines of different origin, including breast, lung and colon carcinoma, as well as lung fibrosarcoma and osteosarcoma. In the majority of the cell lines, we observed a decline of HdmX (as well as Wip1, which will be discussed later) after 8 h of RITA treatment, while the osteosarcoma SJSA and the colon carcinoma RKO were exceptions (Figure 2a). In these cells, HdmX levels did not decrease even after 24 h of treatment (Figure 2b).
Next, we tested whether there is a correlation between the decrease of HdmX and RITA sensitivity. In line with previously published data, we observed that the majority of wild-type p53-expressing human tumour cell lines underwent apoptosis after RITA treatment, as evidenced by the induction sub-G1 fraction (Figure 2c). However, we did not detect a significant induction of apoptosis in SJSA and RKO lines (Figure 2c). Thus, HdmX downregulation correlated with the apoptosis induction by RITA in human tumour cell lines.
The results presented above suggest that HdmX downregulation contributes to the efficient apoptosis induction by RITA. To test this further, we assessed whether depletion of HdmX by shRNA enhanced the biological effect of RITA. In line with Linares et al.,15 HdmX depletion led to an elevated p53 level. This results in stronger p53 induction upon RITA treatment (Figure 2d). Moreover, HdmX depletion significantly enhanced apoptosis induction by p53 upon treatment with RITA (Figure 2e). We also observed augmented growth suppression in a long-term growth suppression assay (Figures 2f and 6c). Taken together, these results clearly show that the potent depletion of HdmX contributes to robust apoptosis induction and thereby aids the anti-tumour effects conferred by RITA-activated p53.
The decrease of HdmX protein levels occurs through a proteasome-dependent mechanism independently of Hdm2
As HdmX is a long-lived protein (see Figure 3a), the reduction of its level observed already 4 h after treatment suggests enhanced protein degradation. Therefore, we examined whether RITA treatment leads to decreased HdmX protein stability. The half-life of HdmX was significantly shortened upon RITA treatment, indicating active degradation (Figure 3a). Next, we studied whether inhibition of the proteasome by MG132 could stabilize HdmX, and indeed the protein levels were rescued (Figure 3b).
Figure 3.
The reduction of HdmX levels is mediated by the proteasome and is Hdm2 independent. Protein levels were assessed by western blotting. (a) HdmX protein half-live is drastically decreased upon RITA treatment in MCF7 cells. Cells were treated with DMSO or RITA, followed by a cycloheximide chase. (b) HdmX degradation is proteasome dependent, since it is rescued by pretreatment with the proteasome-inhibitor MG132 for 3 h before RITA treatment of HCT116 and MCF7 cells. (c) HdmX degradation induced by RITA was not rescued by an siRNA-mediated knockdown of Hdm2 in HCT116 cells, as assessed by western blotting
Hdm2 is the most prominent negative regulator of HdmX. It is an E3 ligase that, upon certain signals, forms a heterodimer with HdmX, leading to its ubiquitination and subsequent degradation by the proteasome.4, 5, 32 To address whether Hdm2 is involved in HdmX downregulation after RITA treatment, we depleted Hdm2 by means of siRNA. As expected, HdmX and p53 protein levels were elevated upon Hdm2 knockdown. RITA-induced downregulation of HdmX levels was, however, not compromised in the absence of Hdm2 (Figure 3c). Similarly, inhibition of Hdm2 by overexpression of dominant negative Hdm2 mutants did not rescue the level of HdmX (data not shown). We conclude that RITA-induced downregulation of HdmX is proteasome-dependent, but is not mediated by Hdm2.
ATM is essential for HdmX downregulation by RITA
Several studies have demonstrated that DNA-damage signalling induces degradation of HdmX.4, 5 RITA has been shown to induce p53-dependent DNA damage signalling, including ATM activation.33 As ATM directly phosphorylates and thereby destabilizes HdmX, we tested whether ATM is involved in HdmX downregulation. ATM depletion did not influence p53 induction by RITA, suggesting that DNA damage signalling is downstream of p53 (Figure 4a). However, HdmX degradation was substantially inhibited (Figure 4a). Further, pretreatment with the selective ATM-inhibitor KU55933 partially prevented the degradation of HdmX (Figure 4b), suggesting ATM involvement. Since the rescue was incomplete, we cannot rule out the possibility of involvement of other kinases such as ATR.34
Figure 4.
The downregulation of HdmX is ATM dependent. Protein levels were assessed by immunoblotting. (a) HdmX degradation was prevented by siRNA-mediated ATM depletion prior to RITA treatment of HCT116 and MCF7 cells. (b) The ATM-inhibitor KU55933 partially prevented HdmX downregulation by RITA in HCT116 cells. (c) RITA treatment induces phosphorylation of ATM and its downstream target KAP-1 in HCT116 and MCF7 cells. (d) RITA-induced ATM phosphorylation was confirmed in HCT116 TP53+/+ cells by immunofluorescence. (e) HA-HdmX or the non-phosphorylatable mutant HA-HdmX-TM was expressed in MCF7 cells. After RITA treatment for 8 h, immunoprecipitation was performed using anti-HA or non-specific control antibodies. RITA induces phosphorylation of wild-type HdmX at S367 and S403 but not of HdmX-TM. (f) HdmX-TM that cannot be phosphorylated by ATM is protected from degradation induced by RITA treatment. HdmX levels were assessed in MCF7 cells stably expressing a non-phophorylatable triple mutant of HdmX pretreated with RITA for 3 h, followed by cycloheximide treatment for the indicated time spans
To confirm ATM involvement, we investigated its phosphorylation upon RITA treatment. ATM phosphorylation was increased, as demonstrated by western blotting and immunocytochemistry (Figures 4c and d), supporting the notion of ATM activation. In line with this, HdmX phosphorylation at S367 and S403 increased (Figure 4e). To further address the importance of these phosphorylations for RITA-induced HdmX degradation, we assessed whether mutation of serine residues can rescue HdmX levels. In contrast to wild-type HdmX (Figure 3a), the half-life of non-phosphorylatable triple mutant HdmX-TM carrying S342A, S367A and S403A substitutions did not decrease after RITA treatment (Figure 4f). These data demonstrate that signalling via ATM is essential for RITA-induced depletion of HdmX.
p53-dependent downregulation of Wip1 mRNA and protein
Activation of ATM in cancer cells is frequently neutralized by p53-activated Wip1,23 which inhibits ATM and p53 by removing activating phosphorylations.24 Microarray analysis of gene expression induced by RITA in MCF7 cells revealed potent downregulation of PPM1D, while treatment with nutlin3a led to its induction (Figure 5a). This was confirmed by quantitative real-time PCR (qPCR) (Figure 5b). In addition, treatment with the p53 activator 5-FU also resulted in PPM1D induction (Figure 5b). Consistent with these results, the level of Wip1 protein was induced by nutlin3a and 5-FU, and drastically decreased upon RITA treatment (Figure 5c).
Figure 5.
Wip1 is downregulated by RITA-reactivated p53 on mRNA and protein level. (a) Microarray analysis of MCF7 cells revealed the downregulation of PPM1D mRNA upon RITA treatment and upregulation upon nutlin3a treatment. MCF7 cells were treated with 1 μM RITA or 10 μM nutlin3a, or DMSO as a control, for the indicated times, mRNA was collected and resulting cDNA hybridized to Affimetrix microarrays. The microarray data were analysed using the computer language R. (b) Downregulation of PPM1D mRNA upon RITA and upregulation upon nutlin3a and 5-FU was detected by qPCR. HCT116 and MCF7 cells were treated with RITA, nutlin3a or 5-FU for 8 h and mRNA levels of PPM1D (Wip1) were assessed by qPCR (mean±S.E.M.; n=3). (c) The protein level of Wip1 (PPM1D) was downregulated in MCF7 cells treated with RITA, but upregulated by nutlin3a or 5-FU, as assessed by western blot after 8 h of treatment. (d) Wip1 was not downregulated in the two cell lines RKO and SJSA, as shown by western blotting. The cells were treated with RITA or DMSO as a control for the indicated time spans. (e) The downregulation of Wip1 is p53 dependent. HCT116 cells and their isogenic p53-null derivate HCT116TP53−/− were treated with DMSO or RITA for 8 h. Wip1 level was decreased only in the p53-positive cells
We found a strong correlation between the decrease of HdmX and Wip1 and the induction of apoptosis in the panel of cancer cell lines (Figures 2a and c). Importantly, neither HdmX nor Wip1 were downregulated after RITA treatment in RKO and SJSA cells, (Figures 2a and 5d). Similar to HdmX, the effect on Wip1 was p53 dependent, and was not observed in p53-null cells (Figure 5e).
Wip1 depletion results in HdmX downregulation and aids apoptosis induction by p53
The important role of Wip1 for maintaining a high level of HdmX was underscored by substantially decreased HdmX expression upon Wip1 knockdown by means of shRNA (Figure 6a). The HdmX level was further decreased upon RITA treatment, along with Wip1, most probably due to incomplete depletion of Wip1 by shRNA (Figure 6a).
Figure 6.
Wip1 depletion contributes to HdmX downregulation and p53-induced apoptosis. (a) Wip1 depletion results in low HdmX level, which was further decreased upon treatment with RITA, as assessed by western blotting. Wip1 was stably knocked down by lentiviral-expressed Wip1 shRNA. shRNA against GFP was used as a control. (b) Wip1 depletion leads to the destabilization of HdmX, as evidenced by the restoration of low HdmX levels in PPM1D knockdown cells by exposure to the proteasome-inhibitor MG132. Parental HCT116 cells were used as a reference. (c) Wip1 depletion sensitizes MCF7 cells to nutlin3a and as well as to RITA as tested by a growth suppression assay. Cells were seeded at low density, treated with 10 μM nutlin3a, 1 μM RITA or DMSO for 48 h and stained with crystal violet. (d) Overexpression of FLAG-Wip1 prevented downregulation of HdmX upon RITA treatment in U2OS cells. Fourty eight hours after transfection with Flag-Wip1 encoding or empty vector, cells were treated with RITA for 8 h to detect protein levels. (e) Model of p53-dependent downregulation of HdmX. (Left panel) p53 activation is blunted by a combination of two negative feedback loops, including the negative feedback between p53 and Hdm2/HdmX, and the negative feedback between p53 and ATM mediated by the phosphatase Wip1. Upon RITA treatment, repression of Wip1 disrupts both negative feedback loops, ultimately leading to ATM activation and HdmX downregulation and thus activating p53 by three different ways, namely (1) reversing the repression by Wip1, (2) abolishing the inhibition by HdmX and (3) inducing activation by ATM
Our results presented above (Figure 3b) showed clear proteasome dependence of RITA-induced HdmX downregulation. Thus, we addressed the question, whether HdmX decrease observed upon Wip1 depletion shows the same characteristics. Using parental HCT116 cells as a reference, we estimated that HCT116-Wip1 shRNA cells exhibit 65% of HdmX protein levels. Blocking the proteasome with MG132 up to 8 h gradually restored HdmX levels to 95% (Figure 6b).
Our results suggest that Wip1 inhibition is important for the p53-mediated biological response in cancer cells. As shown in Figure 6c, downregulation of Wip1 by means of shRNA facilitated the growth suppression effect of nutlin3a and RITA in MCF7 cells, assessed by a long-term growth suppression assay. Since Wip1 decrease seems to be crucial for HdmX degradation, we expressed ectopic Wip1 in U2OS cells. We found that Wip1 overexpression prevents HdmX degradation after RITA treatment (Figure 6d).
Taken together, our data show that Wip1 is efficiently downregulated in a p53-dependent manner upon RITA treatment. This has a crucial role in the decrease of HdmX protein levels and contributes to the efficient growth suppression by pharmacologically activated p53.
Discussion
Diverse alterations, which inactivate the p53 tumour suppressor function in cancers with non-mutated TP53, converge on two p53 inhibitors, Hdm2 and HdmX. Since HdmX and Hdm2 regulate p53 independently of each other and act synergistically, inhibition of both Hdm2 and HdmX is required for full p53 activation.35 This makes both proteins important targets for cancer therapy. Several small molecules inhibiting Hdm2 have been identified; however, these do not affect HdmX, impeding their efficiency, in particular in tumours with overexpressed HdmX.16 An inhibitor of the HdmX/p53 interaction, SJ-172550 has been described recently;19 it does not kill cancer cells on its own, but has an additive effect when combined with Hdm2 inhibitors. However, simultaneous targeting of Hdm2 and HdmX by a single molecule has not yet been reported.
In the present study, we found that in addition to preventing the p53/Hdm2 interaction, the small molecule RITA9 induces a potent destabilization of HdmX protein in cancer cells in vitro and in vivo, in a p53-dependent manner.
Our results suggest that active ATM is required for HdmX downregulation by RITA. Several studies have demonstrated that DNA damage signalling regulates HdmX and Hdm2 protein stability.3, 4 The checkpoint kinase ATM and its downstream kinase Chk2 induce phosphorylation of Hdm22 and HdmX,5 leading to their degradation. Our data showing partial rescue of HdmX level upon RITA treatment by both ATM knockdown and the ATM-inhibitor KU55933 supports the involvement of ATM in HdmX downregulation. This notion was further strengthened by our finding that the HdmX mutant that cannot be phosphorylated by ATM was rescued from RITA-induced degradation.
p53 induces DNA damage signalling upon activation by RITA.33 Notably, it appears that this p53 induction is not dependent on DNA damage signalling, since ATM inhibition did not prevent it. These data therefore suggest that induction of DNA damage signalling occurs downstream of p53.
Further elucidation of the factors involved in DNA damage signalling upon RITA treatment allowed us to identify the involvement of the Wip1 phosphatase in HdmX depletion. It has been reported that Wip1 can directly reverse ATM-mediated phosphorylation of HdmX, which leads to its stabilization.24 We observed substantial Wip1 downregulation on protein and mRNA levels upon p53 activation by RITA. Further, our results showed that inhibition of Wip1 has a causal role in the decrease of HdmX, suggesting that the low level of Wip1 helps to unleash HdmX degradation following p53 activation by RITA.
Based on our results we suggest a model (Figure 6e) in which inhibition of Wip1 by pharmacologically activated p53 creates a positive feedback loop uncoupling p53 from its negative regulators Hdm2 and HdmX. RITA-activated p53 represses the expression of its own inhibitor Wip1, which confers robust activation of p53 by acting on several levels. First, it restores active ATM, which phosphorylates p53, thereby activating it. Second, ATM-mediated phosphorylation of HdmX induces its degradation, negating HdmX-mediated inhibition of p53. Further, in the absence of Wip1 the activating phosphorylation of p53 and inhibitory phosphorylation of HdmX by ATM cannot be removed, which amplifies the signal, triggering full-scale p53 activation.
Normal cell homeostasis requires p53 activity to be kept at bay in the absence of stress. This is achieved by a combination of two negative feedback loops, the one between p53 and Hdm2/HdmX, and that between p53, ATM and Chk2, mediated by Wip1. Both these feedback loops are corrupted in cancers, as evidenced by overexpression and amplification of Hdm2, HdmX and Wip1 in numerous cancers that do not carry TP53 mutations. We demonstrate that pharmacological reactivation of p53 by RITA is able to counteract this imbalance and restore an appropriate p53 response, ultimately leading to apoptosis in tumour cells.
As inhibition of Wip1, Hdm2 and HdmX by a single small molecule is unprecedented, RITA might open a novel therapeutic approach for tumours that are dependent on the overexpression of these factors. In line with this notion, high HdmX levels have been shown to correlate with RITA sensitivity in a panel of breast cancer lines.36 Simultaneous inhibition of HdmX, Hdm2 and Wip1 might be an essential component for anti-cancer therapies targeting p53. This capability of p53 might allow us to manage with the daunting challenge of anti-cancer therapy – diverse genetic abnormalities in individual cancers.
It is worth noting that in accordance with the absence of p53 activation we did not observe inhibition of HdmX in non-transformed cells by RITA, suggesting that the possibility of toxic effects of RITA in normal cells due to HdmX inhibition is low.
Our finding that Wip1 was repressed upon RITA treatment in a p53-dependent manner is very intriguing. It implies that the p53-mediated negative feedback loops insuring survival of cells following stress signalling could be reversed under certain conditions. We have previously shown that RITA-induced p53 activation can block the pause-and-repair survival pathway mediated by induction of p21.28 In this study, we demonstrated that the p53/Wip1 negative feedback loop that negates p53 activation could be converted to a positive feedback loop that potentiates p53 activity. This opens a possibility to irreversibly shift the survival–death balance and to produce a robust apoptotic outcome, leading to elimination of tumour cells.
Notably, treatment with nutlin3a did not lead to downregulation of HdmX or Wip1 levels in HCT116 and MCF7 cells. In order to address the mechanism underlying the differential regulation of Wip1 by RITA and nutlin3a, that is, repression versus induction, respectively, we analysed the contribution of several p53 cofactors that are differentially regulated by RITA and nutlin3a, including hnRNP K28 and HIPK2.29 However, our data did not support the involvement of these factors (data not shown). An interesting example of a transcription factor, which can both activate and repress the same genes upon binding to their promoters is MyoD. Depending on its interaction with HDAC1 or LSD1 at the promoter of its target genes, MyoD can repress or activate genes, respectively.37, 38 A similar mechanism could be engaged in the switch between p53-mediated activation and repression of its target gene Wip1. Since there are >50 known p53 partner proteins, which can modulate p53-mediated regulation of gene expression, a high throughput proteomic approach is required to identify such a cofactor(s).
We showed that HdmX and Wip1 downregulation contribute to the induction of apoptosis by p53. As previously reported, nultin3a induces growth arrest in cell lines with high HdmX expression.28 Upon downregulation of HdmX by shRNA or by combinational treatment with DNA damaging drugs that induce HdmX degradation, nutlin3a induces apoptosis,16 suggesting that neutralization of the inhibitory effect of HdmX is crucial for the induction of apoptosis by p53. This notion is corroborated by our results showing that shRNA-mediated depletion of HdmX induced p53 levels and enhanced apoptosis upon RITA treatment. Further, Wip1 depletion facilitated growth suppression by nutlin3a, supporting the notion that simultaneous targeting of both Wip1 (this study) and HdmX16 is required for a full-scale induction of apoptosis upon inhibition of Hdm2.
Two cell lines in which RITA treatment did not induce apoptosis, namely, RKO and SJSA, showed neither Wip1 and HdmX downregulation, nor Hdm2 oscillation. It is plausible that certain undefined defects in the p53 pathway may account for the difference in response to RITA in these cell lines. Further studies will help to elucidate these issues.
In conclusion, our study provides several lines of evidence showing that in addition to blocking the inhibitory effects of Hdm2 on p53, activation of p53 by RITA simultaneously abrogates the two oncogenes, Wip1 and HdmX, creating a positive feedback loop, which potentiates apoptosis induction by p53. These findings open an approach for the development of anti-cancer drug(s) with a broader spectrum than drugs that target only Hdm2.
Materials and Methods
Cell lines and plasmids
The colon carcinoma cell line HCT116 and its p53-null derivative, HCT116 TP53−/−, were a gift from B Vogelstein, John Hopkins University, USA. The Wip1 shRNA encoding plasmid and the Flag-Wip1 encoding construct were a gift from René H Medema, Utrecht, The Netherlands. The siRNA for ATM was bought from Santa Cruz Biotechnologies (Santa Cruz, CA, USA). Knockdown for HdmX was performed as previously published.36
In vitro assays
Western blotting was performed according to the standard procedures. The proteins were detected using the following antibodies: anti-p53 DO-1 (Santa Cruz Biotechnologies, Santa Cruz, CA, USA), anti-Hdm2 2A10 supernatant (a gift from M Perry, NIH, Bethesda, MA, USA), anti-HdmX, anti-p403-HdmX, KAP1, pKAP1 (all from Bethyl Laboratories, Montgomery, TX, USA), anti-Wip1, anti-ATM mat3 (kind gift from Yosef Shiloh), anti pATM (Rockland Immunochemicals, Gilbertsville, PA, USA), anti-pS367-HdmX (kind gift of Koji Okamoto) and anti-actin AC-15 (Sigma, Stockholm, Sweden). The p53-inhibitor, PFT-α, a gift from A Gudkov, Roswell Park Cancer Institute, USA, was used at 10 μM for 3 h prior to RITA treatment. Protein degradation was inhibited by using 30 μM of the proteasomal-inhibitor MG132 (Sigma). To inhibit ATM signalling, cells were pretreated with 10 μM KU55933 (Tocris Bioscience, Bristol, UK) for 2 h. HdmX half-life was detected by blocking protein synthesis using cycloheximide (chx) (Sigma). Cells were pretreated with 1 μM RITA for 3 h, incubated with 30 μg/ml chx and harvested at indicated time points after chx treatment. For densitometrical analysis, the programme Image J was used.
Animal experiments
The Northern Stockholm Animal Ethical Committee approved all animal studies, and animal care was in accordance with Karolinska Institute guidelines. Animal experiments were performed as described in Grinkevich et al.27
Affimetrix microarrays
MCF7 cells were treated with DMSO, 1 μM RITA or 10 μM nutlin3a for the indicated time. Raw data from the hgu219 chips were normalized using the robust multiarray average procedure, implemented in the R package affy.39 Per gene hypothesis testing was performed using Benjamini and Hochberg method implemented in R package LIMMA using linear models for microarray data.40
Quantitative PCR
Cells were treated with 1 μM RITA, 10 μM nutlin3a (Merck, Darmstadt, Germany) or 100 μM 5-FU (Sigma) for 8 h. mRNA was isolated and reverse transcribed to cDNA according to the manufacturer's instructions (5 Prime, Hamburg, Germany; Invitrogen Stockholm, Sweden). For qPCR, the following concentrations were used: 150 nM primers; 10 ng cDNA; 7.5 μl 2 × master mix (Bio-Rad, Sundbyberg, Sweden); water to a total of 15 μl; Primers used: GAPDH forward 5′-TCA-TTT-CCT-GGT-ATG-ACA-ACG-3′ reverse 5′-ATG-TGG-GCC-ATG-AGG T-3′ PPM1D forward 5′-ATA-CCT-GAA-CCT-GAC-TGA-C-3′ reverse 5′-CTC-CTC-CAG-TGA-CTT-GAC-3′ data were analysed using the Bio-RadiQ5 system software (Bio-Rad, Sundbyberg, Sweden).
Growth suppression assays
For fluorescence-activated cell sorting (FACS) analysis of sub-G1 fraction, cells were seeded in six-well plates and treated with RITA. After 48 h, cells were harvested by trypsinization, fixed with 70% ethanol, treated with RNaseA (0.25 mg/ml) and stained with propidium iodide (0.02 mg/ml).
For long-term viability assay, 200 cells/well were seeded into six-well plates and treated with 1 μM RITA or nutlin3a for 24 h, the medium was changed and cells were allowed to grow for 7 days, unless indicated otherwise. The colonies were stained with crystal violet.
Acknowledgments
This study was funded by the Swedish Cancer Society, the Swedish Research Council, the Cancer Society of Stockholm, and supported by EC/FP6 projects ‘Mutant p53', ‘Active p53' and ‘Net2Drug'. We are indebted to all colleagues who shared reagents and cell lines with us, and grateful to Y Shi for technical assistance.
Glossary
- RITA
reactivation of p53 and induction of tumour cell apoptosis
- 5-FU
5-fluorouracil
- Wip1
wild-type p53-induced phosphatase 1
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related
- qPCR
quantitative real-time reverse transcription polymerase chain reaction
- FACS
fluorescent-activated cell sorting
- Chx
cycloheximide
- DMSO
dimethyl sulfoxide
- PFT-α
pifithrin-α
- NHDF
normal human diploid fibroblasts
- Hdm2
human double minute 2
- HdmX
human double minute X
- siRNA
short inhibitory ribonucleic acid
- shRNA
short hairpin ribonucleic acid
- mRNA
messenger ribonucleic acid
- DNA
desoxyribonucleic acid
- cDNA
copy desoxyribonucleic acid
- nM
nanomolar
- μM
micromolar
- mM
millimolar
- h
hour
The authors declare no conflict of interest.
Footnotes
Edited by M Oren
References
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci USA. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci USA. 2005;102:5056–5061. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 2005;24:3411–3422. doi: 10.1038/sj.emboj.7600812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos YF, Stad R, Attema J, Peltenburg LT, van der Eb AJ, Jochemsen AG. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res. 2001;61:1839–1842. [PubMed] [Google Scholar]
- Riemenschneider MJ, Buschges R, Wolter M, Reifenberger J, Bostrom J, Kraus JA, et al. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 1999;59:6091–6096. [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- Hedstrom E, Issaeva N, Enge M, Selivanova G. Tumor-specific induction of apoptosis by a p53-reactivating compound. Exp Cell Res. 2009;315:451–461. doi: 10.1016/j.yexcr.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JT, Mayo LD, Singhi AD, Gudkov AV, Stark GR, Jackson MW. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 2006;66:3169–3176. doi: 10.1158/0008-5472.CAN-05-3832. [DOI] [PubMed] [Google Scholar]
- Wang YV, Wade M, Wong E, Li YC, Rodewald LW, Wahl GM. Quantitative analyses reveal the importance of regulated Hdmx degradation for p53 activation. Proc Natl Acad Sci USA. 2007;104:12365–12370. doi: 10.1073/pnas.0701497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazgier M, Liu M, Zou G, Yuan W, Li C, Li C, et al. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc Natl Acad Sci USA. 2009;106:4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed D, Shen Y, Shelat AA, Arnold LA, Ferreira AM, Zhu F, et al. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem. 2010;285:10786–10796. doi: 10.1074/jbc.M109.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Nguyen TA, Moon SH, Darlington Y, Sommer M, Donehower LA. The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008;27:123–135. doi: 10.1007/s10555-008-9127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov ON, Timofeev O, Lwin HN, Kek C, Appella E, Bulavin DV. Wip1 phosphatase regulates p53-dependent apoptosis of stem cells and tumorigenesis in the mouse intestine. Cell Stem Cell. 2007;1:180–190. doi: 10.1016/j.stem.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, et al. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci USA. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lin L, Guo H, Yang J, Jones SN, Jochemsen A, et al. Phosphorylation and degradation of MdmX is inhibited by Wip1 phosphatase in the DNA damage response. Cancer Res. 2009;69:7960–7968. doi: 10.1158/0008-5472.CAN-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Nguyen TA, Donehower LA. Reversal of the ATM/ATR-mediated DNA damage response by the oncogenic phosphatase PPM1D. Cell Cycle. 2005;4:1060–1064. [PubMed] [Google Scholar]
- Belova GI, Demidov ON, Fornace AJ, Jr, Bulavin DV. Chemical inhibition of Wip1 phosphatase contributes to suppression of tumorigenesis. Cancer Biol Ther. 2005;4:1154–1158. doi: 10.4161/cbt.4.10.2204. [DOI] [PubMed] [Google Scholar]
- Grinkevich V, Nikulenkov F, Shi Y, Enge M, Bao W, Maljukova A, et al. Ablation of key oncogenic pathways by RITA-reactivated p53 is required for efficient apoptosis. Cancer Cell. 2009;15:441–453. doi: 10.1016/j.ccr.2009.03.021. [DOI] [PubMed] [Google Scholar]
- Enge M, Bao W, Hedstrom E, Jackson SP, Moumen A, Selivanova G. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell. 2009;15:171–183. doi: 10.1016/j.ccr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Rinaldo C, Prodosmo A, Siepi F, Moncada A, Sacchi A, Selivanova G, et al. HIPK2 regulation by MDM2 determines tumor cell response to the p53-reactivating drugs nutlin-3 and RITA. Cancer Res. 2009;69:6241–6248. doi: 10.1158/0008-5472.CAN-09-0337. [DOI] [PubMed] [Google Scholar]
- Lev Bar-Or R, Maya R, Segel LA, Alon U, Levine AJ, Oren M. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc Natl Acad Sci USA. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- de Graaf P, Little NA, Ramos YF, Meulmeester E, Letteboer SJ, Jochemsen AG. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem. 2003;278:38315–38324. doi: 10.1074/jbc.M213034200. [DOI] [PubMed] [Google Scholar]
- Yang J, Ahmed A, Poon E, Perusinghe N, de Haven Brandon A, Box G, et al. Small-molecule activation of p53 blocks hypoxia-inducible factor 1alpha and vascular endothelial growth factor expression in vivo and leads to tumor cell apoptosis in normoxia and hypoxia. Mol Cell Biol. 2009;29:2243–2253. doi: 10.1128/MCB.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12:342–354. doi: 10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Lam S, Lodder K, Teunisse AF, Rabelink MJ, Schutte M, Jochemsen AG. Role of Mdm4 in drug sensitivity of breast cancer cells. Oncogene. 2010;29:2415–2426. doi: 10.1038/onc.2009.522. [DOI] [PubMed] [Google Scholar]
- Choi J, Jang H, Kim H, Kim ST, Cho EJ, Youn HD. Histone demethylase LSD1 is required to induce skeletal muscle differentiation by regulating myogenic factors. Biochem Biophys Res Commun. 2010;401:327–332. doi: 10.1016/j.bbrc.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK.Linear models and empirical bayes methods for assessing differential expression in microarray experiments Stat Appl Genet Mol Biol 20043(Article 3). [DOI] [PubMed] [Google Scholar]