Abstract
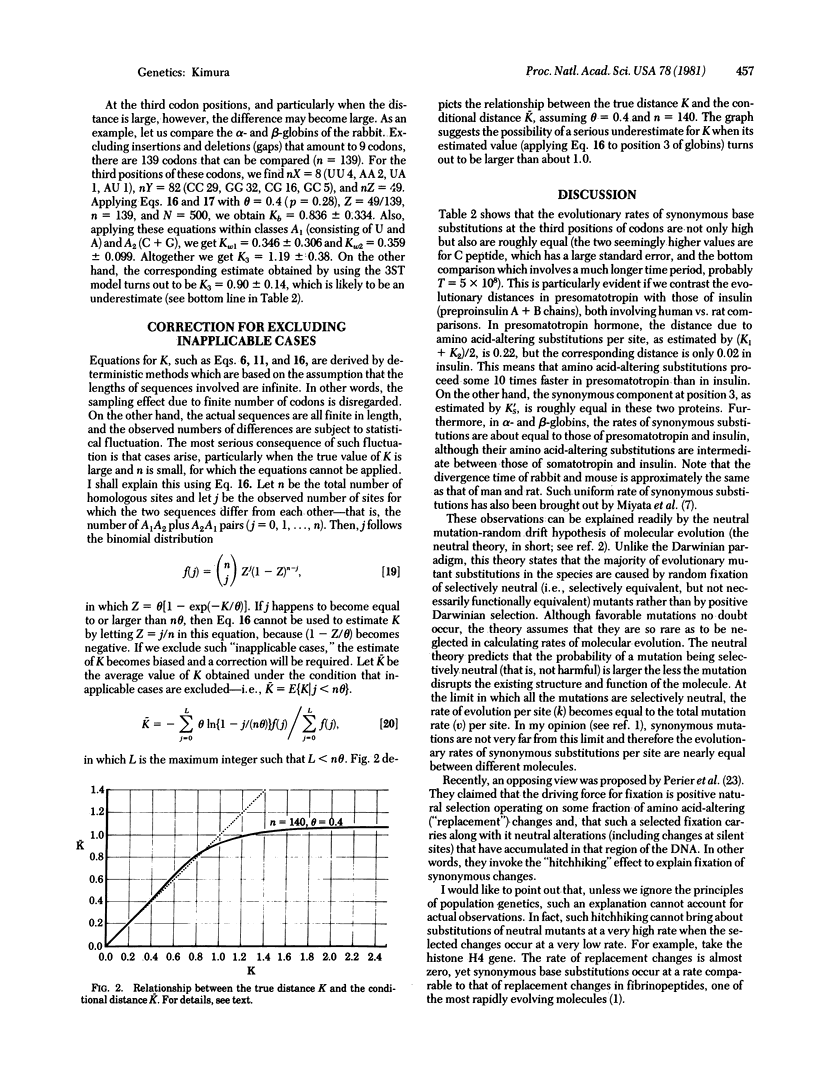
By using two models of evolutionary base substitutions--"three-substitution-type" and "two-frequency-class" models--some formulae are derived which permit a simple estimation of the evolutionary distances (and also the evolutionary rates when the divergence times are known) through comparative studies of DNA (and RNA) sequences. These formulae are applied to estimate the base substitution rates at the first, second, and third positions of codons in genes for presomatotropins, preproinsulins, and alpha- and beta-globins (using comparisons involving mammals). Also, formulae for estimating the synonymous component (at the third codon position) and the standard errors are obtained. It is pointed out that the rates of synonymous base substitutions not only are very high but also are roughly equal to each other between genes even when amino acid-altering substitution rates are quite different and that this is consistent with the neutral mutation-random drift hypothesis of molecular evolution.
Full text
PDF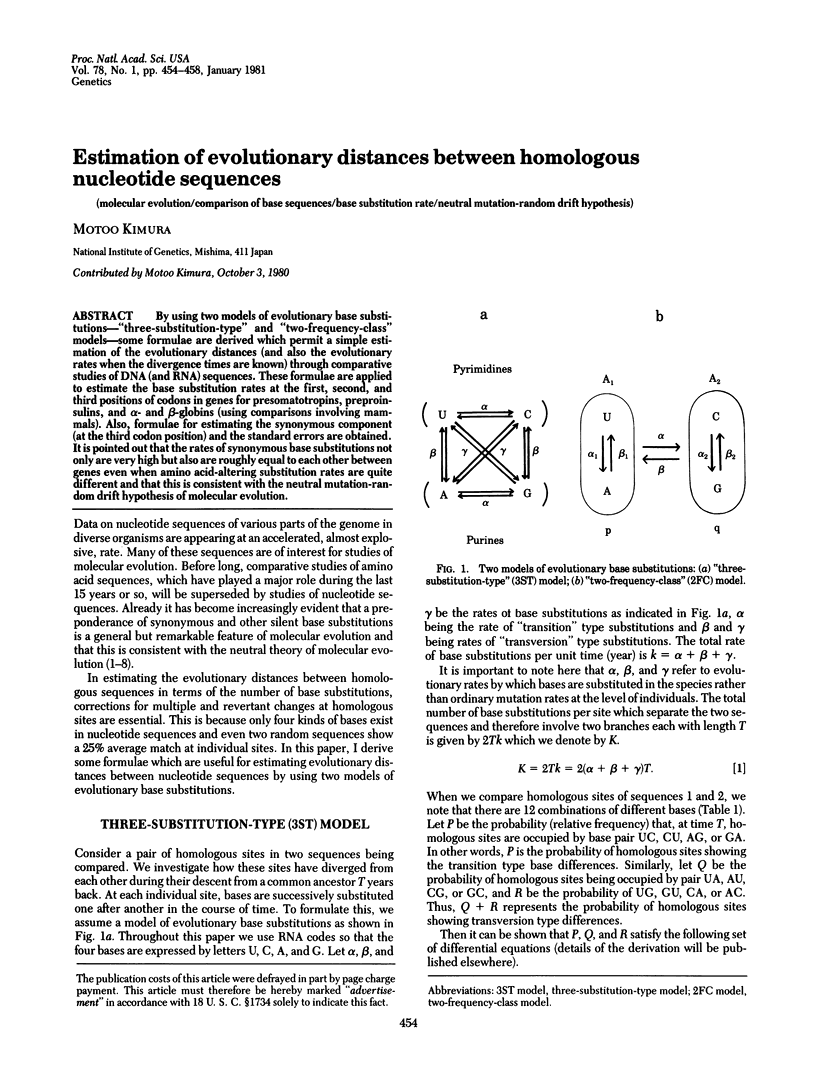



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell G. I., Pictet R. L., Rutter W. J., Cordell B., Tischer E., Goodman H. M. Sequence of the human insulin gene. Nature. 1980 Mar 6;284(5751):26–32. doi: 10.1038/284026a0. [DOI] [PubMed] [Google Scholar]
- Cordell B., Bell G., Tischer E., DeNoto F. M., Ullrich A., Pictet R., Rutter W. J., Goodman H. M. Isolation and characterization of a cloned rat insulin gene. Cell. 1979 Oct;18(2):533–543. doi: 10.1016/0092-8674(79)90070-9. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Kafatos F. C., Maniatis T. The primary structure of rabbit beta-globin mRNA as determined from cloned DNA. Cell. 1977 Apr;10(4):571–585. doi: 10.1016/0092-8674(77)90090-3. [DOI] [PubMed] [Google Scholar]
- Heindell H. C., Liu A., Paddock G. V., Studnicka G. M., Salser W. A. The primary sequence of rabbit alpha-globin mRNA. Cell. 1978 Sep;15(1):43–54. doi: 10.1016/0092-8674(78)90081-8. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., King J. L. Evolutionary nucleotide replacements in DNA. Nature. 1979 Oct 18;281(5732):605–606. doi: 10.1038/281605a0. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. Neutral changes during divergent evolution of hemoglobins. J Mol Evol. 1978 Aug 2;11(3):267–269. doi: 10.1007/BF01734488. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. Silent nucleotide substitutions and the molecular evolutionary clock. Science. 1980 Nov 28;210(4473):973–978. doi: 10.1126/science.7434017. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. Silent nucleotide substitutions during evolution. Naturwissenschaften. 1980 Nov;67(11):534–539. doi: 10.1007/BF00450662. [DOI] [PubMed] [Google Scholar]
- Kimura M., Ota T. On the stochastic model for estimation of mutational distance between homologous proteins. J Mol Evol. 1972 Dec 29;2(1):87–90. doi: 10.1007/BF01653945. [DOI] [PubMed] [Google Scholar]
- Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977 May 19;267(5608):275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Konkel D. A., Tilghman S. M., Leder P. The sequence of the chromosomal mouse beta-globin major gene: homologies in capping, splicing and poly(A) sites. Cell. 1978 Dec;15(4):1125–1132. doi: 10.1016/0092-8674(78)90040-5. [DOI] [PubMed] [Google Scholar]
- Lomedico P., Rosenthal N., Efstratidadis A., Gilbert W., Kolodner R., Tizard R. The structure and evolution of the two nonallelic rat preproinsulin genes. Cell. 1979 Oct;18(2):545–558. doi: 10.1016/0092-8674(79)90071-0. [DOI] [PubMed] [Google Scholar]
- Martial J. A., Hallewell R. A., Baxter J. D., Goodman H. M. Human growth hormone: complementary DNA cloning and expression in bacteria. Science. 1979 Aug 10;205(4406):602–607. doi: 10.1126/science.377496. [DOI] [PubMed] [Google Scholar]
- Miyata T., Yasunaga T., Nishida T. Nucleotide sequence divergence and functional constraint in mRNA evolution. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7328–7332. doi: 10.1073/pnas.77.12.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols B. P., Yanofsky C. Nucleotide sequences of trpA of Salmonella typhimurium and Escherichia coli: an evolutionary comparison. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5244–5248. doi: 10.1073/pnas.76.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka Y., Leder A., Leder P. Unusual alpha-globin-like gene that has cleanly lost both globin intervening sequences. Proc Natl Acad Sci U S A. 1980 May;77(5):2806–2809. doi: 10.1073/pnas.77.5.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perler F., Efstratiadis A., Lomedico P., Gilbert W., Kolodner R., Dodgson J. The evolution of genes: the chicken preproinsulin gene. Cell. 1980 Jun;20(2):555–566. doi: 10.1016/0092-8674(80)90641-8. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Shine J., Martial J. A., Baxter J. D., Goodman H. M. Nucleotide sequence and amplification in bacteria of structural gene for rat growth hormone. Nature. 1977 Dec 8;270(5637):486–494. doi: 10.1038/270486a0. [DOI] [PubMed] [Google Scholar]
- Sures I., Goeddel D. V., Gray A., Ullrich A. Nucleotide sequence of human preproinsulin complementary DNA. Science. 1980 Apr 4;208(4439):57–59. doi: 10.1126/science.6927840. [DOI] [PubMed] [Google Scholar]
- van Ooyen A., van den Berg J., Mantei N., Weissmann C. Comparison of total sequence of a cloned rabbit beta-globin gene and its flanking regions with a homologous mouse sequence. Science. 1979 Oct 19;206(4416):337–344. doi: 10.1126/science.482942. [DOI] [PubMed] [Google Scholar]


