Abstract
Muscleblind-like 1 (MBNL1) is a splicing factor whose improper cellular localization is a central component of myotonic dystrophy. In myotonic dystrophy, the lack of properly localized MBNL1 leads to missplicing of many pre-mRNAs. One of these events is the aberrant inclusion of exon 5 within the MBNL1 pre-mRNA. The region of the MBNL1 gene that includes exon 5 and flanking intronic sequence is highly conserved in vertebrate genomes. The 3′-end of intron 4 is non-canonical in that it contains a predicted branch point that is 141 nucleotides from the 3′-splice site and an AAG 3′-splice site. Using a minigene that includes exon 4, intron 4, exon 5, intron 5, and exon 6 of MBNL1, we showed that MBNL1 regulates inclusion of exon 5. Mapping of the intron 4 branch point confirmed that branching occurs primarily at the predicted distant branch point. Structure probing and footprinting revealed that the highly conserved region between the branch point and 3′-splice site is primarily unstructured and that MBNL1 binds within this region of the pre-mRNA. Deletion of the MBNL1 response element eliminated MBNL1 splicing regulation and led to complete inclusion of exon 5, which is consistent with the suppressive effect of MBNL1 on splicing.
Keywords: RNA, RNA-binding Protein, RNA Splicing, RNA Structure, Zinc
Introduction
Splicing of pre-mRNAs is an important event that contributes to a diverse proteome as well as the regulation of gene expression. It is estimated that more than 90% of human genes undergo alternative splicing (1, 2). To produce a functional mRNA, non-coding regions must be accurately removed, and the coding regions must be ligated together. Splicing occurs via two transesterification reactions that result in removal of the intron and ligation of the exons. This splicing mechanism relies on pre-mRNA sequences, proteins, and small nuclear RNAs (snRNAs) that are necessary for intron and exon definition and the two transesterification reactions. Cis-sequences that are important for splicing include the 5′-splice site (ss),2 the branch point sequence, the polypyrimidine (PY) tract, and the 3′-ss. These canonical intronic motifs, plus additional regulatory splicing motifs found in exons and introns, are recognized by splicing factors and small nuclear ribonucleoproteins (U1, U2, U4, U5, and U6) to form the spliceosome, which catalyzes intron removal (for a review, see Ref. 3).
There are many splicing factors that are only involved in a subset of splicing decisions. These include the human muscleblind-like family of RNA-binding proteins: MBNL1/2/3 also known as MBNL/EXP, MBLL/MPL1, and MBXL/CHCR, respectively. The founding member of this family, muscleblind (Mbl), was discovered in Drosophila and was shown to be important for photoreceptor differentiation and terminal differentiation of muscles (4, 5). Subsequently, MBNL proteins were found to associate with expanded CUG repeats (located in the 3′-untranslated region of the DMPK gene) that have been shown to act as a toxic RNA and are at least partially responsible for causing myotonic dystrophy (DM) type 1 (for reviews, see Refs. 6–8). The expanded CUG repeats sequester MBNL proteins into nuclear foci, leading to loss of active protein (9, 10). Expanded CCUG repeats within the first intron of ZNF9 also sequester MBNL proteins, and this is thought to be at least partially responsible for causing DM type 2 (for reviews, see Refs. 6–8). The sequestration of MBNL1 leads to missplicing of developmentally regulated events, and a few of these events have been linked directly to symptoms in DM types 1 and 2, such as the missplicing of the chloride channel (CLCN1) leading to myotonia (11). Increased levels of CUGBP1 have also been shown to result in missplicing of certain pre-mRNA transcripts and are linked to causing the heart defects found in DM patients (12, 13).
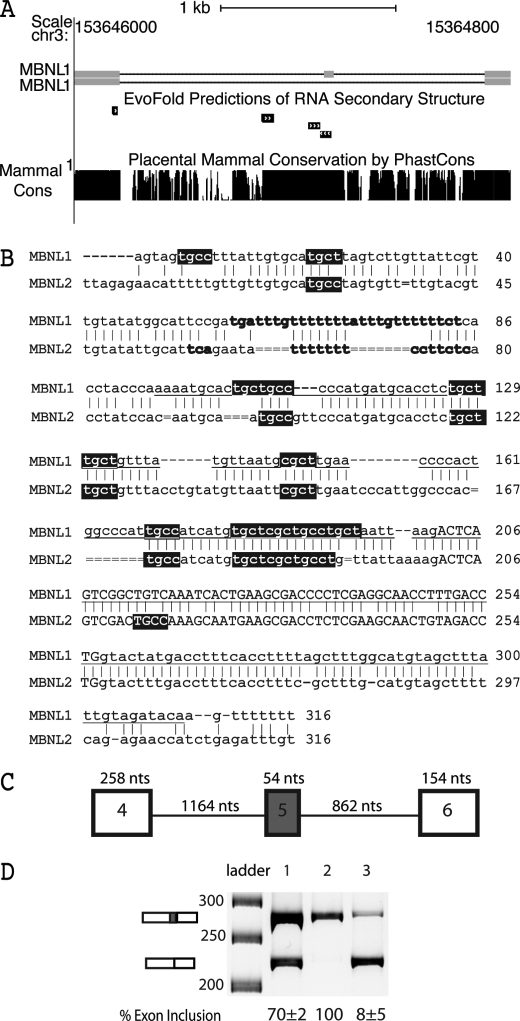
Exon 5 of the MBNL1 pre-mRNA and the paralogous exon in the MBNL2 pre-mRNA are misspliced in DM (14, 15). These paralogous exons are embedded within ultraconserved regions of the genome (16). Fig. 1A shows an edited version of the UCSC Genome Browser showing the conservation upstream, downstream, and within exon 5 of MBNL1 (The ultraconserved element described by Bejerano et al. (16) is underlined in Fig. 1B.). This conserved exon in MBNL1 and MBNL2 encodes 18 amino acids that are C-terminal of the fourth and final zinc finger RNA binding domain. The inclusion of exon 5 causes MBNL1 and MBNL2 to be localized primarily in the nucleus, whereas isoforms of MBNL1 and MBNL2 lacking these amino acids are found both in the nucleus and cytoplasm (14). The MBNL3 gene differs from MBNL1 and MBNL2 in that it lacks a paralogous exon.
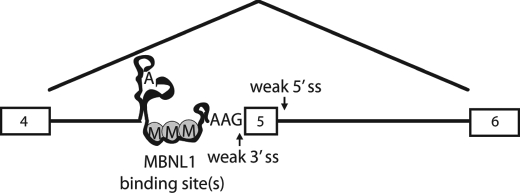
FIGURE 1.
MBNL1 autoregulated splicing. A, two MBNL1 isoforms are depicted with exons as gray boxes. The top isoform contains exon 5. The MBNL1 pre-mRNA contains an ultraconserved region spanning exon 5 through intron 4 shown by a PhastCons plot of placental mammal conservation. RNA structural elements upstream and within exon 5 were predicted by EvoFold (19) and are represented by black boxes with arrows. This figure was edited from the UCSC Genome Browser (GRCh37/hg19 assembly) (44). B, sequence alignment between MBNL1 and MBNL2 shows the 3′-end of intron 4, exon 5, and the highly conserved region of intron 5. Intronic sequence is lowercase, and exon 5 is capitalized. Possible MBNL1/MBNL2 pre-mRNA distant branch points and PY tracts are in bold, and YGCY (MBNL1 binding sites) motifs are highlighted in black. The underlined sequence is the ultraconserved element described by Bejerano et al. (16). Intron 4 of MBNL1 contains a weak AAG 3′-ss, and intron 5 of MBNL1 contains a weak GTACTA 5′-ss. C, schematic of the wild type MBNL1 minigene containing exon 4, intron 4, exon 5, intron 5, and exon 6. D, in vivo splicing of the MBNL1 minigene showing that exon 5 exclusion of the wild type minigene is MBNL1-dependent. Lane 1 shows the wild type splicing of the MBNL1 minigene, lane 2 shows the splicing results from co-transfection of the MBNL1 minigene and CUG960 repeat plasmids, and lane 3 shows the results from the co-transfection of the MBNL1 minigene and MBNL-eGFP plasmids. nts, nucleotides.
We recently identified YGCY as a minimal MBNL1 RNA binding site (17) and demonstrated that insertion of multiple copies of this motif into an intron adjacent to an exon that is not normally regulated by MBNL1 is sufficient for regulation by MBNL1. In general, the location of the YGCY motifs correlates with the effect (e.g. enhancement or suppression) that MBNL1 has on splicing. When YGCY motifs are located upstream of the exon, MBNL1 binding generally leads to exon exclusion; when YGCY motifs are located downstream of an exon, MBNL1 binding generally leads to exon inclusion (17, 18). Twelve YGCY motifs occur within the first 200 nucleotides of the upstream acceptor sequence in MBNL1, but none are found in exon 5 or the donor region (Fig. 1B). The intronic sequence upstream of exon 5 in MBNL2 contains nine YGCY motifs, whereas exon 5 and the intronic region downstream (first 200 nucleotides) only contain one YGCY motif. The location of these YGCY motifs is consistent with MBNL1 proteins acting as repressors of exon 5 in MBNL1 and MBNL2. The location of most of the YGCY motifs upstream of exon 5 in MBNL1 and MBNL2 are conserved (Fig. 1B). Several of the YGCY motifs are found in regions predicted to contain RNA structure elements based on EvoFold (Fig. 1A, gray boxes). It has been proposed that ultraconserved regions could be the result of a requirement for both sequence and RNA structure for function (19).
Previously, we showed that one mechanism through which MBNL1 acts as a repressor is to compete with the basal splicing factor U2AF65 for binding at the 3′-end of the intron (20). In this example, the MBNL1 binding site overlaps with the PY tract (U2AF65 binding site) in intron 4 of the TNNT2 pre-mRNA. We showed that MBNL1 binds intron 4 in the context of an RNA stem-loop and that this complex blocks U2AF65 binding, resulting in inhibition of U2 small nuclear ribonucleoprotein binding, ultimately leading to exon skipping.
The proposed MBNL1 binding sites within intron 4 for the MBNL1 pre-mRNA do not appear to overlap with any of the canonical intronic splicing signals as we observed in the regulation of the TNNT2 pre-mRNA. The architecture of these introns is unique compared with most mammalian introns. For instance, intron 4 of MBNL1 contains a predicted distant branch point sequence (TGAT; Fig. 1B, bold text) that is 141 nucleotides upstream of the 3′-ss. For MBNL2, it is more difficult to predict the branch point because there is not an obvious match to the mammalian branch point consensus sequence CTRAY (21), and the PY tract is shorter in MBNL2 compared with MBNL1 (Fig. 1B). In most mammalian introns, the branch point is found between 20 and 40 nucleotides from the 3′-ss (22). Introns with distant branch point sequences typically lack AG dinucleotides between the branch point sequence and 3′-ss, and this region has been termed an AG exclusion zone (AGEZ). Introduction of an AG in this zone can lead to its use as a cryptic 3′-ss. Introns containing long AGEZs (100 nucleotides or more) are associated with higher rates of alternative splicing, suggesting that these regions contain regulatory elements (23). Interestingly, many genes containing introns with AGEZs of 150 nucleotides or longer are within genes that are either known to be associated with disease or are of biomedical interest (23). Intron 4 in MBNL1 and the paralogous intron in MBNL2 contain an AGEZ. The predicted MBNL1 binding sites are found in the 173-nucleotide AGEZ of intron 4 of MBNL1 and in the 141-nucleotide AGEZ of the MBNL2 intron. The fact that the MBNL1 and MBNL2 introns contain AGEZs and an AAG 3′-ss instead of the YAG consensus 3′-ss found in ∼90% of mammalian introns (24, 25) defines these introns as non-canonical introns.
To study the autoregulated splicing of MBNL1, we created a minigene that contains exons 4, 5, and 6 of MBNL1 and its intervening introns (Fig. 1C). We showed that the MBNL1 protein can regulate a non-canonical intron by binding a mostly unstructured 90-nucleotide response element within the AGEZ upstream of exon 5. Smaller deletions within the MBNL1 response element did not eliminate the ability of MBNL1 to regulate exon 5 exclusion. We determined that intron 4 primarily uses the distant branch point, and deletion of this branch point causes exon 5 skipping.
EXPERIMENTAL PROCEDURES
Sequence Alignment
The sequence alignment of MBNL1 and MBNL2 was made using DNA Strider. Minor adjustments were made to align the 3′-splice site and YGCY motifs.
Labeling of RNAs for Gel Mobility Shift Assay
RNA was transcribed from purified PCR product using T7 RNA polymerase and [α-32P]CTP. The RNA sequence for the 90-nucleotide MBNL1 response element including the T7 site is 5′-GAUAAUACGACUCACUAUAGGGUGCUGCCCCCAUGAUGCACCUCUGCUUGCUGUUUAUGUUAAUGCGCUUGAACCCCACUGGCCCAUUGCCAUCAUGUGCUCGCUGCCUGCU-3′. The T7 site and the three added guanosines for efficient transcription are underlined. The Del 4Δ18, Del 5Δ19, and Del 5Δ19 no YGCY RNAs were transcribed as described for the MBNL1 response element RNA. The Del 5Δ19 no YGCY RNA was created by mutating all GC motifs to AC in the Del 5Δ19 RNA.
Gel Mobility Shift Assay
The assay was performed as described previously (17) except for the following changes. The Del 4Δ18, Del 5Δ19, and Del 5Δ19 no YGCY RNA reactions were run on a 5% polyacrylamide gel (0.5× Tris borate-EDTA, 5% 37.5:1 acrylamide:bisacrylamide) for 2 h at 4 °C at 170 V. The 90-nucleotide MBNL1 response element RNA was run on gels containing 4% glycerol.
Construction of Splicing Reporter Constructs
The MBNL-eGFP construct was obtained from the laboratory of Maury Swanson, and the DMPK-CUG960 plasmid was obtained from the laboratory of Thomas Cooper. The wild type MBNL1 minigene was made by amplifying regions of the MBNL1 gene containing 51 nucleotides from the 3′-end of intron 3, exon 4, intron 4, exon 5, intron 5, exon 6, and 33 nucleotides of the 5′-end of intron 6 from HeLa genomic DNA using PCR primers with unique restriction sites. The forward primer (5′-CCACAGGATCCGCTTCTTCTTCTTCATGTTGACTAAACCTCATG-3′) contained a BamHI site, and the reverse primer (5′-ATTCTTATGCGGCCGCCAGATTCATTTATTAAGAAACCCCACCCC-3′) contained a NotI site. The amplified genomic DNA was cut with BamHI and NotI, inserted into a pcDNA3 plasmid, and sequenced.
The Δbp minigene was made by deleting five nucleotides using standard PCR techniques. The Δbp minigene was created in two segments. The first segment was created by using the forward primer 5′-CCACAGGATCCGCTTCTTCTTCTTCATGTTGACTAAACCTCATG-3′ and the reverse primer 5′-GGGTAGGTGAGAAAAAACAAATAAAAAAACAACGGAATGCCATAACAACGAATAACAAG-3′. The second segment was made using the forward primer 5′-TTGTTTTTTTATTTGTTTTTTCTCACCTACCCAAAAATGCACTGCTGCCCCC-3′ and the reverse primer 5′-ATTCTTATGCGGCCGCCAGATTCATTTATTAAGAAACCCCACCCC-3′. Both segments were then used in the same PCR, and the forward primer 5′-CCACAGGATCCGCTTCTTCTTCTTCATGTTGACTAAACCTCATG-3′ and the reverse primer 5′-ATTCTTATGCGGCCGCCAGATTCATTTATTAAGAAACCCCACCCC-3′ were used to PCR amplify the minigene. The PCR product was cut with BamHI and NotI, ligated into a pcDNA3 plasmid, and sequenced.
The Δ90 minigene was made in two segments. The first segment was made using the forward primer 5′-CCACAGGATCCGCTTCTTCTTCTTCATGTTGACTAAACCTCATG-3′ and the reverse primer 5′-GGCTTTCAATTGGTGCATTTTTGGGTAGGTGAGAAAAAACA-3′. The second segment was made using the forward primer 5′-GGCTTTCAATTGAATTAAGACTCAGTCGGCTGTCAAATCAC-3′ and the reverse primer 5′-ATTCTTATGCGGCCGCCAGATTCATTTATTAAGAAACCCCACCCC-3′. Segment 1 was cut with MfeI and BamHI, and segment 2 was cut with MfeI and NotI. Segments 1 and 2 were then ligated into a pcDNA3 plasmid and sequenced.
The Del 1Δ18 minigene was made in two segments. The first segment was made using the forward primer 5′-CCACAGGATCCGCTTCTTCTTCTTCATGTTGACTAAACCTCATG-3′ and the reverse primer 5′-CATTAACATAAACAGCAAGCAGAGGGTGCATTTTTGGGTAGG-3′. The second segment was made using the forward primer 5′-CCTCTGCTTGCTGTTTATGTTAATGCGCTTGAACC-3′ and the reverse primer 5′-ATTCTTATGCGGCCGCCAGATTCATTTATTAAGAAACCCCACCCC-3′. The two segments were ligated using standard PCR techniques, inserted into a pcDNA3 plasmid, and sequenced.
The Del 2Δ16, Del 3Δ18, Del 4Δ18, and Del 5Δ19 minigenes were made using the PCR techniques described for the Del 1Δ18 minigene. All Del minigenes used the same forward primer for the first segment and the same reverse primer for the second segment. The Del 2Δ16 minigene used the reverse primer 5′-GGTTCAAGCGCATTAACATGCATCATGGGGCAGC-3′ for the first segment and the forward primer 5′-TGTTAATGCGCTTGAACCCCACTGGCCATTGC-3′ for the second segment. The first segment of the Del 3Δ18 minigene was made using the reverse primer 5′-CATGATGGCAATGGGCCAGTGGTAAACAGCAAGCAGAGG-3′, and the second segment was made using the forward primer 5′-CCACTGGCCCATTGCCATCATGTGCTCGC-3′. The first segment of the Del 4Δ18 minigene was made using the reverse primer 5′-GCAGGCAGCGAGCACATGGGTTCAAGCGCATTAAC-3′. The second segment of the Del 4Δ18 minigene was made using the forward primer 5′-CATGTGCTCGCTGCCTGCTAATTAAGACTCAGTCGG C-3′. The first segment of the Del 5Δ19 minigene was made using the reverse primer 5′-GACAGCCGACTGAGTCTTAATTATGGCAATGGGCCAGTGG-3′. The second segment of the Del 5Δ19 minigene was made using the forward primer 5′-AATTAAGACTCAGTCGGCTGTCAAATCACTGAAGCGACCCC-3′.
Cell Culture and Transfection
HeLa cells were cultured and transfected as described previously (17) except for the following changes. 1× antibiotic-antimycotic (Invitrogen) was added to DMEM GlutaMAX media (Invitrogen). Cells were harvested 16–24 h after transfection.
In Vivo Splicing
Splicing assays were done as described previously (26) except for the following changes. All reporters were reverse transcribed using a pcDNA3 plasmid-specific antisense primer, 5′-AGCATTTAGGTGACACTATAGAATAGGG-3′. The −RT reactions were treated the same as the +RT reactions except that no SuperScript II was added to the −RT reactions. The cDNA from the RT reaction (2 μl) was subjected to 26 rounds of PCR (within linear range) in a 20 μl reaction. PCR amplification for all splice products was done using the sense primer 5′-GATCAAGGCTGCCCAATACCAG-3′ and the antisense primer 5′-ATTCTTATGCGGCCGCCAGATTCATTTATTAAGAAACCCCACCCC-3′. The PCR products were resolved on a 6% native polyacrylamide gel (40% 19:1 acrylamide:bisacrylamide) using SYBR Green (Applied Biosystems). The SYBR Green was diluted 1000× in 6× dye. Quantification of bands was done using the Alpha Imager HP Software from Alpha Innotech. Percent exon inclusion was calculated by dividing the amount of the band indicating inclusion by the total amount of splice product (bands indicating inclusion and exclusion). Background was taken from the space between the two bands. All splicing experiments were done in triplicate, and the average with S.D. is shown below gels in the figures.
Branch Point Mapping
The assay was done as described previously (21) except for the following changes. MBNL1 wild type minigene was transfected into HeLa cells, isolated, and treated with DNase as described above. Antisense primer C (5′-GAATAGCTTGTAGTCAGATATAGTTGCTC-3′) was used for reverse transcription using Super Script III. Lariat PCR was done using primers C and sense primer D (5′-GCAGACTCTCTCCTCCTCTCTTCC-3′), and nested PCR was done using antisense primer A (5′-GCTTTTCTGACTGCTAACAAGGAGAGAGC-3′) and sense primer B (5′-TAATTAACTACAAAGAGGAGTTATCCTCCC-3′). Nested lariat RT-PCR products were purified using PCR Cleanup (Qiagen), and individual lariats were isolated by TOPO cloning (Invitrogen) and sequenced.
Protein Purification
The MBNL1 protein construct, which includes amino acids 1–260 and contains an N-terminal GST tag, was expressed and purified as described previously (27) except for the following changes in the protocol. For the lysis of bacterial cells, 10 μg/ml DNase was added with the lysozyme (1 mg/ml), and the cell extract was centrifuged for 30 min at 15,000 rpm. The supernatant was loaded onto GST beads and washed with PBS-T buffer (1× PBS and 1% Triton X-100) and eluted with elution buffer (10 mm reduced glutathione, 50 mm Tris, pH 9.5).
Transcription of Unlabeled RNA
The RNA used for structure probing and footprinting was transcribed from DNA amplified from the MBNL1 pcDNA3 construct used to transfect HeLa cells using the sense primer 5′-GATAATACGACTCACTATAGGGACAACTCAGTAGTGCCTTTATTGTGCATGCTTAGTCTTGTTATTCGTTGTATATGGCATTCCG-3′ (T7 site and additional guanosines are underlined) and the antisense primer 5′-GGGCTGCTGGGCTTTC-3′. To amplify the template, 35 rounds of PCR were done. The template was subjected to PCR cleanup using the Qiagen PCR Cleanup kit. To transcribe DNA, 100–500 ng of DNA was added to each transcription reaction containing 40 mm Tris, pH 8, 10 mm MgCl2, 2 mm spermidine, 0.01% Triton, 1× rNTP, 10 mm DTT, 1 unit of RNasin (Promega), 40% PEG, and T7 DNA polymerase at 37 °C for 1.5–2 h. 1 unit of DNase was added to the transcription and incubated at 37 °C for 1 h. The RNA sample underwent phenol-chloroform extraction and was run on an 11% denaturing gel (7.5 m urea, 11% 29:1 acrylamide:bisacrylamide, 1× Tris borate-EDTA). RNA was eluted using an Elutrap, and the RNA was ethanol-precipitated. The RNA pellet was resuspended in low Tris-EDTA and quantified using Qubit (Invitrogen). RNA was stored at −80 °C.
Footprinting and Structure Probing with T1 and RNase 1
RNA, Structure Buffer, and heparin were snap annealed. The mixture was aliquoted, and different concentrations of GST-tagged MBNL1 were incubated with the RNA sample for 15 min at room temperature. Then 0.25 unit of RNase T1 or 0.3 unit of RNase 1 was added, and the sample was incubated for an additional 15 min (structure probing experiments were done without MBNL1). A final concentration of 81 nm RNA, 0.48 μg of heparin in RNase T1 Structure Buffer (final concentration of 7.7 mm Tris, pH 7, 77 mm KCl, 7.7 mm of MgCl2) and R1 RNase Structure Buffer (final concentration of 7.7 mm Tris, pH 7, 77 mm NaCl, 7.7 mm MgCl2) was ethanol-precipitated by adding 20 μl of inactivation buffer (0.7 m sodium acetate, pH 5.2 in ethanol) and 1 μl of 20 mg/ml glycogen. Samples were incubated on ice for 5 min and spun at maximum speed for 15 min at 4 °C. Pellets were resuspended in a final concentration of 18 nm radiolabeled reverse primer (5′-CAGGTCAAAGGTTGCCTCG-3′) and 0.83 mm dNTPs (the reverse primer was phosphorylated using polynucleotide kinase and [γ-32P]ATP). Samples were incubated at 65 °C for 5 min, at 35 °C for 5 min, and on ice for 1 min. Reverse transcription was carried out by adding appropriate amounts of 5× First Strand buffer, 0.1 m DTT, and 200 units of SuperScript III. Samples were incubated at 52 °C for 1 h and at 70 °C for 15 min. Samples were then phenol-chloroform-extracted and ethanol-precipitated, and the pellet was resuspended in 20 μl of low Tris-EDTA. An equal volume of 2× denaturing dye was added to the samples, which were then incubated at 95 °C for 2 min and run on an 8% denaturing gel (8% 19:1 acrylamide:bisacrylamide, 1× Tris borate-EDTA, 7.5 m urea).
The selective 2′-hydroxyl acylation by primer extension (SHAPE) assay (28) was done as described above except that RNA and heparin were snap annealed in HE buffer (10 mm HEPES, pH 8.0, 1 mm EDTA, pH 8.0). 1× folding mixture buffer (recipe for 3× buffer is 333 mm HEPES, pH 8.0, 20 mm MgCl2, 333 mm NaCl) was added, and the RNA sample was incubated at 37 °C for 20 min. RNA samples were aliquoted, and 1 μl of N-methylisatoic anhydride in neat dimethyl sulfoxide was added to samples. Neat dimethyl sulfoxide was added to a sample as a control (free RNA). Reactions were incubated at 37 °C for 45 min. Inactivation buffer was added, and reverse transcription was done as described above. All RNase T1, RNase R1, and SHAPE gels that were quantitated were done in triplicate. The RNase T1 gel with GST-MBNL1 titration shown in supplemental Fig. 1 was done once.
Quantification and Normalization of T1, RNase 1, and SHAPE Gels
Footprinting and structure probing data were quantified using the SAFA program (29). Data were normalized for each lane as described previously (30). The sequence and text files were imported to the RNAStructure programs (31). The slope (m) and intercept (b) chosen were 2.6 and −0.8 kcal/mol, respectively.
Difference plots were made by subtracting normalized reactivities of the protein lane from the no protein lane. The average of the difference was then added, and the values were plotted. The sequence and SHAPE text files were prepared as described previously (30).
RESULTS
Minigene Recapitulates Autoregulation of MBNL1 Exon 5
The minigene constructed to study splicing of exon 5 contained the full-length sequences of exon 4, intron 4, exon 5, intron 5, and exon 6 (Fig. 1C). The minigene was transfected into HeLa cells to determine whether MBNL1 could regulate splicing of exon 5 in this context. As shown in Fig. 1D, when a second plasmid that expresses 960 CUG repeats (32) was co-transfected into the cells, the inclusion of exon 5 increased to 100% compared with 70% (Fig. 1D). This change is presumably the result of the CUG repeats sequestering the endogenous MBNL proteins. The inclusion of exon 5 could be almost completely blocked (8% inclusion; Fig. 1D) by the overexpression of MBNL1 from a co-transfected plasmid. These results show that protein levels of MBNL1 play a significant role in the regulation of MBNL1 exon 5 and that we can recapitulate MBNL1-regulated splicing in this HeLa cell system.
Characterization of Highly Conserved 3′-End of MBNL1 Intron 4
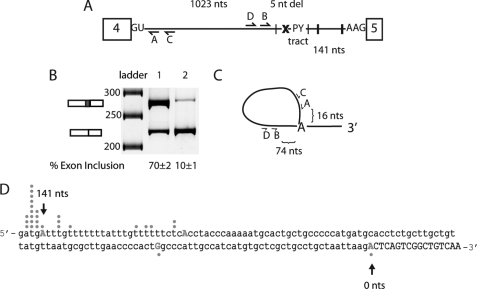
To determine whether the putative distant branch point sequence discussed in the Introduction was important for the splicing of intron 4, the sequence ATGAT (the proposed branch site adenosine is underlined) was deleted (Fig. 2A; referred to as Δbp). As shown in Fig. 2B, the deletion of this motif reduced exon 5 inclusion to 10%. Deletion of the putative branch point sequence caused the splicing machinery to skip this 3′-ss and select the branch point and 3′-ss of intron 5, resulting in skipping of exon 5. These results show that this putative branch point sequence is necessary for high levels of exon 5 inclusion presumably because it contains the branch site adenosine.
FIGURE 2.
Determination of branch point of intron 4 in MBNL1 pre-mRNA. A, schematic of the MBNL1 pre-mRNA showing the predicted MBNL1 binding sites (black hatch marks indicate instances of YGCY motifs); the x represents the deletion of the predicted branch point (ATGAT) in intron 4. The minigene containing the deletion of the predicted branch point was termed Δbp. The locations of the four primers used for branch point mapping are shown (A–D). B, splicing of the wild type and Δbp minigene. Lane 1 contains the wild type MBNL1 minigene, and lane 2 contains the Δbp minigene. C, schematic representation of the primers used for the RT-PCR to isolate lariats. Primer C was used for RT-PCR, primer pairs C and D were used for the initial PCR, and primer pairs A and B were used for the final nested PCR. Distances between primer B and the distant branch point and primer A and the distant branch point are shown. D, sequence for the 3′-end of intron 4 (in lowercase) and part of exon 5 (capitalized) are shown. 145 nucleotides of intronic region upstream exon 5 are shown, and the first 17 nucleotides of exon 5 are shown. Dots correlate with the number of clones for the lariats that were sequenced to the particular nucleotide. The first position in the exon is numbered zero (0) with upstream nucleotides being negative and downstream nucleotides from the first nucleotide of the exon being positive. nt(s), nucleotide(s).
To directly determine whether the adenosine at position −141 within intron 4 functioned as the branch point for this intron, branch point mapping was performed. The strategy to capture intron 4 lariats is shown in Fig. 2C, and previous published protocols were followed (21). Nested PCR was used to decrease background. Primers C and D were used for the first PCR, and primers A and B were used for the second reaction (Fig. 2C). From 45 sequences, 21 mapped to the distant TGAT branch point (−141 nucleotides), five mapped to the end of what we have portrayed as the PY tract (−115 nucleotides), one mapped to a region between the PY tract and 3′-ss (−34 nucleotides), and one mapped to the first nucleotide of exon 5 (0 nucleotides) as shown in Fig. 2C. Dots above and below the nucleotides in Fig. 2D show the likely branch point adenosine for each lariat that was sequenced. The remaining 17 sequences either contained multiple templates, sequences that suggested no lariat formation, or sequences that did not map to intron 4. These results are consistent with the Δbp splicing results (Fig. 2B) and indicate that the distant branch point is the primary branch point for intron 4.
MBNL1 Binds a Primarily Unstructured Region between Distant Branch Point and 3′-Splice Site
With the identification of ultraconserved elements in the genome came the prediction that this conservation could be partly due to RNA structure (16). The program EvoFold predicts three RNA structure elements in the MBNL1 ultraconserved sequence (19) (shown in Fig. 1A). More recently, it has been proposed that these ultraconserved regions have evolved to lack RNA structure (33). To determine the RNA structure of the MBNL1 ultraconserved region and the role of RNA structure if any in MBNL1 binding, structure probing and footprinting experiments were performed.
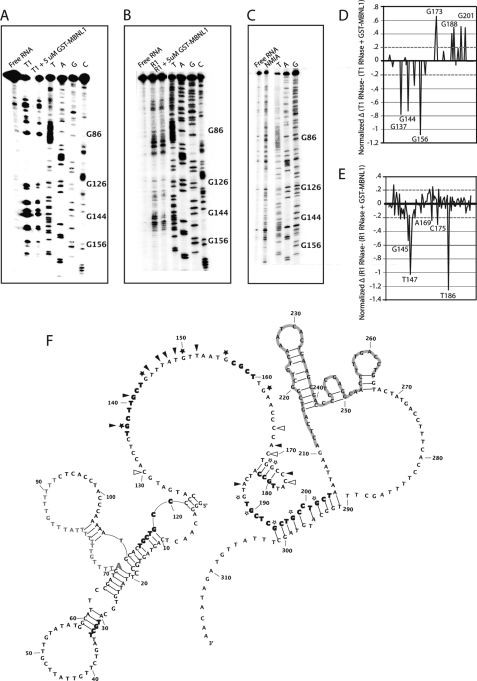
The structure probing and footprinting experiments were done with a 491-nucleotide RNA that contains the last 207 nucleotides of intron 4, all 54 nucleotides of exon 5, and the first 227 nucleotides of intron 5. This RNA included the 212-nucleotide ultraconserved element, which encompasses exon 5 and upstream and downstream intronic sequences (Fig. 1B, underlined). Additional sequence upstream and downstream of the ultraconserved element was used to favor the native secondary structure. Because a larger stretch of sequence upstream of exon 5 is highly conserved and contains proposed MBNL1 binding sites, we focused our footprinting studies on this region of the RNA.
Both SHAPE and RNases were used to determine the secondary structure of the 3′-end of intron 4. RNase T1 cleaves after single-stranded guanosine residues, RNase 1 cleaves after single-stranded nucleotides with a bias for single-stranded pyrimidines, and SHAPE uses N-methylisatoic anhydride to form 2′-O-adducts. N-Methylisatoic anhydride reacts with all nucleotides, and the extent of the modification is dependent on the flexibility of the nucleotide (34). Shown in Fig. 3, A–C, are representative gels of all three assays. The quantified (see “Experimental Procedures”) readout from these experiments was fed into RNAStructure (31) to create the secondary structure shown in Fig. 3F (nucleotides 42–311 were quantified). The RNA appears to have a structured 5′-end, a primarily unstructured linker region containing several of the proposed MBNL1 binding sites, and two more structured regions that contain the end of intron 4 and most of exon 5. The distant branch point sequence is at the junction of two helices with a large bulge that contains the PY tract. The 3′-ss is at the junction of RNA structural elements, and the 5′-splice site is at the end of a helix.
FIGURE 3.
Structure probing and footprinting of 3′-end of intron 4 of MBNL1 pre-mRNA. A, RNase T1 (cleaves single-stranded guanosines) was used for structure probing and footprinting purposes. Lane 1 is the free RNA, lane 2 has RNA and RNase T1, and lane 3 includes RNA, MBNL1, and RNase T1. B, RNase 1 (cleaves single-stranded pyrimidines) was used for structure probing and footprinting purposes. Lanes are similar to that described in A except RNase 1 is used. Lane 3 shows a footprint between nucleotides G126 and G162. C, SHAPE (used to indicate flexible nucleotides) was used to determine the secondary structure of the ultraconserved region. D, a difference plot of the RNase T1 data was created by subtracting the footprinting data (lane 3) from the structure probing data (lane 2) of the RNase T1 gels. E, a difference plot of the R1 RNase data was created in the same way as for D. The difference plots in D and E focus on the region between nucleotides G126 and G210. Nucleotides above 0.2 and below −0.2 (dashed lines shown) were considered to have increased or decreased cleavage, respectively, in the presence of MBNL1. F, experimentally derived secondary structure of the 3′-end of intron 4, exon 5, and the 5′-end of intron 5. Nucleotides 42–311 contain secondary structure data from RNase T1, RNase 1, and SHAPE gels. The structure shown is the compilation of the quantified data of the three structure probing and footprinting assays. Nucleotides that are highlighted in gray are in exon 5. The adenosine in gray with increased font size is the distant branch point, and adjacent nucleotides in gray are the PY tract. Bold nucleotides are potential MBNL1 binding site (YGCY motifs). Symbols placed next to nucleotides show changes in RNA secondary structure due to MBNL1. Black stars and arrowheads indicate nucleotides that are protected in the presence of MBNL1 in the RNase T1 and R1 RNase assays, respectively. Open stars and arrowheads indicate nucleotides that displayed enhanced cleavage in the presence of MBNL1 in the RNase T1 and RNase R1 footprinting assays.
To determine where MBNL1 binds within this RNA, footprinting with T1 and R1 RNases were performed. A titration of MBNL1 protein in the presence of RNase T1 was done to determine the MBNL1 protein concentration required for footprinting (supplemental Fig. 1). MBNL1 was titrated from 0.3 to 10 μm and showed a gradual change in the protection pattern with the end of intron 4 showing protection at higher concentrations. 5 μm MBNL1 was selected for quantitative studies because it showed the most significant and widespread footprint.
The differences in cleavage (see Fig. 3, A and B, lane 3) were quantified by subtracting the averages of the reactivity profiles in the presence and absence of MBNL1 and normalizing the data (Fig. 3, D and E) as described under “Experimental Procedures.” Only nucleotides that showed a difference of more than 0.2 for RNase T1 and RNase 1 are shown in Fig. 3F. Symbols placed next to nucleotides in Fig. 3F show changes in secondary structure only in the presence of MBNL1. For example, G144 showed a large reduction in cleavage by RNase T1, suggesting that MBNL1 interacts with this nucleotide. G137, which is located within a YGCY motif, and G156 (in a UGCG motif) also showed significant protection by MBNL1 (Fig. 3, D and F). It is interesting that of the 10 YGCY motifs within the last 141 nucleotides of intron 4 only two YGCY motifs showed a footprint (nucleotides G137 and G141). However, nucleotides near YGCY motifs, such as G144–G150, G156, C175, and A196, also showed an increased amount of protection, suggesting that MBNL1 interacts with these nucleotides. Although it appears that there are differences in the RNase T1 cleavage upstream of G86 (Fig. 3A, lanes 2 and 3), quantification of three different gels showed no significant difference. Interestingly, some nucleotides were cleaved more in the presence of MBNL1, such as nucleotides G150, C167, C172, G173, G176, G198, C190, G194, G197, and G201. Nucleotides with enhanced cleavage near protected sites could be due to MBNL1 affecting the local secondary structure of the RNA.
Identification of 90-nucleotide MBNL1 Regulatory Element in 3′-End of Intron 4
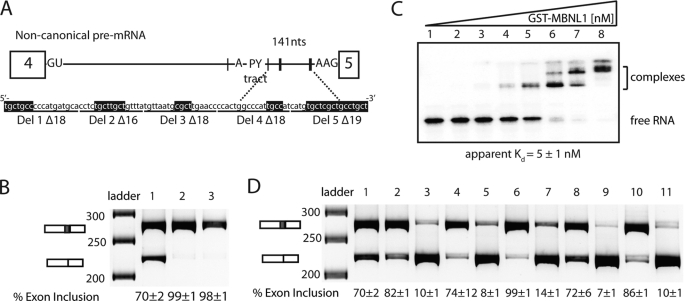
To determine whether the region of intron 4 protected by MBNL1 is required for MBNL1 to negatively regulate exon 5, this section of RNA was deleted from the MBNL1 minigene and replaced by an MfeI-cut site. 90 nucleotides between the distant branch point and 3′-ss were deleted, resulting in an intron 4 lacking all YGCY motifs except two located upstream of the branch point. This deletion (Δ90) also resulted in an intron that was more similar to canonical introns in which the PY tract and branch point are found closer to the 3′-ss (Fig. 4A). Splicing of the Δ90 minigene resulted in almost complete inclusion of exon 5 (Fig. 4B), and the overexpression of MBNL1 did not alter the splicing pattern (Fig. 4B), indicating that this region of the intron (MBNL1 response element) is required for regulation by MBNL1. Interestingly, this element does not appear to contain any essential positive splicing signals for exon 5.
FIGURE 4.
90 nucleotides between PY tract and 3′-splice site are required for MBNL1 autoregulation. A, a schematic representation of the non-canonical intron 4 of MBNL1 and sequence of the 90-nucleotide MBNL1 response element. Black hatch marks indicate instances of YGCY motifs on the schematic with thickness of the hatch mark indicating clustered YGCY motifs. YGCY motifs are highlighted in black on the sequence. The 90-nucleotide MBNL1 response element was deleted from the MBNL1 minigene (Δ90 construct). Shorter deletions within this response element are indicated under the sequence (Del 1–5 with Δ indicating the length of the deletion). B, splicing of Δ90 minigene in HeLa cells in the absence (lane 2) and presence of overexpressed MBNL1-eGFP (lane 3) where lane 1 shows the wild type splicing of the MBNL1 minigene. C, gel shift of the MBNL1 response element. Concentrations of the protein increase from left to right: 0, 0.026, 0.2, 0.9, 6, 30, 200, and 1200 nm. D, splicing of Del 1–5 in HeLa cells in the absence (first of set) and presence (second of set) of overexpressed MBNL1-eGFP. Lane 1 shows the wild type splicing of the MBNL1 minigene, and lanes 2–11 are sets of the following deletions: Del 1, lanes 2 and 3; Del 2, lanes 4 and 5; Del 3, lanes 6 and 7; Del 4, lanes 8 and 9; and Del 5, lanes 10 and 11. nts, nucleotides.
To characterize the binding of MBNL1 to this response element, a gel shift assay was performed with this RNA. MBNL1 bound this RNA with high affinity (apparent Kd of 5 nm; Fig. 4C). It appears that several MBNL1 proteins bind this RNA because three different complexes could be distinguished (Fig. 4C). This result is not surprising given that this 90-nucleotide RNA contains 10 YGCY motifs.
In an effort to determine whether the MBNL1 response element could be pared down to a more minimal element, smaller deletions in this region were made. Del 1Δ18 eliminated the two YGCY motifs closest to the PY tract (the Δ represents the total number of nucleotides deleted in each construct), Del 2Δ16 eliminated the next two downstream YGCY motifs, Del 3Δ18 eliminated one YGCY motif, Del 4Δ18 eliminated one YGCY motif, and Del 5Δ19 eliminated a string of four YGCY motifs (Fig. 4A). Del 1Δ18, Del 3Δ18, and Del 5Δ19 all resulted in an increase of exon 5 inclusion (Fig. 4D) where levels ranged from 82 to 99% inclusion, whereas Del 2Δ16 and Del 4Δ18 both resulted in wild type levels of exon 5 inclusion (∼70% inclusion). In all deletions, MBNL1 was still able to inhibit exon 5 inclusion, and the effect of MBNL1 overexpression was strong in all cases (levels ranged from 7 to 14% inclusion of exon 5). These results indicate that the loss of one to four YGCY motifs in the MBNL1 response element is not sufficient to abrogate the ability of MBNL1 to regulate exon 5 splicing.
To determine how the deletions in the MBNL1 response element affected MBNL1 binding, two RNAs (Del 4Δ18 and Del 5Δ19; 72- and 71-nucleotide RNAs, respectively) were tested in the gel shift assay. Del 4Δ18 was bound by MBNL1 with an apparent Kd of 108 ± 27 nm, and Del 5Δ19 was bound by MBNL1 with an apparent Kd of 70 ± 12 nm (supplemental Fig. 2). These RNAs bound much more weakly to MBNL1 compared with the 90-nucleotide response element, which bound MBNL1 with 5 nm affinity. The approximate 50-fold decrease in binding due to the deletion of a single YGCY motif in Del 4Δ18 was surprising and suggests that this motif and the surrounding sequence are important for high affinity binding by MBNL1. Alternatively, the deletion of the 18 nucleotides could have affected the other MBNL1 sites negatively by altering the RNA structure or spacing of the sites. As expected, when all of the GC motifs in Del 5Δ19 were mutated to AC, MBNL1 did not bind the RNA (supplemental Fig. 2).
DISCUSSION
Polypyrimidine Tract-binding Protein 1 (PTB1) and MBNL1 Negatively Regulate Splicing of Their Pre-mRNAs through Introns Containing Distant Branch Points
PTB1, also known as heterogeneous nuclear ribonucleoprotein I, is an alternative splicing factor that regulates many different splicing events (35). Like MBNL1 and MBNL2, this factor also autoregulates the splicing of its own pre-mRNA through the usage of a predicted distant branch point (36). The distant branch point is contained within a 351-nucleotide AGEZ in intron 10 of the PTB1 pre-mRNA (36). The autoregulation of PTB1 leads to exon 11 exclusion, resulting in a premature termination codon that is predicted to induce the nonsense-mediated decay pathway. This autoregulation of splicing allows PTB1 to tightly control its own protein levels. MBNL1 and MBNL2 differ in that their autoregulation leads to different protein isoforms. Presumably, these different isoforms of MBNL1 and MBNL2 have different functions, but currently, the major known difference between the isoforms is that those lacking exon 5 are found in both the nucleus and cytoplasm, whereas the isoforms containing exon 5 are primarily nuclear (14, 37).
Non-canonical Intron in MBNL1 Pre-mRNA
The 3′-end of MBNL1 intron 4 is different from the 3′-end of most other introns. First, the 3′-end of intron 4 is contained within an ultraconserved element longer than 200 nucleotides and is 100% conserved between human, rat, and mouse genomes (16). Second, this intron is unique because it contains a distant branch point and an AAG 3′-splice site. Most human introns contain a predicted branch point in the last 20–40 nucleotides of the intron and a YAG 3′-ss. In the canonical intron architecture, U2AF35 binds the 3′-ss, U2AF65 binds the PY tract, and U2 small nuclear ribonucleoprotein binds at the branch point (38). It is not clear how these factors recognize introns containing distant branch point sequences. However, it has been suggested that the second step in splicing may involve a mechanism in which the spliceosome performs a linear scan downstream from the distant branch point and PY tract until it reaches the first AG dinucleotide (39, 40). Antibodies to the polypyrimidine tract-binding protein were used to analyze components that assemble on alternatively spliced pre-mRNAs that use a distant branch point (41). Of two pre-mRNAs studied (α- and β-TM), a common set of uncharacterized proteins was identified in addition to PTB1 that assemble on alternatively spliced pre-mRNAs with distant branch point sequences. However, it is unclear how these proteins function in alternative splicing.
One possible mechanism is that the 141-nucleotide RNA linker of MBNL1 contains binding sites for proteins that interact with U2AF35, U2AF65, and U2 small nuclear ribonucleoprotein that facilitate their binding and spliceosome formation. Alternatively, this RNA linker may be sequestered out of the way (likely bound by heterogeneous nuclear ribonucleoproteins) in some manner. It has been shown that the presence of a stem-loop between a distant branch point and 3′-ss inhibits the second step of splicing (39). It is possible that like the stem-loop MBNL1 binding to this region of the intron results in a structure that blocks scanning to the 3′-ss (Fig. 5). The inability to splice this pre-mRNA in vitro blocked our efforts to determine at which step MBNL1 is regulating splicing.
FIGURE 5.
Model for MBNL1 binding and excluding exon 5. MBNL1 binds the 90-nucleotide regulatory region in the AGEZ to regulate exon 5 exclusion. By binding within the AGEZ, MBNL1 appears to inhibit the ability of the spliceosome to locate the 3′-ss. The M enclosed in a circle represents MBNL1 proteins binding the pre-mRNA.
A recent bioinformatics study suggests that highly conserved regions are actually less structured compared with other regions of the pre-mRNA (33), and our structure probing data for MBNL1 are consistent with this hypothesis. It is possible that this lack of structure within highly conserved regions make them more accessible to splicing factors for regulation.
MBNL1 Negatively Regulates Splicing through Multiple Mechanisms
Studies of where MBNL1 binds its pre-mRNA targets to regulate alternative splicing suggest that MBNL1 may not regulate exon exclusion through a conserved mechanism. In intron 4 of the MBNL1 pre-mRNA, we observed that MBNL1 regulates exon 5 exclusion by binding a response element located within an AGEZ just downstream of a distant branch point and PY tract. In the CLCN1 pre-mRNA, MBNL1 represses exon 7A inclusion by binding the 5′-end of exon 7A (which contains an exonic splicing enhancer) and flanking intronic regions (42). Previously, we showed that MBNL1 and U2AF65 compete for binding at the 3′-end of intron 4 of the TNNT2 pre-mRNA to regulate exon 5. MBNL1-regulated TNNT2 exon 5 exclusion may involve MBNL1 binding the RNA in a looped conformation based on a model from a crystal structure of MBNL1 in complex with a short RNA, whereas U2AF65 interacts with the RNA in a single-stranded conformation (43).
The binding of multiple MBNL1 proteins to the MBNL1 intron 4 linker may result in the RNA adopting a conformation that inhibits formation of a functional splicing complex at this 3′-ss. Footprinting and structure probing data showed that MBNL1 binds a mostly unstructured part of the RNA (nucleotides 137–162) and causes more cleavage of nucleotides just downstream (nucleotides 167–201), suggesting that upon binding MBNL1 may cause downstream RNA to become unstructured, allowing more MBNL1 proteins to bind (Fig. 3C). In the presence of MBNL1, the 3′-ss of intron 4 was not accessible to the spliceosome, resulting in the exclusion of exon 5 (Fig. 5). The MBNL1 response element does not overlap with the branch point, PY tract, or 3′-splice site; therefore, direct competition between MBNL1 and constitutive splicing factors does not appear to be a likely mechanism for the regulation by MBNL1. It is possible that additional MBNL1 proteins bind outside of the response element to compete with constitutive splicing factors, or alternatively, the presence of MBNL1 blocks the ability of the splicing machinery to locate (via scanning) the 3′-ss from the distant branch point.
Supplementary Material
Acknowledgments
We thank Stacey Wagner, Rodger Voelker, and other members of the Berglund laboratory for helpful discussions and comments on the manuscript. We are grateful to Amy Mahady for her assistance with cloning the deletion minigenes and to Dr. Cooper and Dr. Swanson for sending us the CUG960 and MBNL1 plasmids, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grant AR053903 (to J. A. B.) and Training Grant GM-07759 (to D. P. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- ss
- splice site
- MBNL
- muscleblind-like
- DM
- myotonic dystrophy
- PY
- polypyrimidine
- SHAPE
- selective 2′-hydroxyl acylation by primer extension
- PTB1
- polypyrimidine tract-binding protein 1
- AGEZ
- AG exclusion zone.
REFERENCES
- 1. Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. (2008) Nature 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J. (2008) Nat. Genet. 40, 1413–1415 [DOI] [PubMed] [Google Scholar]
- 3. Wahl M. C., Will C. L., Lührmann R. (2009) Cell 136, 701–718 [DOI] [PubMed] [Google Scholar]
- 4. Artero R., Prokop A., Paricio N., Begemann G., Pueyo I., Mlodzik M., Perez-Alonso M., Baylies M. K. (1998) Dev. Biol. 195, 131–143 [DOI] [PubMed] [Google Scholar]
- 5. Begemann G., Paricio N., Artero R., Kiss I., Pérez-Alonso M., Mlodzik M. (1997) Development 124, 4321–4331 [DOI] [PubMed] [Google Scholar]
- 6. Ranum L. P., Day J. W. (2004) Am. J. Hum. Genet. 74, 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ranum L. P., Cooper T. A. (2006) Annu. Rev. Neurosci. 29, 259–277 [DOI] [PubMed] [Google Scholar]
- 8. Osborne R. J., Thornton C. A. (2006) Hum. Mol. Genet. 15, R162–R169 [DOI] [PubMed] [Google Scholar]
- 9. Fardaei M., Larkin K., Brook J. D., Hamshere M. G. (2001) Nucleic Acids Res. 29, 2766–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fardaei M., Rogers M. T., Thorpe H. M., Larkin K., Hamshere M. G., Harper P. S., Brook J. D. (2002) Hum. Mol. Genet. 11, 805–814 [DOI] [PubMed] [Google Scholar]
- 11. Charlet-B N., Savkur R. S., Singh G., Philips A. V., Grice E. A., Cooper T. A. (2002) Mol. Cell 10, 45–53 [DOI] [PubMed] [Google Scholar]
- 12. Ladd A. N., Taffet G., Hartley C., Kearney D. L., Cooper T. A. (2005) Mol. Cell. Biol. 25, 6267–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee J. E., Cooper T. A. (2009) Biochem. Soc. Trans. 37, 1281–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Terenzi F., Ladd A. N. (2010) RNA Biol. 7, 43–55 [DOI] [PubMed] [Google Scholar]
- 15. Lin X., Miller J. W., Mankodi A., Kanadia R. N., Yuan Y., Moxley R. T., Swanson M. S., Thornton C. A. (2006) Hum. Mol. Genet. 15, 2087–2097 [DOI] [PubMed] [Google Scholar]
- 16. Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W. J., Mattick J. S., Haussler D. (2004) Science 304, 1321–1325 [DOI] [PubMed] [Google Scholar]
- 17. Goers E. S., Purcell J., Voelker R. B., Gates D. P., Berglund J. A. (2010) Nucleic Acids Res. 38, 2467–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du H., Cline M. S., Osborne R. J., Tuttle D. L., Clark T. A., Donohue J. P., Hall M. P., Shiue L., Swanson M. S., Thornton C. A., Ares M., Jr. (2010) Nat. Struct. Mol. Biol. 17, 187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pedersen J. S., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E. S., Kent J., Miller W., Haussler D. (2006) PLoS Comput. Biol. 2, e33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Warf M. B., Diegel J. V., von Hippel P. H., Berglund J. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9203–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao K., Masuda A., Matsuura T., Ohno K. (2008) Nucleic Acids Res. 36, 2257–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reed R. (1989) Genes Dev. 3, 2113–2123 [DOI] [PubMed] [Google Scholar]
- 23. Gooding C., Clark F., Wollerton M. C., Grellscheid S. N., Groom H., Smith C. W. (2006) Genome Biol. 7, R1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mount S. M. (1982) Nucleic Acids Res. 10, 459–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang M. Q. (1998) Hum Mol. Genet. 7, 919–932 [DOI] [PubMed] [Google Scholar]
- 26. Murray J. I., Voelker R. B., Henscheid K. L., Warf M. B., Berglund J. A. (2008) Genome Biol. 9, R97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Warf M. B., Berglund J. A. (2007) RNA 13, 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Merino E. J., Wilkinson K. A., Coughlan J. L., Weeks K. M. (2005) J. Am. Chem. Soc. 127, 4223–4231 [DOI] [PubMed] [Google Scholar]
- 29. Das R., Laederach A., Pearlman S. M., Herschlag D., Altman R. B. (2005) RNA 11, 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Low J. T., Weeks K. M. (2010) Methods 52, 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reuter J. S., Mathews D. H. (2010) BMC Bioinformatics 11, 129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ho T. H., Charlet-B N., Poulos M. G., Singh G., Swanson M. S., Cooper T. A. (2004) EMBO J. 23, 3103–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sathirapongsasuti J. F., Sathira N., Suzuki Y., Huttenhower C., Sugano S. (2011) Nucleic Acids Res. 39, 1967–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilkinson K. A., Merino E. J., Weeks K. M. (2006) Nat. Protoc. 1, 1610–1616 [DOI] [PubMed] [Google Scholar]
- 35. Llorian M., Schwartz S., Clark T. A., Hollander D., Tan L. Y., Spellman R., Gordon A., Schweitzer A. C., de la Grange P., Ast G., Smith C. W. (2010) Nat. Struct. Mol. Biol. 17, 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wollerton M. C., Gooding C., Wagner E. J., Garcia-Blanco M. A., Smith C. W. (2004) Mol. Cell 13, 91–100 [DOI] [PubMed] [Google Scholar]
- 37. Tran H., Gourrier N., Lemercier-Neuillet C., Dhaenens C. M., Vautrin A., Fernandez-Gomez F. J., Arandel L., Carpentier C., Obriot H., Eddarkaoui S., Delattre L., Van Brussels E., Holt I., Morris G. E., Sablonnière B., Buée L., Charlet-Berguerand N., Schraen-Maschke S., Furling D., Behm-Ansmant I., Branlant C., Caillet-Boudin M. L., Sergeant N. (2011) J. Biol. Chem. 286, 16435–16446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Black D. L. (2003) Annu. Rev. Biochem. 72, 291–336 [DOI] [PubMed] [Google Scholar]
- 39. Smith C. W., Chu T. T., Nadal-Ginard B. (1993) Mol. Cell. Biol. 13, 4939–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen S., Anderson K., Moore M. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grossman J. S., Meyer M. I., Wang Y. C., Mulligan G. J., Kobayashi R., Helfman D. M. (1998) RNA 4, 613–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kino Y., Washizu C., Oma Y., Onishi H., Nezu Y., Sasagawa N., Nukina N., Ishiura S. (2009) Nucleic Acids Res. 37, 6477–6490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Teplova M., Patel D. J. (2008) Nat. Struct. Mol. Biol. 15, 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fujita P. A., Rhead B., Zweig A. S., Hinrichs A. S., Karolchik D., Cline M. S., Goldman M., Barber G. P., Clawson H., Coelho A., Diekhans M., Dreszer T. R., Giardine B. M., Harte R. A., Hillman-Jackson J., Hsu F., Kirkup V., Kuhn R. M., Learned K., Li C. H., Meyer L. R., Pohl A., Raney B. J., Rosenbloom K. R., Smith K. E., Haussler D., Kent W. J. (2011) Nucleic Acids Res. 39, D876–D882 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.