Abstract
Chromosomal replication is initiated from the replication origin oriC in Escherichia coli by the active ATP-bound form of DnaA protein. The regulatory inactivation of DnaA (RIDA) system, a complex of the ADP-bound Hda and the DNA-loaded replicase clamp, represses extra initiations by facilitating DnaA-bound ATP hydrolysis, yielding the inactive ADP-bound form of DnaA. However, the mechanisms involved in promoting the DnaA-Hda interaction have not been determined except for the involvement of an interaction between the AAA+ domains of the two. This study revealed that DnaA Leu-422 and Pro-423 residues within DnaA domain IV, including a typical DNA-binding HTH motif, are specifically required for RIDA-dependent ATP hydrolysis in vitro and that these residues support efficient interaction with the DNA-loaded clamp·Hda complex and with Hda in vitro. Consistently, substitutions of these residues caused accumulation of ATP-bound DnaA in vivo and oriC-dependent inhibition of cell growth. Leu-422 plays a more important role in these activities than Pro-423. By contrast, neither of these residues is crucial for DNA replication from oriC, although they are highly conserved in DnaA orthologues. Structural analysis of a DnaA·Hda complex model suggested that these residues make contact with residues in the vicinity of the Hda AAA+ sensor I that participates in formation of a nucleotide-interacting surface. Together, the results show that functional DnaA-Hda interactions require a second interaction site within DnaA domain IV in addition to the AAA+ domain and suggest that these interactions are crucial for the formation of RIDA complexes that are active for DnaA-ATP hydrolysis.
Keywords: DNA Replication, Multifunctional Protein, Protein Assembly, Protein-DNA Interaction, Protein Domains, Protein-Protein Interactions, AAA+ Family, Regulation of DNA Replication
Introduction
The initiation of chromosomal replication is strictly controlled to ensure that chromosomal DNA is replicated only once per cell cycle in all cellular organisms. In Escherichia coli, ATP-bound DnaA (ATP-DnaA), but not ADP-bound DnaA (ADP-DnaA), forms a specific multimeric complex on the chromosomal replication origin (oriC) to form an initiation complex (1, 2). DiaA, a DnaA-binding protein that promotes DnaA multimerization and subsequent duplex unwinding within oriC, facilitates formation of an open complex (3). DnaB helicase is then loaded onto the unwound region, in a DnaA- and DnaC-mediated manner, which expands the single-stranded region to allow loading of DnaG primase and the DNA polymerase III holoenzyme (4–6). The holoenzyme consists of subcomplexes of the core, clamp (β subunit homodimer), and clamp loader (DnaX complex). The clamp forms a ring-shaped structure and is loaded onto the primed DNA by the DnaX complex to tether the core on DNA during DNA synthesis. Only the clamp remains on the nascent DNA after Okazaki fragment synthesis, and the core and the DnaX complex are recycled (4).
E. coli cells have at least four systems to prevent extra DNA replication initiations (7, 8). The first is the timely sequestration of oriC from ATP-DnaA by SeqA (9–12), the second is DnaA titration by the chromosomal datA locus (13), and the third is transcriptional regulation of the dnaA gene (14–16). The fourth system is called regulatory inactivation of DnaA (RIDA).2 Here, a nucleoprotein complex consisting of the DNA-loaded clamp and the ADP-bound Hda protein promotes the hydrolysis of DnaA bound-ATP (DnaA-ATP) to yield inactive ADP-DnaA (see Fig. 1A) (17–19). The requirement for the DNA-loaded clamp in RIDA ensures timely inactivation of DnaA coupled with the progress of the chromosomal replication cycle (17, 20). The cellular level of ATP-DnaA increases prior to initiation and decreases during chromosomal replication (21). Introduction of an hda-deficient mutant allele or a RIDA-resistant dnaA mutant allele results in a higher level of cellular ATP-DnaA and extra DNA replication initiations (18, 22–25).
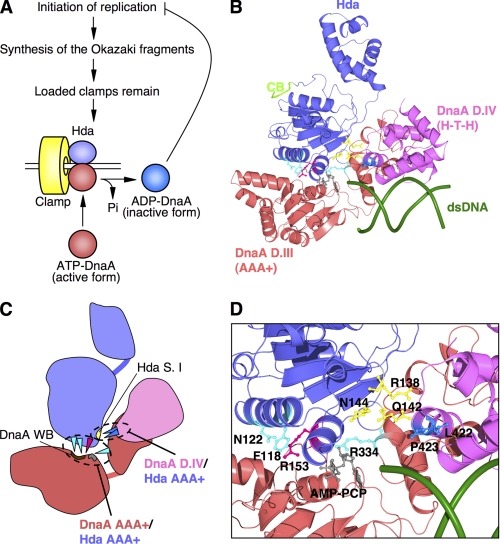
FIGURE 1.
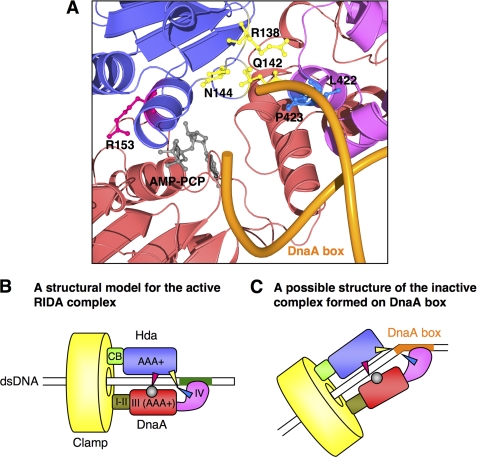
Structural model of Hda·DnaA domains III-IV·dsDNA complex. A, a simplified RIDA model. ATP-DnaA, the initiation-active form, is hydrolyzed by Hda complexed with the DNA-loaded clamp during DNA replication, yielding the inactivated ADP-DnaA, which results in repression of extra DNA replication initiations. Hda (purple), clamp (yellow), ATP-DnaA (red), and ADP-DnaA (blue) are indicated. Pi, inorganic phosphate. B, a structural model of E. coli DnaA domain III (DnaA D.III) was obtained from SWISS-MODEL Workspace using an AMPPCP-bound DnaA structure (Protein Data Bank code 2HCB). Crystal structures of E. coli DnaA domain IV (DnaA D.IV) and a 12-mer dsDNA were obtained from the Protein Data Bank (codes 1J1V and 1D28, respectively). A DnaA domain IV·dsDNA complex was constructed using the structure of an E. coli DnaA domain IV·DnaA box complex. A structural model of E. coli Hda was obtained from SWISS-MODEL Workspace using an S. amazonensis Hda structure (Protein Data Bank code 3BOS). The Hda (purple)·DnaA domain III (red)·domain IV (pink)·dsDNA (green) complex model was constructed using the AMPPCP-bound DnaA oligomer structure with MolFeat software (FiatLux). The indicated amino acid residues and AMPPCP are shown as ball-and-stick models. The Hda Arg finger (Arg-153) and AMPPCP are red and gray, respectively. Hda Phe-118, Asn-122, and DnaA sensor II Arg-334 residues are cyan. Hda Arg-138, Gln-142, and Asn-144 residues are yellow. DnaA Leu-422 and Pro-423 residues are blue. The clamp-binding motif (CB) of Hda is light green. C, schematic representation of B. DnaA AAA+-Hda AAA+ interaction and DnaA domain IV-Hda AAA+ interaction regions are indicated. The DnaA Walker B (WB) motif is brown. The Hda sensor I (S. I) motif, including Hda Arg-138, Gln-142, and Asn-144 residues, is yellow. D, a close-up representation of the interaction of DnaA-Hda amino acid residues as given in B.
The fundamental concept of RIDA, which provides clamp-dependent feedback regulation of replication initiation, is conserved in other bacteria and eukaryotes (8). In Bacillus subtilis, which is evolutionarily distant from E. coli, the YabA protein, a functional homologue of Hda, forms multimeric complexes containing the cognate DnaA and clamp at replication forks and represses extra DNA replication initiations (26–28). In addition, HdaA, an Hda orthologue of Caulobacter crescentus, colocalizes with the clamp, which results in the repression of extra DNA replication initiations (29). Cdt1, a eukaryotic initiation protein, is required for formation of prereplicative complexes at origin sites to load the MCM2–7 replicative helicase (7). Cdt1 protein is ubiquitylated for targeted degradation by complexes of the DNA-loaded clamp (proliferating cell nuclear antigen) and the E3 ubiquitin ligase cullin 4·DNA damage-binding protein 1 complex, thereby repressing reinitiation of replication (30–32).
E. coli Hda harbors a conserved clamp-binding motif (QL(S/D)LF) at its N terminus (33, 34) that is required for clamp binding and RIDA (34, 35). In addition to this motif, Hda contains an AAA+ domain with the Walker-type nucleotide-binding motif and several conserved amino acid sequence motifs (18, 36). Hda AAA+ does not bind ATP but specifically and stably binds to ADP to yield the activated monomeric form of Hda (19).
DnaA consists of four functional domains (1). The N-terminal domain I interacts with several proteins, including DiaA and DnaB (37–39). Domain II is a flexible linker (40, 41). Domain III is an AAA+ domain that shares amino acid sequence similarity with Hda (18, 36). The C-terminal domain IV is a DNA-binding region that contains a helix-turn-helix motif (42–44). This domain specifically binds to the 9-mer DnaA boxes that are present at oriC, datA locus, and many other sites on the chromosome.
A model of the RIDA intermediate complex in which DnaA domain I interacts with the Hda·clamp complex, DnaA domain IV interacts with the DNA flanking the clamp, and DnaA domain III interacts with the Hda AAA+ domain was proposed (20, 22, 34, 45). Several amino acid residues within the Hda AAA+ Box VI and Box VII motifs, such as Hda Arg-153 (Arg finger), Phe-118 (H-finger), and Asn-122 (E-finger), play a role in DnaA-ATP hydrolysis and in the DnaA-Hda interaction (see Fig. 1, B–D). In addition, DnaA Arg-334 within the AAA+ sensor II motif is important for DnaA-intrinsic ATPase activity, the Hda interaction, and DnaA-ATP hydrolysis in RIDA (22, 45). In a model of the DnaA·Hda complex, DnaA Arg-334 resides near the Hda AAA+ domain and the nucleotide-interacting pocket of the DnaA AAA+ domain (see Fig. 1, B–D) (22, 45). However, interactions between Hda and DnaA in the RIDA system have not been identified except for those between the AAA+ domains of the two (45). As only a limited region containing the Hda Arg finger plays a crucial role in specific interactions with DnaA domain III (45), a region in another DnaA domain might also be required for sustaining strict specificity in binding to Hda.
A structural model for a DnaA·Hda·DNA complex was constructed, and potential interactions between DnaA domain IV and Hda were identified. DnaA Leu-422 and Pro-423 residues within DnaA domain IV were required for RIDA activity and interaction with Hda in vitro, which is consistent with the results of in vivo analyses. By contrast, these amino acid residues did not play a crucial role in the process of DNA replication initiation. These findings suggest that cross-talk between DnaA domain IV and the Hda AAA+ domain is required for the formation of an active RIDA complex.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
The E. coli K12 derivatives KH5402-1 (thyA) and WM433 (dnaA204) have been described previously (17, 22). YT411 (rnhA::cat), KA451 (dnaA::Tn10 rnhA::cat), KA429 (rnhA::cat ΔoriC1071::Tn10), and KA450 (ΔoriC1071::Tn10 dnaA17 (Am) rnhA199 (Am)) are derivatives of KH5402-1 (22). KP7364 (ΔdnaA::spec rnhA::kan) has been described (17). The plasmids pKA234, pING1, M13E10, M13KEW101, pBSoriC, pSN306, and pHSL99 were described previously (22, 46). For the construction of pL422A, pL422G, and pP423A plasmids, a base substitution was introduced into the wild-type dnaA allele carried on pKA234 using a QuikChange site-directed mutagenesis kit (Stratagene) using pairs of mutagenic primers (for L422A, 5′-GAGCTGACTAACCACAGCGCTCCGGAGATTGGCGATGCG-3′ and its complementary strand; for L422G, 5′-GAGCTGACTAACCACAGTGGGCCCGAGATTGGCGATGCG-3′ and its complementary strand; and for P423A, 5′-GCGAAAGAGCTGACTAACCACTCGCTAGCGGAGATTGGCGATGCG-3′ and its complementary strand). For the construction of pSNL422A, pSNL422G, and pSNP423A plasmids, a base substitution was introduced into the wild-type dnaA allele carried on pSN306 as described above.
Purification of Mutant DnaA Proteins
Mutant DnaA proteins were purified from KA450 cells bearing pL422A, pL422G, or pP423A as described previously for wild-type and other mutant DnaA proteins (39).
ATP and ADP Binding Assays
The ATP and ADP binding activities of DnaA proteins were determined by a filter retention assay as described previously (39).
Reconstituted RIDA System
The reconstituted staged RIDA assay was performed essentially as described previously (19). First, the DNA-loaded clamps were isolated using a gel filtration spin column as described previously (34). Next, [α-32P]ATP-DnaA (0.25 pmol) was incubated at 30 °C for 20 min in the presence of 30 μm ADP, 10 ng of the isolated DNA-loaded clamps, and the indicated amounts of the C-terminal hexahistidine-fused Hda (Hda-cHis) in RIDA buffer (12.5 μl) containing 20 mm Tris-HCl (pH 7.5), 8 mm dithiothreitol, 8 mm magnesium acetate, 0.01% Brij-58, 10% glycerol, 0.1 mg/ml bovine serum albumin, and 120 mm potassium glutamate. Nucleotides bound to DnaA were recovered on a nitrocellulose filter, separated by thin-layer chromatography, and quantified by a BAS2500 imaging analyzer (Fujifilm).
Intrinsic ATPase Activity
The DNA-dependent intrinsic ATPase activity of DnaA proteins was assessed as described previously (34). Briefly, [α-32P]ATP-DnaA (0.5 pmol) was incubated at 30 °C for the indicated time in RIDA buffer (25 μl) containing 15 ng of φX174 replicative form II DNA (4.3 fmol as a circle). Nucleotides bound to DnaA were monitored by thin-layer chromatography as described for the RIDA reaction.
DNA Binding Activity
The DNA binding activity of DnaA proteins was determined by surface plasmon resonance (SPR) analysis as described previously (46, 47). Binding of DnaA to DnaA box was performed as described previously (46). For analysis of DnaA binding to a nonspecific DNA, biotinylated dsDNA bearing a sequence that has no specific affinity for DnaA (48) was prepared by PCR using the primers 5′-bio-TCCTTGAACTATATCGGGCAG-3′ (where bio is biotin) and its non-biotinylated complementary strand. This DNA (∼500 resonance units) was immobilized on the sensor chip SA (Biacore) according to the manufacturer's instructions. DnaA protein was incubated on ice for 15 min in buffer containing 25 mm Hepes-KOH (pH 7.5), 150 mm potassium acetate, 1 mm magnesium acetate, 0.005% Surfactant P20 (Biacore), 0.1 mm ATP, 0.025 μg/ml poly(dA-dT)·poly(dA-dT), and 0.025 μg/ml poly(dI-dC)·poly(dI-dC). A sample was injected into the Biacore X flow cells filled with the same buffer at 22 °C at a flow rate 40 μl/min.
DnaA-Hda Interaction Using Pulldown Assay
The pulldown assay was performed as described previously (45). Hda-cHis (5 pmol) was incubated on ice for 15 min in 12.5 μl of NP200-20 buffer containing 5 mm Tris-HCl (pH 7.5), 10% glycerol, 200 mm potassium glutamate, 0.01% Brij-58, 8 mm 2-mercaptoethanol, 8 mm magnesium acetate, and 20 mm imidazole in the presence of 0.1 mm ADP and Co2+-conjugated magnetic beads (40 μg; Invitrogen) equilibrated with the same buffer. The DNA-loaded clamps (1 pmol as the clamp) and the indicated amounts of ATP-DnaA were included, and the mixture was incubated on ice for 10 min. The beads and bound proteins were collected and washed with NP200-20 buffer (25 μl). Proteins were eluted in 3 μl of NP200-1000 buffer (the same as NP200-20 except for 1 m imidazole) and analyzed by SDS-12% PAGE and silver staining. Band intensities were measured with ImageJ software.
DnaA-Hda Interaction Using SPR Analysis
The Hda binding activity of DnaA proteins was determined by SPR analysis using an NTA sensor chip (Biacore). Buffers used for this analysis were as follows: (i) regeneration buffer (10 mm Hepes-KOH (pH 8.3), 150 mm NaCl, 350 mm EDTA, and 0.005% Surfactant P20; (ii) wash buffer (10 mm Tris-HCl (pH 7.6), 300 mm NaCl, 1 mm EDTA, and 0.05% SDS; (iii) nickel buffer (10 mm Hepes-KOH (pH 7.6), 150 mm NaCl, 50 μm EDTA, 0.01% Brij-58, and 5 mm NiCl2; and (iv) eluent buffer (10 mm Hepes-KOH (pH 7.6), 150 mm potassium acetate, 5 mm magnesium acetate, 50 μm EDTA, 0.01% Brij-58, 8 mm 2-mercaptoethanol, and 0.1 mm ADP). All reactions were performed at 20 °C. The sensor chip contained two flow cells, and one flow cell was used as a reference cell with the N-terminal hexahistidine-fused green fluorescent protein (His-GFP). After washing with regeneration buffer and wash buffer, the NTA surface was saturated with Ni2+ by loading nickel buffer into the two flow cells at a flow rate of 20 μl/min for 30 s. Hda-cHis and His-GFP proteins were each injected into one of the flow cells in eluent buffer at 2 μl/min until 2500 resonance units was obtained. DnaA protein was diluted in eluent buffer without ADP and with 0.1 mm ATP. Using the KInject command, samples containing DnaA protein were injected at 20 μl/min. The association time was 210 s, and the dissociation time was 420 s.
In Vitro Minichromosome Replication Systems
The DnaA-dependent replication assay using the minichromosome M13E10 replicative form I DNA and a protein extract prepared from WM433 was performed as described previously (22). The reconstituted replication assay using the minichromosome pBSoriC replicative form I DNA and purified DNA replication proteins was also performed as described previously (46).
P1 Nuclease Assay for Open Complex Formation
The P1 nuclease assay was performed as described previously (39).
In Vivo Analysis of Nucleotide-bound DnaA Forms
This in vivo analysis was performed as described previously (22). Cells were grown at 37 °C in a phosphate labeling medium containing 0.4 mCi/ml [32P]orthophosphate until the A660 of the culture reached 0.2 and then further incubated at the same temperature for 90 min in the presence of 1 mm isopropyl β-d-(−)-thiogalactopyranoside. Cleared lysates (750 μl) from aliquots (2 ml) of the culture were prepared and mixed with anti-DnaA antiserum (5 μl) that had been preincubated on ice for 30 min in a cleared lysate (60 μl) of KP7364. Protein A-Sepharose (60 μl; 50% slurry) was added, suspended at 4 °C for 30 min, and washed repeatedly in chilled buffers. After removal of the final washing solution, immunoprecipitates were extracted in a solution (20 μl) containing 1 m HCOOH and 5 mm each ATP, ADP, and AMP. Radiolabeled nucleotides were separated using thin-layer chromatography and quantified using an imaging analyzer.
RESULTS
Construction of Structural Model for DnaA Domains III-IV·Hda·dsDNA Complex
To identify a novel region required for the DnaA-Hda interaction, a homology model of the E. coli DnaA domains III-IV·Hda complex was constructed using the Aquifex aeolicus AMPPCP-DnaA oligomer structure as a starting point (Fig. 1, B and C) (49). The resulting model was consistent with the results of our previous analysis of the DnaA domain III·Hda complex structure that suggested specific interactions between Hda Arg-153 Arg finger and DnaA-bound ATP, between Hda Ser-152 and DnaA Arg-334 Arg finger, and between Hda Phe-118 and the DnaA Walker B motif (Fig. 1, B and D) (45). After consideration of the structure of E. coli DnaA domain IV complexed with DnaA box DNA (43), dsDNA was arranged to bind specifically with DnaA domain IV.
The resulting model showed that DnaA Leu-422 and Pro-423 residues within DnaA domain IV would reside on an interface with the Hda AAA+ domain (Fig. 1, B–D). These residues are located in the L3 loop and α4 helix of the DnaA domain IV structure (43), and could interact with a downstream Hda sensor I motif, including Hda Arg-138, Gln-142, and Asn-144 residues.
DnaA L422A/G and P423A Are Insensitive to RIDA
To determine whether DnaA Leu-422 and Pro-423 play an important role in RIDA, we substituted those amino acid residues with alanine. DnaA Leu-422 was also substituted with glycine because glycine is generally considered to be more effective in causing functional change than alanine. Using methods similar to those for wild-type DnaA, DnaA L422A, L422G, and P423A mutant proteins were overproduced in a dnaA-null host strain and purified to >90% purity as judged by SDS-PAGE and Coomassie Brilliant Blue staining (Fig. 2A). Mutant protein binding activities for ATP and ADP were comparable with those of wild-type DnaA (Fig. 2B).
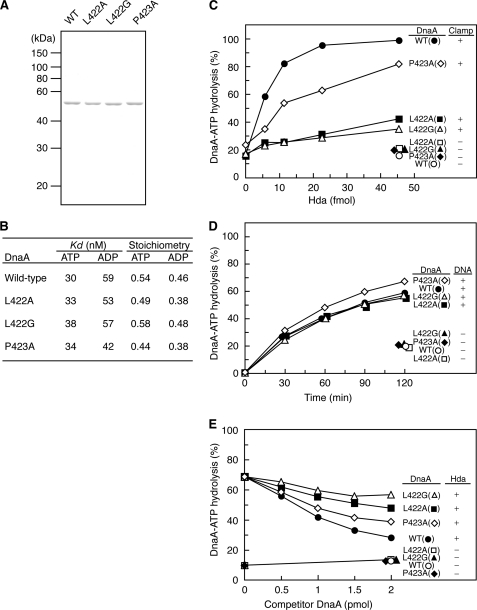
FIGURE 2.
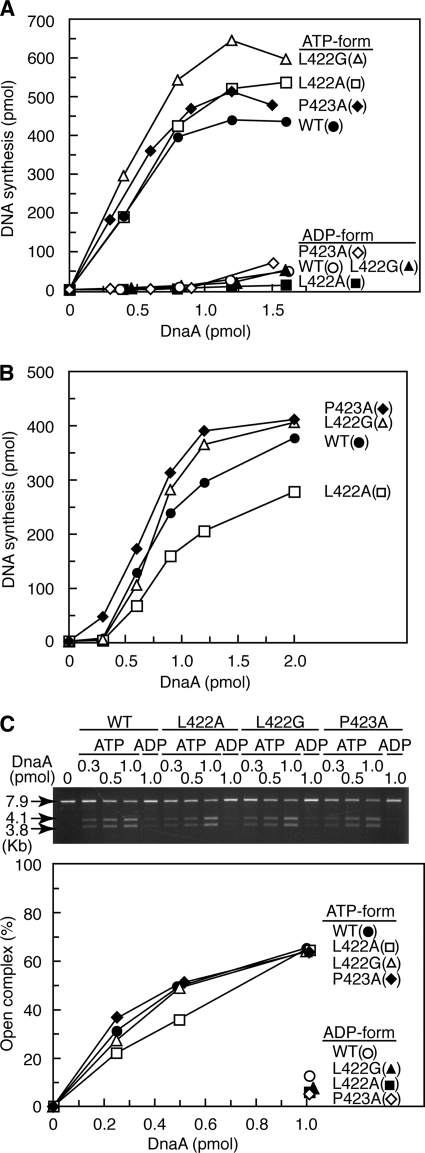
Analysis of DnaA mutant proteins and RIDA activity. A, purified wild-type and mutant DnaA proteins (500 ng) were stained with Coomassie Brilliant Blue after SDS-PAGE. B, the affinity of DnaA for ATP or ADP was determined using DnaA (2 pmol) and a filter retention assay. Kd and stoichiometry were calculated by Scatchard plot. C, DnaA-ATP hydrolysis was assessed using a staged RIDA reconstituted system. [α-32P]ATP-DnaA (0.25 pmol) was incubated at 30 °C for 20 min in buffer containing the indicated amounts of Hda protein in the presence or absence of the DNA-loaded clamps (10 ng). The ratio of ADP-DnaA to total ATP-/ADP-DnaA is shown as a percentage. D, intrinsic ATPase activity. [α-32P]ATP-DnaA (0.5 pmol) was incubated at 30 °C for the indicated time in the presence or absence of φX174 DNA (15 ng). E, DnaA competition assay in RIDA. [α-32P]ATP-DnaA (0.25 pmol) was incubated at 30 °C for 20 min in buffer containing the DNA-loaded clamps (0.4 ng) and the indicated amounts of wild-type or mutant DnaA proteins in the presence or absence of Hda (0.55 pmol).
To determine the sensitivity of DnaA mutant proteins to RIDA in vitro, a RIDA assay using a reconstituted staged system was used. The DNA-loaded clamps were isolated by gel filtration and incubated in the presence of [α-32P]ATP-DnaA and Hda. The results indicated that, unlike wild-type DnaA, DnaA L422A and L422G were not sensitive to RIDA (Fig. 2C). The sensitivity of DnaA P423A to RIDA was decreased (Fig. 2C). Thus, the DnaA Leu-422 residue was crucial for RIDA in vitro. However, the three mutant proteins had intrinsic DNA-dependent ATPase activity levels that were similar to that of wild-type DnaA (Fig. 2D). These results suggested that DnaA Leu-422 and Pro-423 residues specifically functioned in RIDA-specific DnaA-ATP hydrolysis.
To determine whether DnaA mutant proteins interact with the DNA-loaded clamp·Hda complex, a RIDA reaction competition assay was performed (45). A small amount of the DNA-loaded clamps was incubated with [α-32P]ATP-DnaA, high amounts of Hda, and the indicated amounts of non-radiolabeled competitor wild-type or mutant ATP-DnaA. The competitive inhibition of the RIDA reaction caused by DnaA L422A and L422G was less than that caused by wild-type DnaA (Fig. 2E). DnaA P423A inhibited the reaction moderately, but this was still less than the inhibition caused by wild-type DnaA. These results suggested that DnaA Leu-422 and Pro-423 residues functioned in DnaA interaction with the DNA-loaded clamp·Hda complex.
DNA Binding Activities of DnaA Mutants
DnaA domain IV is a DNA-binding region that has a high affinity for the DnaA box. In the co-crystal structure reported previously (43), DnaA Leu-422 and Pro-423 residues interact with a 13-mer dsDNA fragment (5′-TGTTATCCACAGG-3′) containing the DnaA box R1 (bold). In that structure, the main chain NH group of Leu-422 is thought to interact with the phosphate group of T (T11) that is a complementary base to the 11th A (in the 13-mer shown above). The side chain of Pro-423 is postulated to interact with the methyl group of T11 by van der Waals force. In addition, the 1H-15N heteronuclear single quantum correlation spectrum of DnaA domain IV in NMR shows a prominent chemical shift perturbation on Leu-422 in the presence of the DnaA box (50). Thus, to determine whether the mutant proteins are affected in their affinities for the DnaA box, they were analyzed with 21-mer dsDNA fragments containing the DnaA box R1 or a nonspecific DNA sequence (nonsense DNA) as a control using SPR.
These results showed that L422G and P423A mutant proteins bound the DnaA box with kinetics similar to the binding observed with wild-type DnaA (Fig. 3A, C, and D). DnaA L422A had a slightly reduced affinity (Fig. 3B), which was consistent with the data of NMR analysis (50). The sustained binding activity of DnaA L422A/G mutant proteins might be caused by the interaction of the Leu-422 main chain (but not the side chain) with the DnaA box. The activity of Pro-423 could be complemented by the substitution of alanine bearing a side chain. Otherwise, the small contribution of Pro-423 to DnaA box binding could be explained by specific interactions supported by van der Waals force.
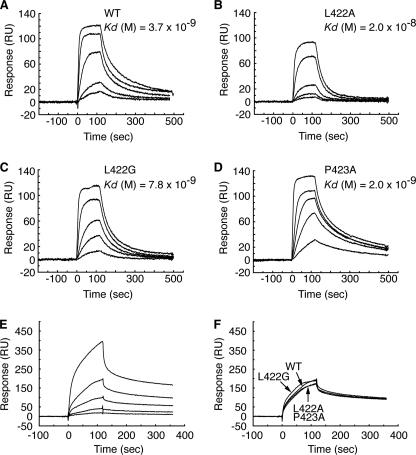
FIGURE 3.
DNA binding activities of DnaA mutant proteins. A–D, DnaA box binding activity. Affinities for the DnaA box of wild-type DnaA (A), DnaA L422A (B), DnaA L422G (C), and DnaA P423A (D) were determined by SPR analysis. Concentrations of DnaA proteins used were 2.5, 5.0, 10, 25, and 75 nm for all experiments. Kd values were calculated by curve fitting of the sensorgrams to a theoretical 1:1 interaction model (Langmuir binding). RU, resonance units. E and F, nonsense DNA binding activity. Affinity for a nonsense DNA was measured by SPR analysis using wild-type DnaA at concentrations of 9.0, 19, 38, 75, and 150 nm (E). Affinities of DnaA mutant proteins were also compared with those of wild-type DnaA at 75 nm (F).
RIDA is dependent on the clamps loaded on duplex DNA (20), but a specific DNA sequence is not required. Using SPR analysis, the affinity of the DnaA mutants for a nonspecific DNA sequence was determined. Given that DnaA binding to nonspecific DNA is weaker than its binding to DnaA box DNA (48), large amounts of nonsense DNA (21-mer) were immobilized on a sensor chip. Wild-type DnaA protein (9–150 nm) showed a dose-dependent increase in binding to nonsense DNA (Fig. 3E). When analyzed using an intermediate concentration (75 nm), the resonance patterns of DnaA mutant proteins were similar to those of wild-type DnaA (Fig. 3F). Taken together, the nonspecific DNA binding activity of DnaA mutants supports the idea that the RIDA insensitivity of DnaA mutants was not due to a decrease in their affinity for DNA. These results are consistent with the model (Fig. 1, B and D) and the results of NMR analysis (50), which show that these amino acid residues make no contact with nonspecific DNA.
Hda Binding Activities of DnaA Mutants
To more quantitatively analyze the DnaA interaction, pulldown assays using His-tagged Hda and Co2+-conjugated magnetic beads were performed. In this assay, DnaA is recovered in an ADP-Hda dose-dependent manner in the presence of the DNA-loaded clamps (19). Binding specificity is supported by the data showing that DnaA R334A (AAA+ sensor II) and Hda Q6A (the clamp-binding site) mutants (which are defective in Hda interaction and clamp binding, respectively) severely reduce the recovery of DnaA in these assays (45) (Fig. 4A). When DnaA L422A/G proteins were similarly assessed, the recovery of these mutants was severely reduced compared with wild-type DnaA (Fig. 4A), suggesting that the DnaA Leu-422 residue is crucial for Hda interaction. The residual binding of these mutants was 10–20% of wild-type DnaA-binding, which can be explained by possible interactions between AAA+ domains of DnaA and Hda. The recovery of DnaA P423A was roughly 50% of that of the wild-type DnaA (Fig. 4A). Notably, these results are consistent with the results showing that RIDA sensitivities of DnaA L422A/G and P423A are 10–15% and about 50% of that of the wild-type DnaA, respectively (Fig. 2C). Thus, these results are effective for quantitative understanding of DnaA domain IV-dependent interactions with Hda.
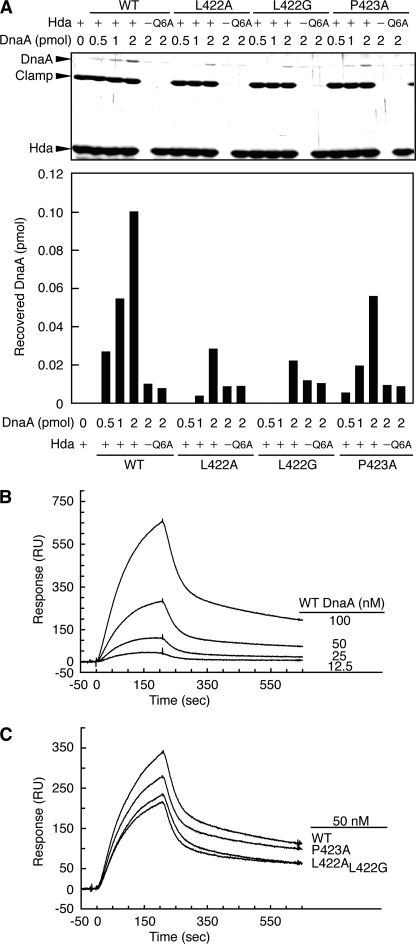
FIGURE 4.
Hda binding activity of DnaA mutant proteins. A, Co2+-conjugated beads were incubated on ice in buffer containing (+) or excluding (−) 5 pmol of Hda-cHis or Hda Q6A-cHis (Q6A) in the presence of ADP followed by incubation on ice for 10 min in the presence of the DNA-loaded clamps (1 pmol as the clamp) and the indicated amounts of the wild-type (WT) and mutant ATP-DnaA proteins (L422A, L422G, and P423A). Bead-bound materials were isolated and analyzed by SDS-PAGE and silver staining. The DnaA bands were quantified by ImageJ software, and the recovered amounts were determined using a standard curve. B, Hda binding activity of wild-type DnaA was measured by SPR analysis using DnaA concentrations of 12.5, 25, 50, and 100 nm. RU, resonance units. C, Hda binding activities of DnaA mutant proteins were also compared with those of wild-type DnaA at 50 nm.
Next, SPR analysis was used to determine direct binding modes of wild-type and mutant DnaA proteins to Hda. His-tagged Hda and His-tagged GFP (used as a background control) were separately bound to a nickel-NTA-coated chip. The DnaA-Hda interaction was detected when the concentration of wild-type DnaA was 12.5–100 nm (Fig. 4B). Because the reaction was not saturated even at 100 nm DnaA, precise kinetic parameters could not be calculated. Thus, in agreement with the pulldown data, the direct binding of DnaA to Hda is probably weak in the absence of DNA-loaded clamps. At 50 nm (within the linear range for the reaction), DnaA L422A and L422G exhibited reduced Hda binding activity relative to that of wild-type DnaA (Fig. 4C). The affinity of DnaA P423A was moderately reduced (Fig. 4C), which was consistent with the RIDA activity data (Fig. 2C). Therefore, Leu-422 and Pro-423 residues had significant and specific roles in the direct DnaA-Hda interaction.
Residual activities of DnaA L422A/G and P423A in Hda binding (Fig. 4, A and C) can be explained by direct interactions between the AAA+ domains of DnaA and Hda. Previous studies show that specific binding of DnaA to Hda requires several amino acid residues within the AAA+ domains of both Hda and DnaA, including Hda Phe-118 (H-finger), Asn-122 (E-finger), Arg-153 (Arg finger), and DnaA Arg-334 (sensor II) (Fig. 1, B–D) (34, 45). These interactions as well as possible nonspecific interactions conceivably cause the low levels of binding activity between DnaA and Hda even in the absence of DNA-loaded clamps. The DnaA Leu-422/Pro-423-mediated interaction might assist or regulate the functional interactions of these residues (see “Discussion”).
DnaA Mutants Are Active in Minichromosome Replication and Formation of Initiation Complexes and ADP Release
The function of the mutant proteins during replication initiation at oriC in vitro was first characterized using an in vitro DnaA complementation system with a crude protein extract and an oriC plasmid. The replication activity and its ATP-dependent regulation of DnaA L422A and P423A were similar to those of wild-type DnaA (Fig. 5A). DnaA L422G exhibited a slightly higher replication activity than wild-type DnaA (Fig. 5A) that might have been caused by inhibition of RIDA activity contained in the crude extract (17). Next, we used a reconstituted replication system using purified replication proteins and found that DnaA L422G and P423A had replication activities that were similar to those of the wild type (Fig. 5B). DnaA L422A had reduced replication activity (Fig. 5B), reflecting the decreased DnaA box binding activity of this protein (Fig. 3B).
FIGURE 5.
Replication and oriC unwinding activities of DnaA mutant proteins. A and B, DnaA activities in replication initiation were assessed using minichromosome replication systems using a crude protein fraction (A) and purified replicative proteins (B). The indicated amounts of the ATP- or ADP-bound forms of wild-type and mutant DnaA proteins were incubated at 30 °C for 20 (A) or 30 min (B). C, oriC unwinding activity. The indicated amounts of the ATP- or ADP-bound forms of wild-type and mutant DnaA proteins were incubated at 38 °C for 3 min in buffer containing M13KEW101 oriC plasmid (7.9 kb) and HU protein followed by further incubation with P1 nuclease. The resultant DNA was purified, digested with AlwNI, and analyzed using 1% agarose gel electrophoresis and ethidium bromide staining. The amounts of 3.8- and 4.1-kb fragments detected were normalized to the amount of total DNA and were plotted as an open complex (%).
Open complex formation was assessed using an oriC plasmid and P1 nuclease, which specifically cleaves single-stranded DNA. If the oriC-unwinding region on the plasmid were cleaved by P1 nuclease, subsequent digestion with AlwNI restriction enzyme would produce two DNA fragments of 3.8 and 4.1 kb (39). The results indicated that, like wild-type DnaA, DnaA L422G and P423A were active and regulated in an ATP binding-dependent manner during open complex formation (Fig. 5C). DnaA L422A exhibited a slightly reduced activity relative to wild-type DnaA (Fig. 5C) consistent with the results of the reconstituted replication assay (Fig. 5B). These results suggested that the DnaA mutants were active in the DNA replication initiation process, including formation of initial complexes and DnaB helicase loading on oriC.
The inactive ADP-DnaA yielded by RIDA is reactivated to ATP-DnaA by the nucleotide exchange activity of DnaA-reactivating sequences (DARS1 and -2) on the chromosome that are required for timely DNA replication initiation during the cell cycle (51). DARSs bear a DnaA box cluster promoting formation of a specific DnaA multimer and dissociation of ADP bound to DnaA (51). DnaA L422A/G and P423A proteins were substantially active in the DARS1-dependent DnaA-ADP dissociation in vitro (data not shown). The combined results support the proposed function of DnaA Leu-422 and Pro-423 residues specifically in RIDA-dependent DnaA-ATP hydrolysis.
DnaA L422A/G and P423A Proteins Inhibit Cell Growth
Certain dnaA mutants, such as dnaAcos and dnaA29 (R334A), are inactive in RIDA and cause oriC-dependent overinitiation of chromosomal replication, resulting in inhibition of cell growth and colony formation (22, 52, 53). In the absence of rnhA, an alternative oriC-independent replication system operates that allows cells without oriC to grow even in the presence of overinitiating dnaA mutant alleles (18, 22, 52, 54). To investigate in vivo RIDA activity of DnaA L422A/G and P423A, we asked whether these DnaA mutants exhibit oriC-dependent inhibition of colony formation (Table 1). Introduction of the dnaA L422A/G and P423G mutant alleles on a pING1 (vector) derivative inhibited or impeded colony formation when the host cells (KH5402-1, YT411, and KA451) were active for chromosomal replication from oriC even in the absence of rnhA (Table 1). In particular, colony formation was severely inhibited in the chromosomal dnaA-disrupted cells (KA451), indicating that the inhibition was caused only by expression of the mutant dnaA alleles but not by co-expression of the wild-type and mutant dnaA alleles.
TABLE 1.
oriC-dependent inhibition of DnaA mutant colony formation
Cells were transformed with pING1 (vector), pKA234 (wild-type dnaA), pL422A (dnaA L422A), pL422G (dnaA L4222G), and pP423A (dnaA P423A) and incubated at 37 °C for 15 h (for KH5402-1 and YT411) or for 30 h (for KA451, KA429, and KA450) on LB medium containing thymine and ampicillin. The growth rate of oriC- and/or dnaA-disrupted cells (i.e. KA451, KA429, and KA450) was about 90 min under these conditions, whereas that of KH5402-1 and YT411 cells was about 40 min. Colonies with a diameter of >∼0.5 mm were counted. Transformation frequencies are indicated as values relative to that of pING1 (vector). Transformation efficiencies of the indicated strains by pING1 were 105–106 cells/μg of DNA. Δ, del-1017; Am, amber mutation.
| Strain | Genotype |
Relative transformation frequency |
||||||
|---|---|---|---|---|---|---|---|---|
| rnhA | dnaA | oriC | pING1 (vector) | pKA234 (WT) | pL422A (L422A) | pL422G (L422G) | pP423A (P423A) | |
| KH5402-1 | + | + | + | 1.0 | 0.84 | <1.7 × 10−3 | <1.7 × 10−3 | 4.7 × 10−2a |
| YT411 | ::cat | + | + | 1.0 | 1.1 | <4.4 × 10−3 | <4.4 × 10−3 | <4.4 × 10−3 |
| KA451 | ::cat | ::Tn10 | + | 1.0 | 2.5 | <8.8 × 10−3 | <8.8 × 10−3 | <8.8 × 10−3 |
| KA429 | ::cat | + | Δ | 1.0 | 2.4 | 1.6 | 3.0 | 0.65 |
| KA450 | Am | Am | Δ | 1.0 | 0.94 | 0.64 | 0.36 | 0.73 |
a Colonies formed were tiny and heterogeneous.
By contrast, introduction of those alleles did not inhibit colony formation when the host cells lacked oriC in the absence of rnhA (KA429 and KA450) (Table 1). In addition, expression levels of wild-type and mutant DnaA proteins in KA450 were comparable (supplemental Fig. 1), suggesting that the inhibition of colony formation was not due to simple overexpression of the mutant proteins.
These results suggested that DnaA L422A, L422G, and P423A mutant proteins specifically affected DNA replication initiation at oriC and were consistent with the idea that the mutants promoted overinitiation in vivo due to a specific defect in RIDA. The dnaA P423A mutant inhibited cell growth at a level that was moderate compared with that caused by dnaA L422A/G. This observation was also consistent with the reduced activity of this allele in RIDA in vitro.
Even when the incubation time after transformation of cells was extended to 20 h, colony formation efficiency of KH5402-1 bearing pL422A or pL422G was only 5% of that by pKA234, and even the colonies that were formed contained cells that could not form new colonies (data not shown). Colony formation of KH5402-1 bearing pP423A was also inhibited (Table 1); cells in the colonies could grow but only at a 2-fold slower rate compared with KH5402-1 cells bearing pING1 or pKA234 (data not shown).
dnaA L422A/G and P423A Mutants Promote Accumulation of ATP-DnaA
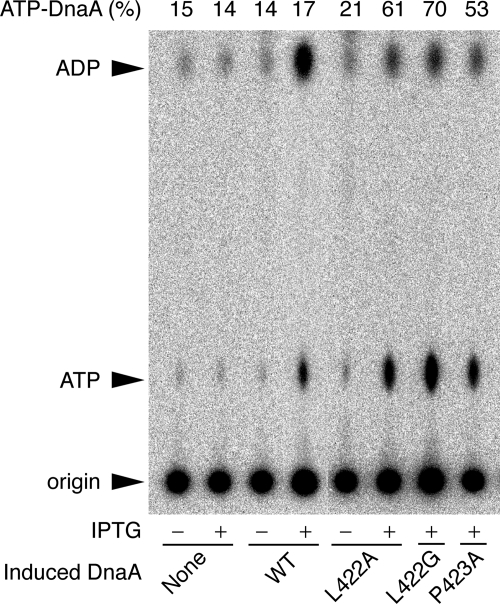
In cultures of wild-type cells, the ATP-bound form of DnaA represents 10–20% of the total ATP/ADP-DnaA content (21, 22). As constitutive expression of the mutant genes inhibits the growth of KH5402-1 host cells (Table 1), controlled expression of the mutant alleles was achieved by placing them downstream of the lac promoter on a pBR322 derivative in lacIq cells. To assess the relative ratio of ATP/ADP forms of the mutant DnaA proteins in vivo, cells were grown in a synthetic medium containing [32P]orthophosphate. DnaA protein was isolated by immunoprecipitation, and the nucleotides bound to the protein were analyzed by thin-layer chromatography (22).
When DnaA L422A expression was induced for 90 min in the presence of 1 mm isopropyl β-d-(−)-thiogalactopyranoside, the proportion of ATP-DnaA increased from 21 to 61% (Fig. 6). Similarly, the levels of the ATP forms of DnaA L422G and P423A increased from basal levels to 70 and 53%, respectively, in the presence of isopropyl β-d-(−)-thiogalactopyranoside (Fig. 6). However, induction of the wild-type protein was not associated with an increase in the level of ATP-DnaA (Fig. 6), which is consistent with previous data (22). These results suggested that the mutant proteins were defective in RIDA in vivo.
FIGURE 6.
Levels of ATP-DnaA in mutants. KH5402-1 bearing pHSL99 (lacIq) was used as the host strain for plasmids pSN300 vector (None), pSN306 bearing wild-type dnaA (WT), pSNL422A bearing dnaA L422A (L422A), pSNL422G bearing dnaA L422G (L422G), or pSNP423A bearing dnaA P423A (P423A). Cells were grown at 37 °C in supplemented TG medium containing [32P]orthophosphate until the A660 of the culture was 0.2 and then incubated at the same temperature for 90 min in the presence (+) or absence (−) of 1 mm isopropyl β-d-(−)-thiogalactopyranoside (IPTG). Aliquots of the cultures were withdrawn, and nucleotide-bound DnaA proteins were recovered by immunoprecipitation using anti-DnaA antiserum. The results of polyethylenimine-cellulose thin-layer chromatography to analyze DnaA-bound nucleotides are shown. The origin and migration positions of ATP and ADP are indicated by arrows. The ratio of ATP-DnaA to total ATP-/ADP-DnaA is shown as a percentage (ATP-DnaA (%)).
DISCUSSION
A structural model of the Hda·DnaA domains III-IV·DNA complex was used to ascertain amino acid residues within DnaA domain IV that specifically interact with Hda. Mutant analyses revealed that DnaA Leu-422 and Pro-423 residues are important for RIDA-dependent ATP hydrolysis, formation of a complex with the DNA-loaded clamp·Hda complex, and Hda interaction (Figs. 2 and 4). Consistently, these amino acid residues were found to be required for DnaA-ATP hydrolysis in vivo (Fig. 6 and Table 1). These mutants were not defective in the initiation of DNA replication, DARS-dependent reactivation of DnaA in vitro, or intrinsic ATPase activity of DnaA (Figs. 2D and 5). Leu-422 and Pro-423 residues in the DNA-binding domain of DnaA are crucial for functional interactions with Hda during RIDA, although DnaA Ser-421, which is adjacent to Leu-422, might make contact with the DnaA AAA+ domain of the adjacent protomer in a DnaA homomultimer (55). Taken together with the results of earlier analyses (22, 34, 45), we suggest that Hda interacts with two DnaA sites, one in domain III and the other in domain IV. This explains specific DnaA-Hda interactions that have low binding affinity but require strict specificity for DnaA-ATP hydrolysis.
C. crescentus HdaA (29) and Shewanella amazonensis Hda (56), orthologues of E. coli Hda, are thought to function in the hydrolysis of ATP-DnaA like E. coli Hda. Some other bacterial species also have Hda homologues with amino acid sequence similarity to E. coli Hda (45). In the DnaA homologues of these species, the amino acid residues that correspond to E. coli DnaA Leu-422 and Pro-423 are highly conserved (data not shown) (57). Hence, the proposed interaction and regulation models might apply to other bacterial species that possess an Hda orthologue.
The proposed structural model indicates that DnaA Leu-422 and Pro-423 residues are located near the Hda sensor I motif (Ser-127 to Pro-140) (Fig. 1, B–D). In particular, Hda Arg-138, Gln-142, and Asn-144 residues might be exposed on the surface of Hda and be able to interact with DnaA Leu-422 and Pro-423. The sensor I motif of AAA+ chaperones, such as Saccharomyces cerevisiae Hsp104 and the p97/valosin-containing protein-like ATPase from Thermoplasma acidophilum, play a specific role in catalyzing ATP hydrolysis (58–60). In addition, the Arg finger (Arg-153) and its spatially neighboring residues in Hda play a crucial role in RIDA and the interaction with ATP-DnaA (most likely with ATP itself and the ATP-binding Walker B motif (34, 45)). Thus, it is plausible that the interaction between DnaA Leu-422/Pro-423 and the areas near Hda sensor I in addition to that between ATP-DnaA domain III and the Hda Arg finger supports a specific conformation of the DnaA·Hda complex that allows DnaA-ATP hydrolysis (Fig. 7B). The interactions might indirectly promote a conformational change of the Hda Arg finger, thereby affecting DnaA-ATP hydrolysis and the affinity of the two proteins.
FIGURE 7.
Model for interaction of DnaA domain IV with Hda in presence of DnaA box. A, a close-up representation of a homology model for the Hda (purple)·DnaA domains III (red)-IV (pink) complex with the DnaA box DNA (orange). A structure for DnaA domain IV·DnaA box complex was obtained from the protein data bank (Protein Data Bank code 1J1V). The indicated amino acid residues and AMPPCP are shown as ball-and-stick models. The Hda Arg finger (Arg-153) and AMPPCP are red and gray, respectively. Hda Arg-138, Gln-142, and Asn-144 residues are yellow, and DnaA Leu-422 and Pro-423 residues are blue. B and C, models for interaction modes of DnaA and Hda on the DNA-loaded clamp in the absence (B) and presence (C) of a DnaA box. DnaA domains (I–IV), Hda domains (clamp-binding (CB) and AAA+ domains), and the clamp are indicated in different colors. Red arrowhead, the Hda Arg finger; yellow arrowhead, the Hda sensor I region, including Arg-138, Gln-142, and Asn-144; gray circle, ATP; blue arrowhead, the DnaA region, including Leu-422 and Pro-423. Nonsense DNA and DnaA box DNA are shown as green and an orange rectangles, respectively.
The x-ray and NMR analyses of the DnaA domain IV·DnaA box DNA complex show that DnaA Leu-422 is involved in specific binding to the DnaA box DNA (43, 50). In the crystal structure, the main chain of Leu-422 also interacts with a phosphate group of the DnaA box DNA, which is mediated by DNA bending at 28° (Fig. 7A) (43). Thus, substitution of the side chain of the Leu residue still allowed DnaA box binding activity (Fig. 3, B and C). DNA bending by DnaA binding to the DnaA box is also observed in solution (48). A model in which the DnaA box DNA acts as a physical obstacle that inhibits the interaction between DnaA Leu-422/Pro-423 residues and the surface of the Hda AAA+ domain is proposed (Fig. 7, A and C). In fact, ATP-DnaA molecules are hydrolyzed by RIDA in vivo, and ∼20% (200–400 molecules) of 1000–2000 DnaA molecules present in the cell are present as ATP-DnaA (21). This could be explained if ATP-DnaA molecules bound to the DnaA box DNA are insensitive to RIDA in vivo. There are ∼300 DnaA boxes on the E. coli chromosome (61). Maintaining a basal level of ATP-DnaA may be relevant to efficient origin firing in the next round of replication initiation and ATP-DnaA-specific transcriptional regulation of genes like dnaA and the nrd operon (47, 62).
There are examples where a DNA-binding region in a protein plays a role in protein-protein interactions other than DNA binding. The DNA-binding region of Cdc6, a replication-licensing factor in eukaryotic and archaeal cells, interacts with and regulates the MCM helicase (63, 64). Additionally, the DNA-binding regions of OmpR and PhoB, which are transcriptional regulators in E. coli, are suggested to contain an interaction site for a subunit of RNA polymerase and to regulate directly RNA polymerase activity (65, 66). The protein-protein interaction on a DNA-binding region may function for the direct recognition of conformational changes within the region induced by DNA binding. In the RIDA reaction, DnaA domain IV likely interacts with the clamp-loaded DNA to form an active RIDA complex (20). The cross-talk between the Hda AAA+ domain and DnaA Leu-422/Pro-423 within domain IV may serve to allow Hda to recognize the interaction of DnaA domain IV with the clamp-loaded DNA.
Supplementary Material
Acknowledgments
We are grateful to Dr. Norie Fujikawa, Dr. Hitoshi Kurumizaka, Dr. Tadashi Ueda, and Dr. Yoshito Abe for discussion on the structural analysis of DnaA domain IV.
This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Technology and Science of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- RIDA
- regulatory inactivation of DnaA
- AAA
- ATPases associated with a variety of cellular activities
- DARS
- DnaA-reactivating sequence
- SPR
- surface plasmon resonance
- NTA
- nitrilotriacetic acid
- MCM
- minichromosome maintenance
- AMPPCP
- adenosine 5′-(β,γ-methylene)triphosphate.
REFERENCES
- 1. Leonard A. C., Grimwade J. E. (2010) Curr. Opin. Microbiol. 13, 766–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaguni J. M. (2006) Annu. Rev. Microbiol. 60, 351–375 [DOI] [PubMed] [Google Scholar]
- 3. Keyamura K., Fujikawa N., Ishida T., Ozaki S., Su'etsugu M., Fujimitsu K., Kagawa W., Yokoyama S., Kurumizaka H., Katayama T. (2007) Genes Dev. 21, 2083–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O'Donnell M. (2006) J. Biol. Chem. 281, 10653–10656 [DOI] [PubMed] [Google Scholar]
- 5. Mott M. L., Erzberger J. P., Coons M. M., Berger J. M. (2008) Cell 135, 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Makowska-Grzyska M., Kaguni J. M. (2010) Mol. Cell 37, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Remus D., Diffley J. F. (2009) Curr. Opin. Cell Biol. 21, 771–777 [DOI] [PubMed] [Google Scholar]
- 8. Katayama T., Ozaki S., Keyamura K., Fujimitsu K. (2010) Nat. Rev. Microbiol. 8, 163–170 [DOI] [PubMed] [Google Scholar]
- 9. Lu M., Campbell J. L., Boye E., Kleckner N. (1994) Cell 77, 413–426 [DOI] [PubMed] [Google Scholar]
- 10. Slater S., Wold S., Lu M., Boye E., Skarstad K., Kleckner N. (1995) Cell 82, 927–936 [DOI] [PubMed] [Google Scholar]
- 11. Nievera C., Torgue J. J., Grimwade J. E., Leonard A. C. (2006) Mol. Cell 24, 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Waldminghaus T., Skarstad K. (2009) Plasmid 61, 141–150 [DOI] [PubMed] [Google Scholar]
- 13. Kitagawa R., Ozaki T., Moriya S., Ogawa T. (1998) Genes Dev. 12, 3032–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zakrzewska-Czerwiñska J., Jakimowicz D., Zawilak-Pawlik A., Messer W. (2007) FEMS Microbiol. Rev. 31, 378–387 [DOI] [PubMed] [Google Scholar]
- 15. Theisen P. W., Grimwade J. E., Leonard A. C., Bogan J. A., Helmstetter C. E. (1993) Mol. Microbiol. 10, 575–584 [DOI] [PubMed] [Google Scholar]
- 16. Riber L., Løbner-Olesen A. (2005) J. Bacteriol. 187, 5605–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Katayama T., Kubota T., Kurokawa K., Crooke E., Sekimizu K. (1998) Cell 94, 61–71 [DOI] [PubMed] [Google Scholar]
- 18. Kato J., Katayama T. (2001) EMBO J. 20, 4253–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Su'etsugu M., Nakamura K., Keyamura K., Kudo Y., Katayama T. (2008) J. Biol. Chem. 283, 36118–36131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Su'etsugu M., Takata M., Kubota T., Matsuda Y., Katayama T. (2004) Genes Cells 9, 509–522 [DOI] [PubMed] [Google Scholar]
- 21. Kurokawa K., Nishida S., Emoto A., Sekimizu K., Katayama T. (1999) EMBO J. 18, 6642–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishida S., Fujimitsu K., Sekimizu K., Ohmura T., Ueda T., Katayama T. (2002) J. Biol. Chem. 277, 14986–14995 [DOI] [PubMed] [Google Scholar]
- 23. Camara J. E., Breier A. M., Brendler T., Austin S., Cozzarelli N. R., Crooke E. (2005) EMBO Rep. 6, 736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fujimitsu K., Su'etsugu M., Yamaguchi Y., Mazda K., Fu N., Kawakami H., Katayama T. (2008) J. Bacteriol. 190, 5368–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Riber L., Fujimitsu K., Katayama T., Løbner-Olesen A. (2009) Mol. Microbiol. 71, 107–122 [DOI] [PubMed] [Google Scholar]
- 26. Noirot-Gros M. F., Dervyn E., Wu L. J., Mervelet P., Errington J., Ehrlich S. D., Noirot P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hayashi M., Ogura Y., Harry E. J., Ogasawara N., Moriya S. (2005) FEMS Microbiol. Lett. 247, 73–79 [DOI] [PubMed] [Google Scholar]
- 28. Noirot-Gros M. F., Velten M., Yoshimura M., McGovern S., Morimoto T., Ehrlich S. D., Ogasawara N., Polard P., Noirot P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2368–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Collier J., Shapiro L. (2009) J. Bacteriol. 191, 5706–5716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arias E. E., Walter J. C. (2006) Nat. Cell Biol. 8, 84–90 [DOI] [PubMed] [Google Scholar]
- 31. Nishitani H., Sugimoto N., Roukos V., Nakanishi Y., Saijo M., Obuse C., Tsurimoto T., Nakayama K. I., Nakayama K., Fujita M., Lygerou Z., Nishimoto T. (2006) EMBO J. 25, 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ralph E., Boye E., Kearsey S. E. (2006) EMBO Rep. 7, 1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dalrymple B. P., Kongsuwan K., Wijffels G., Dixon N. E., Jennings P. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11627–11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Su'etsugu M., Shimuta T. R., Ishida T., Kawakami H., Katayama T. (2005) J. Biol. Chem. 280, 6528–6536 [DOI] [PubMed] [Google Scholar]
- 35. Kurz M., Dalrymple B., Wijffels G., Kongsuwan K. (2004) J. Bacteriol. 186, 3508–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. (1999) Genome Res. 9, 27–43 [PubMed] [Google Scholar]
- 37. Seitz H., Weigel C., Messer W. (2000) Mol. Microbiol. 37, 1270–1279 [DOI] [PubMed] [Google Scholar]
- 38. Felczak M. M., Simmons L. A., Kaguni J. M. (2005) J. Biol. Chem. 280, 24627–24633 [DOI] [PubMed] [Google Scholar]
- 39. Keyamura K., Abe Y., Higashi M., Ueda T., Katayama T. (2009) J. Biol. Chem. 284, 25038–25050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Messer W. (2002) FEMS Microbiol. Rev. 26, 355–374 [DOI] [PubMed] [Google Scholar]
- 41. Abe Y., Jo T., Matsuda Y., Matsunaga C., Katayama T., Ueda T. (2007) J. Biol. Chem. 282, 17816–17827 [DOI] [PubMed] [Google Scholar]
- 42. Roth A., Messer W. (1995) EMBO J. 14, 2106–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., Yokoyama S. (2003) Nucleic Acids Res. 31, 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Asklund M., Atlung T. (2005) J. Mol. Biol. 345, 717–730 [DOI] [PubMed] [Google Scholar]
- 45. Nakamura K., Katayama T. (2010) Mol. Microbiol. 76, 302–317 [DOI] [PubMed] [Google Scholar]
- 46. Kawakami H., Keyamura K., Katayama T. (2005) J. Biol. Chem. 280, 27420–27430 [DOI] [PubMed] [Google Scholar]
- 47. Speck C., Weigel C., Messer W. (1999) EMBO J. 18, 6169–6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schaper S., Messer W. (1995) J. Biol. Chem. 270, 17622–17626 [DOI] [PubMed] [Google Scholar]
- 49. Erzberger J. P., Mott M. L., Berger J. M. (2006) Nat. Struct. Mol. Biol. 13, 676–683 [DOI] [PubMed] [Google Scholar]
- 50. Yoshida Y., Obita T., Kokusho Y., Ohmura T., Katayama T., Ueda T., Imoto T. (2003) Cell. Mol. Life Sci. 60, 1998–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fujimitsu K., Senriuchi T., Katayama T. (2009) Genes Dev. 23, 1221–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Katayama T. (2001) Mol. Microbiol. 41, 9–17 [DOI] [PubMed] [Google Scholar]
- 53. Felczak M. M., Kaguni J. M. (2009) Mol. Microbiol. 72, 1348–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kogoma T. (1997) Microbiol. Mol. Biol. Rev. 61, 212–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Duderstadt K. E., Mott M. L., Crisona N. J., Chuang K., Yang H., Berger J. M. (2010) J. Biol. Chem. 285, 28229–28239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xu Q., McMullan D., Abdubek P., Astakhova T., Carlton D., Chen C., Chiu H. J., Clayton T., Das D., Deller M. C., Duan L., Elsliger M. A., Feuerhelm J., Hale J., Han G. W., Jaroszewski L., Jin K. K., Johnson H. A., Klock H. E., Knuth M. W., Kozbial P., Sri Krishna S., Kumar A., Marciano D., Miller M. D., Morse A. T., Nigoghossian E., Nopakun A., Okach L., Oommachen S., Paulsen J., Puckett C., Reyes R., Rife C. L., Sefcovic N., Trame C., van den Bedem H., Weekes D., Hodgson K. O., Wooley J., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2009) J. Mol. Biol. 385, 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yoshikawa H., Ogasawara N. (1991) Mol. Microbiol. 5, 2589–2597 [DOI] [PubMed] [Google Scholar]
- 58. Hattendorf D. A., Lindquist S. L. (2002) EMBO J. 21, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gerega A., Rockel B., Peters J., Tamura T., Baumeister W., Zwickl P. (2005) J. Biol. Chem. 280, 42856–42862 [DOI] [PubMed] [Google Scholar]
- 60. Hanson P. I., Whiteheart S. W. (2005) Nat. Rev. Mol. Cell Biol. 6, 519–529 [DOI] [PubMed] [Google Scholar]
- 61. Roth A., Messer W. (1998) Mol. Microbiol. 28, 395–401 [DOI] [PubMed] [Google Scholar]
- 62. Gon S., Camara J. E., Klungsøyr H. K., Crooke E., Skarstad K., Beckwith J. (2006) EMBO J. 25, 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shin J. H., Grabowski B., Kasiviswanathan R., Bell S. D., Kelman Z. (2003) J. Biol. Chem. 278, 38059–38067 [DOI] [PubMed] [Google Scholar]
- 64. De Felice M., Esposito L., Pucci B., De Falco M., Manco G., Rossi M., Pisani F. M. (2004) Biochem. J. 381, 645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Makino K., Amemura M., Kawamoto T., Kimura S., Shinagawa H., Nakata A., Suzuki M. (1996) J. Mol. Biol. 259, 15–26 [DOI] [PubMed] [Google Scholar]
- 66. Kondo H., Nakagawa A., Nishihira J., Nishimura Y., Mizuno T., Tanaka I. (1997) Nat. Struct. Biol. 4, 28–31 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.