Abstract
During the orchestrated process leading to mature erythrocytes, reticulocytes must synthesize large amounts of hemoglobin, while eliminating numerous cellular components. Exosomes are small secreted vesicles that play an important role in this process of specific elimination. To understand the mechanisms of proteolipidic sorting leading to their biogenesis, we have explored changes in the composition of exosomes released by reticulocytes during their differentiation, in parallel to their physical properties. By combining proteomic and lipidomic approaches, we found dramatic alterations in the composition of the exosomes retrieved over the course of a 7-day in vitro differentiation protocol. Our data support a previously proposed model, whereby in reticulocytes the biogenesis of exosomes involves several distinct mechanisms for the preferential recruitment of particular proteins and lipids and suggest that the respective prominence of those pathways changes over the course of the differentiation process.
Keywords: Cell Differentiation, Erythropoiesis, Membrane Trafficking, Proteomics, Vesicles, Exosomes, Lipidomics, Membrane Microdomains, Reticulocyte Maturation
Introduction
The maturation of reticulocytes into erythrocytes is the final step of erythropoiesis. Following expulsion of the nucleus, this process of terminal differentiation, which takes place over the course of a few days, occurs first in the erythroblastic islands of the bone marrow and then in the blood circulation (1). It corresponds to a phase of extensive cellular remodeling, which entails the following: (i) changing from large, motile, and irregularly shaped cells of disperse sizes to erythrocytes, which are small and uniformly biconcave discoid cells; (ii) discarding intracellular organelles; (iii) completing the production of hemoglobin and other erythrocyte-specific proteins; and (iv) modifying the protein and lipid composition of the plasma membrane (2–4). One of the major processes involved in the elimination of the cellular components is the release of small proteolipidic vesicles, called exosomes, into the extracellular medium after fusion of the multivesicular bodies issued from endosomal compartments with the plasma membrane (5–7).
Exosomes are small vesicles (50–100 nm) delineated by a proteolipidic bilayer. They were discovered in the 1980s by Johnstone and co-workers (8) during their study of the fate of the transferrin receptor (TfR),3 which is eliminated during the course of the reticulocyte maturation. Since their discovery in the content of the context of reticulocyte maturation, the secretion of exosomes has been observed in various cell lines (9), and additional roles have been uncovered, such as in the context of the immune system as intermediates of intercellular communications (10, 11) and even in the intercellular transfer of material such as mRNA (12). Moreover, the use of exosomes as biomarkers for various pathologies (13) and as vectors for anti-tumoral vaccination (14) has been proposed. Consequently, a sizable number of studies have sought to understand their biogenesis and functions. From analyses of the proteolipidic composition of exosomes in various cell lines, three different “routes” for the sorting of proteins and lipids into exosomes have been proposed, which would act in parallel in reticulocytes (6, 7). The first route relies on cytosolic machineries involving the protein Alix and either ESCRT complexes (for endosomal sorting complex required for transport) (15) and/or lysobisphosphatidic acid associated with a pH gradient (16). The second route is associated with membrane lipid microdomains enriched in ceramides (17, 18). The third route is initiated on the lumenal side of the endosomal compartment by aggregating factors such as lectins (19, 20).
To date, however, no proteomic or lipidomic characterization of exosomes produced by reticulocytes has yet been reported. The mechanisms regulating the processes of proteolipidic sorting resulting in the secretion of exosomes in these cells that are devoid of nuclei remain unexplained. Indeed, although there is no transcription in reticulocytes, the defects in cell surface expression associated with certain erythroid diseases suggest that exosome biogenesis is subject to regulatory mechanisms (7). For those reasons, the complexity of the biogenesis of reticulocyte exosomes is worthy of further investigations. To this end, we have established a protocol for maintaining young reticulocytes in culture in vitro for a week, during which they mature and produce sizable quantities of exosomes. After ensuring that the materials produced indeed had all the physical characteristics of bona fide exosomes, we analyzed their proteolipidic composition over the course of the reticulocytes' maturation process. For this we used mass spectrometry for a thorough characterization of their protein content, whereas the lipid contents were determined by high performance chromatography techniques. This combinatorial approach revealed a specific and sequential sorting of both proteins and lipids in exosomes, suggesting an orchestrated coexistence of the different mechanisms described above. Our study not only contributes to the understanding of the process of exosome biogenesis but also provides hypotheses regarding the involvement of exosomes in particular erythrocytic diseases.
MATERIALS AND METHODS
Animals
For the production of reticulocytes, we used retired Sprague-Dawley male rats (Janvier, France), in which we induced anemia via two injections of an aqueous solution of phenylhydrazine hydrochloride (Sigma, 50 mg/ml) at the dose of 50 mg/kg on days 1 and 2. Rats were then sacrificed on day 5 by terminal exsanguination under pentobarbital anesthesia. Blood clotting was prevented by adding heparin to the syringes used for the collection. All procedures were reviewed and approved by the Animal Care and Use Committee of the Institut de Pharmacologie et de Biologie Structurale.
Cells
Erythrocytes were obtained from the blood of treated and untreated rats. After an initial wash in 1 volume of PBS, blood cells were separated on a Percoll gradient (d: 1.100, 1.105, 1.110, and 1.123 g/ml, 5 min at 500 × g, 5 min at 1500 × g, and 45 min at 3500 × g). Young reticulocytes were collected in the 1.100 density band and washed three times with DPBS, and their purity was assessed by FACS analysis (see supplemental Fig. S1). Purified reticulocytes were placed in culture at 37 °C with 5% CO2 in 33 volumes of RPMI 1640 medium supplemented with 5 mm glutamine (Invitrogen), 5 mm adenosine, 10 mm inosine (Sigma), and 4.5% exosome-free fetal calf serum, obtained by overnight ultracentrifugation at 100,000 × g, 50 units/ml penicillin, and 50 μg/ml streptomycin (Invitrogen). Exosomes were isolated after 36, 84, and 156 h of culture (called D2, D4, and D7, respectively) by differential centrifugation; cells were first separated by gentle centrifugation (1500 × g for 15 min at 4 °C), before placing them back in culture in fresh medium.
Antibodies, Fluorescents Probes, and Reagents
For flow cytometry reagents, monoclonal hybridoma supernatants from F16.4.4 and MRC-OX18 (anti-rat MHCI) and MRC-OX26 (anti-rat CD71) were home-produced; goat anti-mouse Alexa488, Alexa647-conjugated human transferrin, and Syto12 nucleic acid stain were purchased from Invitrogen. 1,6-Diphenyl-1,3,5-hexatriene (DPH) was from Molecular Probes (Eugene OR).
Chemicals
The following chemicals were used: cholesterol, 1,2-palmitoyl-sn-glycero-3-phosphocholine (DPPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), brain sphingomyelin, and EDTA were purchased from Sigma; MOPS, sodium azide (NaN3), and all other chemicals were of analytical grade and were obtained from Fluka (Buchs, Switzerland). The purity of phospholipids and sterols was tested by thin layer chromatography with chloroform/methanol/water (65:25:4, v/v) as migration solvent. Their detection was performed with Dittmer's reagent. Stock solutions of lipids were prepared in chloroform/methanol (9:1, v/v) and stored at 4 °C. Phospholipid concentrations were determined by a standard phosphate assay according to Zak et al. (59). All water used was purified through a Milli-Q system (Millipore, Bedford, MA).
FACS Staining and Analyses
For staining with antibodies (OX26, OX18, and F16.4.4), pellets of 7·106 cells were resuspended in 50 μl of tissue culture supernatant or in FACS buffer (DPBS, 2% fetal bovine serum) for the negative controls and incubated at 4 °C for 45 min. Samples were then washed three times in FACS buffer, before incubation with Alexa488 anti-mouse secondary antibody (1:200, Invitrogen) for 45 min at 4 °C, and three final washes before analysis. Staining with transferrin-Alexa647 (Invitrogen) was carried out with 5 μl of 5 mg/ml stock solution in a final volume of 50 μl, for 90 min at 4 °C, followed by three washes in FACS buffer. Staining with Syto12 was carried out with a final concentration of 5 μm applied 10 min before FACS analysis without rinsing. All FACS acquisitions were carried out on an LSRII flow cytometer (BD Biosciences), and raw data were subsequently analyzed with FlowJo software (Version 7.6; TreeStar Inc.). All analyses were gated on live cells as determined by forward and side scatter. Analyses carried out on separate days were performed using the same staining solutions, the same machine, and exactly the same settings, ensuring each time that identical intensities were obtained for the negative and positive controls (nucleated cells).
Exosome Production and Preparation
Supernatants from the maturing reticulocytes, containing exosomes, were first centrifuged (20,000 × g for 30 min at 4 °C) to remove cellular debris. Exosomes were then recovered from that supernatant by ultracentrifugation (100,000 × g for 120 min at 4 °C). The pellet (exosome fraction) was washed twice by centrifugation (100,000 × g for 1 h at 4 °C) in Hepes 20 mm, NaCl 150 mm, pH 7.2, and the pellet was resuspended in the same buffer with complete antiprotease (Roche Applied Science). The protein concentration was determined using the Bradford method. Phospholipid (PL) concentrations were measured with an optimized Rouser procedure (21).
Sucrose Gradient
Exosomes were layered on top of a 10-ml discontinuous sucrose gradient (0.5–2.5 m sucrose) and spun in a swinging ultracentrifuge rotor (Beckman SW41). Gradients were centrifuged at equilibrium for 16 h at 200,000 × g after which 900-μl fractions were collected from the top of the tubes. Each volume was divided in two, and 900 μl of water was added to each. Samples were then centrifuged for 1 h at 100,000 × g, and each pellet was resuspended in Laemmli buffer before separation by SDS-PAGE. Gels were analyzed by silver or Coomassie Blue staining.
Transmission Electron Microscopy
Specimens were prepared for EM using the conventional negative staining procedure. Drops of purified exosomes (5–10 μl corresponding to 1–5 μg of protein) were absorbed on Formvar carbon-coated grids for 20 min. The drops were then blotted, and the grids were fixed in 2% paraformaldehyde, washed with water, and negatively stained with uranyl acetate (1%) for 10 min. Grids were examined with a transmission electron microscope (Jeol 1200 EX) at 80 kV. Images were acquired using a digital camera (AMT-USA) at a ×40–300,000 magnification. The diameter of exosomes was evaluated using the software AMT Advantage HR camera system. Measurements were performed on 40 pictures taken on two separate preparations of D2, D4, and D7 exosomes. In total, around 150 vesicles were measured for each condition.
Proteomic Analysis
After reduction and alkylation of cysteine residues, each sample, corresponding to 20 μg of proteins, was separated by 4–12% SDS-PAGE. Proteins were visualized by Coomassie Blue staining, and each lane was cut into 10 slices and subjected to in-gel tryptic digestion. For the initial preliminary analysis, the tryptic digests were analyzed by nanoLC-MS/MS using a Qq-Tof mass spectrometer (QSTAR Pulsar XL, Applied Biosystems, Foster City, CA), and the two subsequent analyses were run on an UltiMate 3000 system (Dionex, Amsterdam, The Netherlands) coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) as described previously (22), except that peptides were eluted using a 0–50% gradient of solvent B during 55 min at a 300 nl/min flow rate. Mascot (2.3.01) was used to automatically extract peak lists from .raw files. MS/MS data were searched against Rattus sequences in the public data base UniProt Knowledgebase release 2010_09, which consists of Swiss-Prot release 2010_09 and TrEMBL release 2010_09. Protein hits were automatically validated using in-house software MFPaQ version 4.0. Mascot results were parsed with the in-house developed software Mascot File Parsing and Quantification (MFPaQ) version 4.0 (23), and protein hits were automatically validated if they satisfied one of the following criteria: identification with at least one top ranking peptide with a score greater than the significance threshold score for p < 0.001 or at least two top ranking peptides with a score greater than the significance threshold score for p < 0.05. Validated proteins were submitted to a relative quantification using MS/MS spectral counting, which has been adapted from the method described by Liu et al. (24). This strategy does not require any labeling of proteins but is based on the counting of the total number of MS/MS spectra identified for a protein.
Lipidomic Analysis
For neutral lipid molecular species analysis, lipids, corresponding to 20 nmol of phospholipids, were extracted from D2, D4, and D7 exosomes according to Bligh and Dyer (25) in chloroform/methanol/water (2.5:2.5:2.1, v/v/v) in the presence of the following internal standards, 2 μg of stigmasterol, 2 μg of ceramide NC15 (prepared according to Vieu et al. (26)), 2 μg of cholesteryl heptadecanoate, 2 μg of glyceryl triheptadecanoate. Chloroform phases were evaporated to dryness, and dissolved in 20 μl of ethyl acetate. The lipid extract (1 μl) was analyzed by gas-liquid chromatography on a FOCUS Thermo Electron system using Zebron-1 Phenomenex fused silica capillary columns (5-m × 0.32-mm inner diameter, 0.50-μm film thickness) according to Barrans et al. (27). The raise of oven temperature was programmed from 200 to 350 °C at a rate of 5 °C/min, and the carrier gas was hydrogen (0.5 bar). The injector and the detector were at 315 and 345 °C, respectively.
Cer-SM Analysis
The dried neutral lipid extracts were submitted to a mild alkaline treatment in methanolic NaOH, 0.6 n (1 ml), and then to sialylation in 50 μl of N,O-bis(trimethylsilyl trifluoroacetamide (1% trimethylsilyl chloride)/acetonitrile (1:1, v/v) overnight at room temperature. Crude extracts (5 μl) were directly analyzed by gas-liquid chromatography according to Vieu et al. (26), on a 4890 Hewlett-Packard system using a RESTEK RTX-50 fused silica capillary column (30-m × 0.32-mm inner diameter, 0.1-μm film thickness). Oven temperature was programmed to raise from 195 to 310 °C at a rate of 3.5 °C/min, and the carrier gas was hydrogen (0.5 bar). The injector and the detector were at 310 and 340 °C respectively.
The phospholipid composition of the lipid extracts of D2, D4, and D7 exosomes obtained by the Folch procedure (60) was determined by HPTLC (nano-SILGUR-20, Macherey Nagel) following Yao and Rastetter (28). Plates were labeled with I2 vapor for 1 h, before scanning at 300 dpi resolution. The intensities of the various spots corresponding to the different lipid species were then quantified with the ImageJ software. The ratios of the various lipids were then calculated using these values because we found that, under the conditions used, equivalent amounts of purified standards of dipalmitoylphosphatidylethanolamine, POPC, brain SM, and bovine phosphatidylserine led to very comparable intensities of I2 coloration for these four classes of PL.
Western Blot Analysis
Samples of exosomes containing adjusted amounts of proteins were separated on 4–12% SDS-PAGE (clearPAGETM, CBS Scientific) and then transferred to nitrocellulose (Amersham Biosciences). The membranes were stained with Ponceau Red to validate that all samples contained similar amounts of protein. Primary antibodies were all used at the indicated dilution in TBS 0.1% Tween, 5% nonfat milk/rabbit anti-class I MHC polyclonal serum (1:1000), raised against a peptide corresponding to RT1.Aa cytoplasmic tail (gift from Simon Powis, University of Saint Andrews), rabbit anti-proteasome (Enzo Life Sciences, BML-PW8155, 1:1000), rabbit anti-Hsp90 (Cell Signaling Technology, C4565, 1:1000), mouse monoclonal BB70 anti-Hsc70 (Assay designs, SPA822, 1:1000), rabbit anti-Tsg101 (Abcam®, ab30871, 1:400), rabbit anti-STAM (Abcam, ab76061, 1:300), rabbit anti-Vps36 (Abcam, ab76331, 1:100), and rabbit anti-CHMP4B/CHMP4C (Abcam, ab76334, 1:300). Proteins were visualized by ECL detection system (Perbio Science) using anti-rabbit or anti-mouse HRP-conjugated IgG (Bio-Rad, dilution: 1:10,000).
Preparation of Small Unilamellar Vesicles
Liposomes were prepared by mixing appropriate volumes of lipid stock solutions. Solvent was evaporated under a nitrogen stream, and samples were pumped under vacuum for 2 h (1 torr). The dried lipid samples were hydrated in MOPS buffer (10 mm, 100 mm NaCl, 0.02% (w/v) NaN3, pH 7.4) at room temperature or heated to 60 °C for DPPC preparations. Samples were vortexed for 4 min to obtain multilamellar vesicles (at final concentrations of 0.05 mm) and were then sonicated for 8 min at a temperature above the Tm using a VibraCell tip sonicator (Sonic & Materials Inc, Danby, CT) delivering 12.5 watts to obtain a mixture of unilamellar vesicles that were resized by 11 successive extrusions through a mini-extruder with a pore size of 0.05 μm (Avanti Polar Lipid, Alabaster AL). The size distribution of those small unilamellar vesicles was checked by quasi-elastic light scattering using a DynaPro device (Wyatt Technology Corp., Santa Barbara, CA) and exhibited a monomodal size distribution with a mean diameter of 57 ± 6 nm.
Fluorescent Labeling of Exosomes with DPH
For fluorescence measurements, amounts of exosomes corresponding to 10–20 nmol of phospholipids were resuspended in 1 ml of Hepes buffer containing a protease inhibitor mixture tablet (Roche Applied Science). To ensure that the anisotropy measurements would not be significantly affected, we always checked that the optical density of the mixture was less than 0.08 at 348 nm. DPH labeling was then carried out at a molecular ratio of 1 probe/360 phospholipids, by adding stock solutions of DPH dissolved in dimethylformamide (5 μl maximum). Exosome/DPH suspensions were incubated 15–30 min at room temperature under gentle agitation before the experiment. Small unilamellar vesicles used as controls were labeled using a similar protocol.
Steady-state Fluorescence Polarization Experiments
The experiments were carried out with a homemade automatic system (29). Labeled samples were placed in a closed housing system, thermostatically controlled by Peltier elements. The temperature increased from 18 to 50 ± 0.1 °C at a rate of 1 °C/3 min. Wavelengths of 348 nm for excitation and 426 nm for emission were used. Polarized fluorescence intensities were corrected for the relative sensitivity of the two detection systems by determining, at each measurement point, the G factor according to Lakowicz (30) and shown in Equation 1,
where IVH corresponds to the intensity of horizontally polarized emission after vertically polarized excitation. Fluorescence anisotropy values were determined within an error of 2.5%.
RESULTS
In Vitro Maturation of Reticulocytes
To produce large numbers of reticulocytes of similar maturation states, we chose to use retired male rats because of their relatively large size and to induce anemia over 2 days by injection of high doses of phenylhydrazine (31, 32). This treatment provokes the precipitation of hemoglobin in the circulating red blood cells and thus causes extensive hemolysis, but it is neither painful nor stressful for the rats because, contrary to exsanguinations, it requires minimal intervention. The hematopoietic system then responds via the induction of a massive production of new reticulocytes, which end up leaving the bone marrow for the bloodstream in an immature state. Three days after the end of the treatment, those reticulocytes can amount to as much as 40% of the circulating red blood cells, compared with 2 or 3% in a normal untreated animal. Less dense than mature erythrocytes, those reticulocytes can be enriched via a Percoll gradient. Under our experimental conditions, the reticulocyte purity after this enrichment was always found to exceed 90% both by flow cytometry after staining with Syto12 (supplemental Fig. S1) and by optical microscopy after methylene blue coloration (not shown).
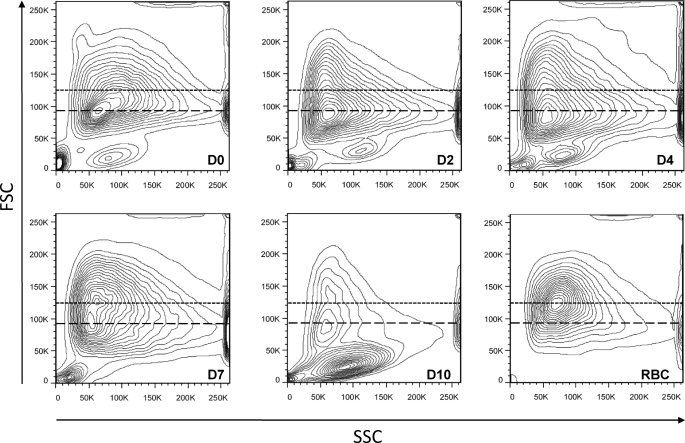
The purified reticulocytes were then placed in culture for up to 10 days, with the medium being changed on D2, D4, and D7, and exosomes were recovered from the cell culture supernatants via ultracentrifugation. When the physical state of the cells was monitored by flow cytometry using forward size scatter (FSC, size) and side size scatter (granulosity) (Fig. 1), there was no sign of significant cell death up to day 7 (Fig. 1, events with FSC <40,000). Furthermore, on day 7, the reticulocytes displayed an increase of FSC, with a sizable proportion of the cells shifting toward a peak of FSC value similar to that of mature red blood cells (Fig. 1, around 125,000). This was accompanied by a very noticeable increase of their hemoglobin content, as detected by visual inspection of the cell pellets (data not shown). Three days later, however, most of the cells showed the much reduced FSC typical of dead cells, which underlines the fact that, in our system, the reticulocytes are all rather well synchronized, but suggesting that, even if the in vitro conditions used allow for their differentiation to progress quite far, certain factors must be missing to generate fully mature long lived red blood cells. Worthy of note, very low amounts of exosomes were recovered in the D10 culture medium, supporting the notion that the materials recovered on previous days are bona fide exosomes that are actively secreted by differentiating reticulocytes and not vesicles released by dying cells.
FIGURE 1.
Monitoring the viability of reticulocytes during their culture in vitro. The physical characteristics of reticulocytes (FSC/side size scatter (SSC) profiles) were recorded just after their purification on a Percoll gradient (D0) and when the medium was changed on D2, D4, D7, and D10. Those were compared with red blood cells (RBC) obtained from an untreated rat. The dotted lines indicate the position of the peak FSC value of RBC (125,000), and the dashed lines indicate that of the peak FSC value of D0 reticulocytes (92,000). Similar results were obtained in three other independent experiments.
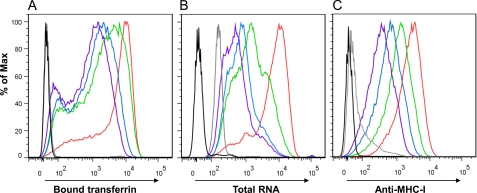
The maturation state of reticulocytes over 7 days was further monitored by flow cytometry on D0, D2, D4, and D7, using three different parameters. First, using fluorescent transferrin, we saw a decrease in the amount of TfR present at the surface of reticulocytes (Fig. 2A), which correlated with a decrease of labeling with anti-TfR antibodies (data not shown). Second, we followed the changes of the RNA content of the reticulocytes as revealed by Syto12 staining, and we found a progressive decrease, with levels of staining approaching those seen in mature red blood cells (Fig. 2B). Finally, because mature red blood cells carry minimal amounts of MHC class I molecules on their surface (albeit more in rodents than in humans), we also stained the maturing reticulocytes with antibodies directed against MHC class I molecules and recorded a similar progressive decrease (Fig. 2C). Although our protocol includes many washing steps, we were initially concerned that remaining traces of phenylhydrazine may hinder the capacity of the reticulocytes to differentiate. In light of our results, however, it seems that, under the cell culture conditions used, the reticulocytes harvested from rats having received this drastic treatment undergo a maturation process that is similar to what happens physiologically in vivo. A further argument lies with the fact that although native hemoglobin is nonfluorescent, the precipitated hemoglobin that results from the phenylhydrazine treatment becomes highly autofluorescent over a broad spectrum (see supplemental Fig. S1 for example). Accordingly, although the hemoglobin contents of the reticulocytes increased progressively over time in culture, we observed no increase of their autofluorescence.
FIGURE 2.
Flow cytometry monitoring of in vitro maturation of reticulocytes. On all three graphs, the different color curves correspond to the following days: D0 (red), D2 (green), D4 (blue), D7 (purple); control, unstained reticulocytes (black); and RBC obtained from an untreated rat (gray). A, TfR expression, as revealed by binding of Alexa647-conjugated human transferrin. B, intracellular RNA, revealed with Syto12 staining. C, cell surface MHC-I, stained with MRC-OX18, followed by secondary anti-mouse antibody conjugated to Alexa488. Staining with F16-4-4 gave comparable results (data not shown). Similar results were obtained in 10 other independent experiments.
Physical Characterization of the Exosomes Released by Reticulocytes
Exosomes were recovered from the cell culture supernatant via ultracentrifugation, following a well defined protocol (33). In our case, two additional washes were added to remove any trace of contaminating soluble proteins that could interfere with the subsequent proteomic analysis (see under “Materials and Methods”). At the end of this procedure, the pellets of exosomes were resuspended in Hepes 20 mm, NaCl 150 mm, and protein and lipid concentrations were evaluated as described under “Materials and Methods.” Typically, from one rat of more than 300 g, we collected 15 ml of blood, corresponding to 5 ml of red blood cells from which 1 ml of purified reticulocytes were recovered after the Percoll gradient. From 1 ml of reticulocyte pellet, the amounts of exosomes recovered decreased progressively over time, with yields of roughly 50 μg on D2, 35 μg on D4, and 30 μg on D7. Remarkably, the ratio of phospholipids over proteins also decreased very noticeably over time (Table 1), suggesting that the composition of the exosomes produced by the reticulocytes changes over the course of their maturation (see below).
TABLE 1.
Phospholipid-to-protein ratios in exosomes (n = 5)
| Ratios | D2 | D4 | D7 |
|---|---|---|---|
| PL/proteina | 1.02 ± 0.09 | 1.06 ± 0.10 | 0.40 ± 0.06 |
| Chol/proteina | 0.43 ± 0.08 | 0.42 ± 0.08 | 0.17 ± 0.04 |
| Chol/chol + PL | 0.30 ± 0.05 | 0.28 ± 0.04 | 0.30 ± 0.04 |
a Nanomoles of PL or chol/μg of proteins are shown.
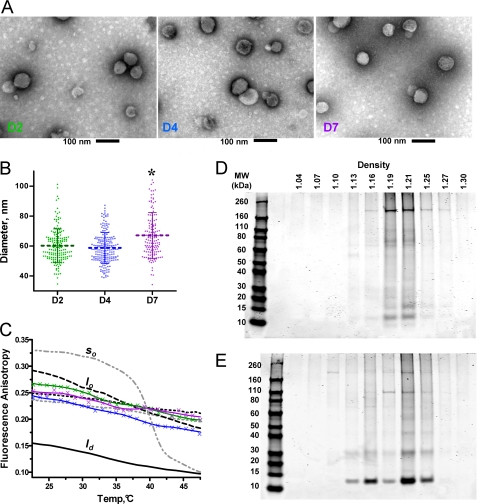
To ensure that the materials recovered at D2, D4, and D7 all corresponded to bona fide exosomes, we compared the morphology and physical characteristics of the different preparations with several complementary approaches as follows: electron microscopy, fluorescence anisotropy, and sedimentation on sucrose gradients. At all three time points, the materials recovered were composed of typical vesicles with the expected aspect and size for exosomes (34) as observed on transmission electron microscopy images (Fig. 3A). Measurements of the diameters of a large number of exosomes on the collected transmission electron microscopy images revealed that D7 vesicles (67 ± 15 nm S.D.) were roughly 10% larger than those harvested on D2 and D4 (60 ± 10 nm S.D.) (Fig. 3B). This difference was found to be significant in terms of S.E. (p < 0.0001).
FIGURE 3.
Morphological and physical characterization of the vesicles harvested on D2, D4, and D7. A, images obtained by transmission electron microscopy on the exosomes collected at D2 (right), D4 (middle), and D7 (left); scale bar, 100 nm. B, distribution and the mean ± S.D. for the diameters of the exosomes harvested at D2, D4, and D7 of culture, measured by the software AMT Advantage HR camera system. Similar results were obtained in two experiments. *, differences between D7 and D4 or D2 were found to be highly significant with a t test (p value < 0.001). C, fluorescence anisotropy values measured for DPH inserted in D2, D4, and D7 exosomes, as well as in various small unilamellar vesicles used as control for the three different physical states encountered in lipid bilayers as follows: gel (so) liquid ordered (lo), and liquid disordered (ld). Color code: exosome preparations: D2 (green), D4 (blue), and D7 (purple); small unilamellar vesicle suspensions: DPPC without chol (gray dash) so > ld transition; POPC without chol (black dash) ld; DPPC with 40% chol (gray dots) stable lo; POPC with 40% chol (black dots) stable lo; POPC with 30% chol and 30% SM (black dash-dot-dash) stable lo. D and E, SDS-PAGE analysis of sucrose gradients fractions isolated at D2 (D) and D7 (E) revealed by Coomassie Blue.
The physical state of the lipid phase of the exosome membranes, expected to be liquid ordered on the basis of previous analyses (35), was characterized by fluorescence anisotropy measurements using DPH, varying the temperature from 20 to 50 °C. The DPH probe reports on the conformational and rotational mobilities of lipid acyl chains and enabled the discrimination between the different possible states of the lipid phase by comparing the measurements with those obtained for model membranes (29). As references, we chose suspensions of lipid vesicles of various compositions as follows: POPC for the disordered liquid (ld), POPC/chol (6:4), DPPC/chol (6:4), and POPC/SM/chol (4:3:3) for the liquid ordered phase (lo), and DPPC for the gel phase (slo) below the transition temperature and liquid disordered phase (ld) above. Over the entire temperature range, fluorescence anisotropy values on D2, D4, and D7 exosomes preparations were reproducibly found to be close to the values obtained for the model vesicles in the lo lipid phase (Fig. 3C).
Finally, we used centrifugation in discontinuous sucrose gradients followed by SDS-PAGE analysis of the different fractions to evaluate the densities of the vesicles. Although D2 and D4 vesicles had comparable densities distributed between 1.16 and 1.21 g/ml (D2, see Fig. 3D; D4, data not shown), D7 vesicles were found between 1.13 and 1.25 g/ml (Fig. 3E). In addition, the protein composition was found to be noticeably different between the D2 and D7 preparations, with those of D4 showing a somewhat intermediate composition between the two (data not shown).
Altogether, our results show that on D2, D4, and D7, the preparations were composed of vesicles that have the physical characteristics (size, density, and membrane fluidity) of typical exosomes (33, 36). Next, to investigate the origin for the modifications in the proteolipidic composition of exosomes over time, we undertook the characterization of their contents by parallel proteomic and lipidomic approaches.
Proteomic Characterization of Exosome Preparations
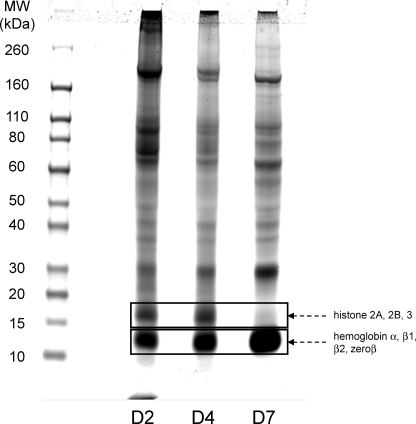
During the SDS-PAGE analysis of the proteins contained in the exosomes recovered after the sucrose gradient, we had already found that D2 and D7 exosomes exhibited different protein patterns (Fig. 3, D and E). As seen on Fig. 4, when larger amounts of unfractionated exosome preparations were separated by SDS-PAGE for their subsequent proteomic analysis, this difference was confirmed, with D4 exosomes showing somewhat intermediate patterns between those of the D2 and D7 samples.
FIGURE 4.
Comparison of the protein patterns of D2, D4, and D7 exosomes. 20 μg of exosomal proteins were separated on an SDS-polyacrylamide gradient gel (4–12%), and stained with Coomassie Blue. Boxes indicate the positions of two types of proteins that were dominating strongly in the mass spectrometry analysis: histone chains around 16 kDa, which decreased very significantly at D7, and hemoglobin chains around 12 kDa, which were most prominent at D7.
The gel shown on Fig. 4 as well as two others prepared with exosome samples obtained through completely independent productions were used for separate proteomic determinations of the protein composition of the exosomes via MS/MS mass spectrometry. Analysis of quantitative changes in protein abundance was performed by a label-free spectral count method using the MFPaQ software developed in-house (22, 23). An initial preliminary analysis was performed on a QSTAR Pulsar XL machine and two others on an LTQ-Orbitrap, a more sensitive machine. Importantly, although they were performed on samples originating from three completely independent preparations, and the QSTAR identified roughly half the number of proteins, those three studies gave very comparable results (supplemental Table S1). More than 700 different proteins were identified in each of the two sets of samples analyzed with the LTQ-Orbitrap, with comparable results between these independent analyses (for the complete list, see the supplemental Table S1). For 95% of the proteins identified by LTQ-Orbitrap (i.e. for 691 out of 729), there was a very good reproducibility of the variations recorded between the preparations harvested on D2, D4, and D7. As a very short summary of the data provided in supplemental Table S1, 522 proteins were consistently found in all exosomes; 101 were found only in D2 or in D2 and D4 exosomes, and 106 were found only in D7 or in D4 and D7 exosomes.
To date, a total of 27 different proteins have been identified in exosomes produced by reticulocytes from either human, sheep, mouse, or rat either via antibody-based methods such as Western blot and FACS staining or via biological tests such as ligand binding or enzyme assays (see review by Blanc and Vidal (6)). Upon comparison of this list with the hundreds of proteins identified by MS, four proteins were found to be missing in our samples, namely caspase-3, acetylcholinesterase, CD58, and calcium-independent phospholipase A2. This underlines the fact that although proteomic approaches can provide the identities of very large numbers of proteins present in complex mixtures, they are by no means absolute and can miss certain polypeptides, which can be detected via more sensitive means such as antibodies or enzymatic assays.
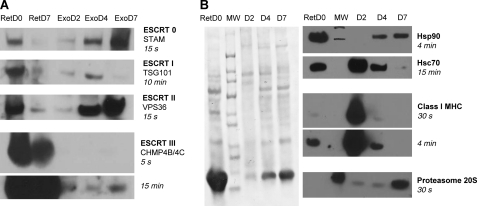
Because the biogenesis of exosomes takes place in late endosomes to end up in multivesicular bodies (MVB), we first checked our lists of proteins for those known to be involved with that particular compartment (Table 2). Reassuringly, we did repeatedly identify the accessory protein Alix, which is among the proteins most often identified in exosomes (36), and the protein Bro1, which was prominent in our early exosome preparations (D2 and D4). ESCRT complexes are also central to MVB biogenesis (37), and various components of the ESCRT complexes were identified by MS, albeit with varied patterns of changes over the course of the maturation process of the reticulocytes. Although the three detected components of the ESCRT-0 complex became more prominent at later time points, CHMP4B, which belongs to the ESCRT-III complex, was rather stable over time. Remarkably, IST1 and VPS4, which are involved in recycling ESCRT-III proteins (38), tended to predominate in the D2 exosomes. Among the members of the ESCRT-I or -II complexes, only TSG101 came up, in only one of our analyses. To reinforce those observations, we then performed Western blot analyses with a panel of antibodies specific for the various ESCRT classes (Fig. 5A). For those ESCRTs that could be detected both by MS and Western blot, the variations recorded showed comparable changes over time with both methods. The one major discrepancy with the MS data was for the ESCRT-II component VPS36, which was not absent as the MS data may have suggested but increased over time to become very prominent in D7 exosomes. This result thus provides a vivid example of the fact alluded to in the previous paragraph that certain proteins can be completely missed by MS analyses even if they are present in sizable quantities in the analyzed samples. For a given protein, however, the efficiency of detection by MS is expected to be the same in different samples, justifying the type of comparisons summarized in supplemental Table S1. Regarding variations over time, the general picture we obtained from Western blots is that all ESCRT complexes are much less present in D7 than in D0 reticulocytes. The high concentration of the components of the ESCRT-0 and -II complexes in D7 exosomes suggests that those may be actively eliminated via exosomes toward the end of the differentiation process, possibly because of the previous elimination of the recycling machinery. For the CHMP4B/4C component of the ESCRT-III complex, the amounts found in exosomes stayed relatively constant over time, in accord with what was seen by MS. Those amounts of CHMP4B/4C found in exosomes were also very low compared with what was found in D0 or even D7 reticulocytes, suggesting that there may not be an active recruitment of that ESCRT-III component into exosomes. Regarding the ESCRT-I component TSG101, its presence appeared to peak at D4 in the experiment shown, as was observed in one of the MS analyses (see supplemental Table S1), but the detection of that protein proved difficult to reproduce, most likely because of poor stability of the antibody. Indeed, in a whole series of subsequent experiments, we systematically failed to detect any specific signal with this antibody, including in the positive control composed of D0 reticulocytes (data not shown).
TABLE 2.
Proteins involved in MVB biogenesis found in reticulocytes exosomes
Empty boxes indicate no detection.
| Identified protein | Gene | Mass | Mean ratio D2/D7 | Ratio D2/D7 S1 | Ratio D2/D7 S2 |
|---|---|---|---|---|---|
| Da | |||||
| Alix | PDC6I | 97141 | 1.57 | 1.75 | 1.38 |
| BRO1 domain-containing protein BROX | BROX | 46676 | 4.69 | 6.00 | 3.39 |
| ESCRT-0 | |||||
| SNAP-25-interacting protein Hrs-2 | HGS | 86819 | 0.32 | 0.55 | 0.08 |
| EGFR pathway substrate 15 isoform B | Q5JC29 | 87189 | D7a | D7a | D7a |
| Putative uncharacterized protein Stam | D3ZI37 | 54890 | 0.22 | 0.38 | 0.05 |
| ESCRT-I | |||||
| Tumor susceptibility gene 101 protein | TS101 | 44221 | 1.00b | 1.00 | |
| ESCRT-II | |||||
| ESCRT-III | |||||
| CHMP4b | D4A9Z8 | 25094 | 1.38 | 1.75 | 1.00 |
| IST1 homolog | IST1 | 40088 | 2.75 | D2a | 2.75 |
| Vacuolar protein sorting-associated protein 4A | VPS4A | 49161 | 1.00 | D2a | 1.00 |
a Data indicate that, in that series, the protein was detected only on the indicated day.
b Data indicate that this value was obtained from a single analysis.
FIGURE 5.
Western blot analysis of D2, D4, and D7 exosomes. Exosome samples, each containing 9 μg of proteins, and whole cell lysates of fresh reticulocytes (50 μg of protein) were separated on 4–12% SDS-PAGE before transferring onto nitrocellulose. A, analysis of selected components of the ESCRT complexes. B, analysis of other cellular components. Left panel, representative Ponceau red staining of one of the three membranes used. Membranes were then incubated with the indicated primary antibodies and then with HRP-conjugated secondary antibodies, before revealing using ECL® with the indicated exposure times. For all panels, similar results were obtained in at least one other experiment.
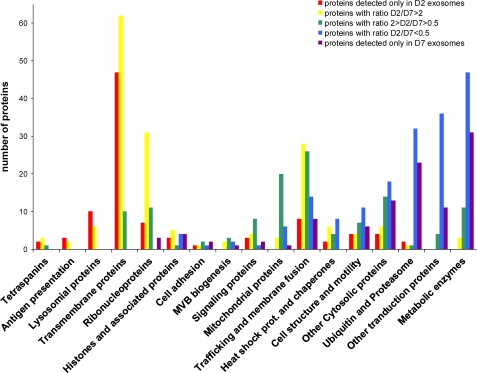
Together with the changes of protein patterns obtained in SDS-polyacrylamide gels (Figs. 3 and 4), those observations strongly suggest that, over the course of maturation of a reticulocyte, there are very significant changes in the contents of the secreted exosomes, most likely because of the sequential activation of different cellular pathways leading to the production of exosomes. To investigate further what those different pathways could be, we thus classified the hundreds of proteins identified in our samples (and listed in supplemental Table S1) into 16 categories related to their cellular roles (Fig. 6). For each of those proteins, we recorded whether their abundance within our exosome preparations went down, up, or stayed relatively constant between D2 and D7. As can be seen on Fig. 6, the overall picture that we obtained is one where classes of proteins fall roughly into three categories, which could potentially correspond to three or more different pathways of exosome biogenesis.
FIGURE 6.
Global analysis of the changes in the types of proteins identified in D2 versus D7 exosomes. Each one of the more than 700 proteins identified in our analyses (see supplemental Table S1) was classified into one of 17 broad families, and the individual D2/D7 ratios were determined based on the number of MS/MS peaks, as described under “Materials and Methods.” The graph shows the number of proteins identified in each of the different families as a function of those D2/D7 ratios.
A first category, which predominates in the early days (with D2/D7 ratio>2), includes primarily proteins found at the plasma membrane (120 proteins identified corresponding to ion channels and transporters, membrane receptors, and antigens such as MHC I and tetraspanins). In addition, nuclear proteins such as histones and many ribosomal proteins (26 subunits of the 40 S and 41 subunits of the 60 S) were also found to predominate in the D2 exosomes, as did all 16 proteins classified as associated with lysosomes. Accordingly, note that on the gel shown on Fig. 4, histones correspond to the band found at 16–17 kDa, which is very prominent at D2 and D4 and essentially absent at D7.
A second clearly defined category, corresponding to those proteins that are eliminated later (with D2/D7 ratio <0.5), includes mostly cytosolic proteins such as metabolic or catabolic enzymes, as well as other proteins involved in cell structure and mobility. Among the most noticeable protein types falling in this category were 20 different tRNA aminoacyl synthetases, 26 components of the 26 S proteasome, and 30 transcription factors. The various chains of hemoglobin, which run at 12 kDa in the gel shown on Fig. 4, are also much more abundant at D7 than at D2 and were indeed found to increase by mass spectrometry (supplemental Table S1), but this presumably reflects their increasing abundance during differentiation rather than their specific elimination.
The last category is the one where levels did not dramatically vary between D2 and D7 (in effect the categories where the green peak is the highest on Fig. 6). In this group, we found proteins from signaling pathways, mitochondrial proteins, and those involved in trafficking and membrane fusion.
Finally, although the pattern obtained for chaperones and heat shock proteins in Fig. 6 was unremarkable, we focused our attention on this group because those are commonly presumed to be involved in specific sorting toward exosomes. Upon closer inspection, we actually found that there were radical changes between the types of chaperones found in D2 and D7 exosomes (Table 3). Indeed, although D2 exosomes contained higher levels of the members of the Hsp40 family and Hsc70, D7 exosomes were enriched for the chaperones belonging to the Hsp90 and TCP-1 families. This observation further supports the view that different pathways for the production of exosomes may be activated sequentially during the course of maturation of a reticulocyte. Western blot analyses allowed us to confirm the prominence of class I MHC heavy chains and Hsc70 in D2 exosomes, whereas Hsp90 and proteasome core proteins were detected almost exclusively in D7 exosomes (Fig. 5B).
TABLE 3.
Modification of specific protein chaperones found in exosomes during reticulocytes maturation
| Identified protein | Gene | Mass | Mean ratio D2/D7 | Ratio D2/D7 S1 | Ratio D2/D7 S2 |
|---|---|---|---|---|---|
| Da | |||||
| DnaJ (Hsp40) homolog | |||||
| Subfamily B, member 1 | B0K030 | 38294 | 3.69 | 4.44 | 2.94 |
| Subfamily B, member 4 | Q5XIP0 | 37969 | 6.00 | 6.00 | Abs |
| Subfamily B member 6 | DNJB6 | 39012 | 2.00 | D2a | 2.00 |
| Subfamily C, member 5 | P60905 | 22885 | D2a | D2a | D2a |
| Putative uncharacterized protein Dnajc13 | D3ZNI6 | 209275 | 0.27 | D7a | 0.53 |
| Hsc70-interacting protein | P50503 | 41424 | 12.25 | D2a | 12.50 |
| Heat shock cognate 71-kDa protein (Hsc 70) | HSP7C | 71055 | 2.26 | 2.45 | 2.06 |
| BAG family molecular chaperone regulator 5 | Q5QJC9 | 51030 | D2a | D2a | D2a |
| Heat shock 70-kDa protein 1A/1B | HSP71 | 70427 | 1.17 | D2a | 1.17 |
| Stress-induced-phosphoprotein 1 | STIP1 | 63157 | 5.57 | 4.00 | 7.14 |
| 78-kDa glucose-regulated protein, Hspa5 | GRP78 | 72473 | 2.06 | 2.35 | 1.76 |
| Heat shock 70 kDa protein 4 | HSP74 | 94057 | 0.18 | 0.05 | 0.32 |
| Endoplasmin | ENPL | 92998 | 0.69 | 0.71 | 0.67 |
| Hsp90 α | HS90A | 85160 | 0.39 | 0.26 | 0.51 |
| Hsp90 β | HS90B | 83571 | 0.22 | D7a | 0.44 |
| T-complex protein 1 | |||||
| α | TCPA | 60834 | 0.47 | 0.22 | 0.71 |
| β | TCPB | 57764 | 0.54 | 0.23 | 0.85 |
| δ | TCPD | 58775 | 0.56 | 0.25 | 0.86 |
| ϵ | TCPE | 59954 | 0.49 | 0.20 | 0.78 |
| γ | TCPG | 61178 | 0.38 | 0.12 | 0.64 |
| Chaperonin subunit 6a (ζ) | Q3MHS9 | 58437 | 0.50 | 0.21 | 0.78 |
| η (RCG55994, isoform CRA_c) | D4AC23 | 60133 | 0.53 | 0.26 | 0.81 |
a Data indicate that, in that series, the protein was detected only on the indicated day.
Lipid Analyses of Exosome Preparations
We first examined the PL composition of the various exosome preparations by high performance thin layer chromatography (Table 4 and supplemental Fig. S2). Although phosphatidylcholine remained around 50%, other lipid classes were found to undergo significant variations as follows: phosphatidylethanolamine increased from 20 to 33%; SM decreased from 20 to 13%, and phosphatidylinositol + phosphatidylserine decreased from 10 to 5%.
TABLE 4.
Phospholipid compositions of D2, D4, and D7 exosomes (n = 4)
Phospholipid compositions, expressed as percentages, after HPTLC on lipid extracts were obtained by the Folch procedure (60) and staining by I2 vapor (supplemental Fig. S2). The data shown are means ± S.E. of values obtained in three independent experiments. PI is phosphatidylinositol, and PC is phosphatidylcholine. The statistical significance of the differences between D2 and D7 values was evaluated with a t test, *, p < 0.05; **, p < 0.01.
| D2 | D4 | D7 | |
|---|---|---|---|
| PE | 19.3 ± 2.5 | 26.0 ± 3.8 | 32.5 ± 2.6** |
| PI + PS | 10.0 ± 1.5 | 9.8 ± 2.9 | 4.7 ± 0.8* |
| PC | 50.6 ± 2.5 | 48.5 ± 5.0 | 51.3 ± 3.0 |
| SM | 20.0 ± 2.1 | 15.3 ± 3.9 | 12.5 ± 2.2* |
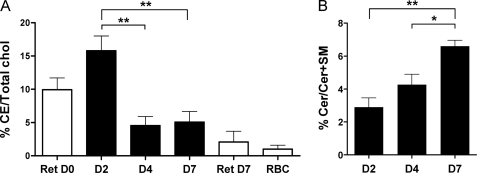
The free cholesterol (FC) content was determined by gas-liquid chromatography after lipid extraction. No significant variation of FC relative to the PL content was observed between the D2, D4, and D7 exosomes (see Table 1), the average FC/(FC + PL) ratios remaining around values of 0.30 ± 0.05. Remarkably, however, the ratios of cholesterol esters (CE) over total cholesterol (CE + FC) were consistently found to be around 4-fold higher in D2 than in D4 and D7 exosomes (Fig. 7A). We thus measured the CE/(CE + FC) ratios in total lipid extracts from reticulocytes at D0 and D7. Accordingly, the proportion of CE was markedly decreased in D7 reticulocytes, approaching the value measured in fully mature red blood cells, thus providing additional proof that D7 reticulocytes are near full maturation (Fig. 7A). This indicates that CE are efficiently eliminated from reticulocytes by exosomes during the early stages of their maturation. Because CE are known to be mainly present in lysosomes and lipid droplets (39), their abundance in D2 exosomes probably originates from the removal of lysosomes carried out by exosome release, which is supported by the observation that lysosomal proteins were also predominantly found in D2 exosomes (Fig. 6).
FIGURE 7.
Lipidomic analysis of D2, D4, and D7 exosomes. The lipid content of exosomes was determined as detailed under “Materials and Methods.” A, ratios of CE over total cholesterol were dramatically increased in D2 exosomes. B, proportion of ceramides over (ceramides + SM) increased over time. The statistical significance of the differences observed was evaluated with a t test. *, p < 0.05; **, p < 0.01).
To test for the implication of the “lipid route,” which involves the formation of ceramides (Cer), we also evaluated the content of exosomes in Cer relative to that of SM, and we found it to be reproducibly increased over 2-fold at D7 compared with D2 (Fig. 7B).
Altogether, our analyses of the lipid contents of exosomes released over the course of the in vitro maturation of reticulocytes are consistent with the proteomic results in suggesting that different pathways are sequentially involved, with the one eliminating ceramides dominating at later stages.
DISCUSSION
The fact that exosomes were actually discovered in reticulocytes was due in large part to the important amounts of various materials that these cells need to eliminate during their terminal maturation into erythrocytes. Because exosomes are the result of a process of proteolipidic sorting within biological membranes, it has been proposed that they could be considered as isolated membrane rafts (17, 40, 41). With a view to explore the mechanisms that govern proteolipidic sorting in biological membranes and lead to the formation of membrane microdomains, we were interested in obtaining large amounts of exosomes and thus chose reticulocytes as a natural source. We soon realized that those exosomes were not a homogeneous entity and that their composition changed over time during the maturation taking place over 7 days in our in vitro experimental setup. Importantly, the vesicles contained in all our preparations harvested at D2, D4, or D7 presented all the expected physical characteristics of bona fide exosomes, be it their aspect, size, density, and membrane fluidity (Fig. 3) and not microvesicles or membrane particles, which would be expected to be of lower density, or apoptotic vesicles, which would be composed of much larger elements and not necessarily in a liquid ordered state (36, 42).
To date, using various techniques, a total of 2624 proteins have been identified in exosomes from a broad range of cell lines (43). Among the 25 proteins that are most often identified in exosomes, only two were absent from our samples, class II MHC and CD63, although CD81 was found only in the D2 exosomes of one of our two proteomic analyses. The fact that class II MHC molecules were not identified is not that surprising given that those are not expressed in reticulocytes but only in antigen-presenting cells. For CD63, we may have failed to identify it in our samples because of its molecular weight, which is close to those of histones, because those were very prominent in our D2 exosome preparations, where CD63, being a cell surface protein, would have been expected.
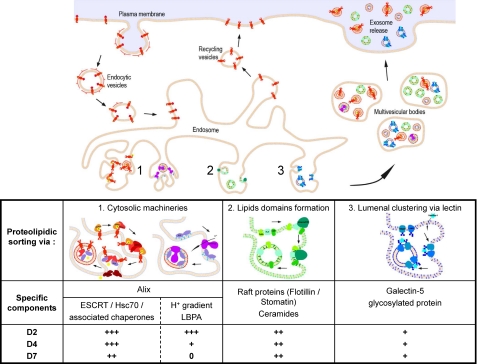
The changes in the protein and lipid contents of exosomes over time, together with the modification of their physical properties, suggested strongly that the contribution of the different mechanisms for the recruitment of cellular components into exosomes may change over the course of maturation of a reticulocyte. This would be in complete agreement with previous reports, based on analyses of exosomes from various cell types, which have suggested that the biogenesis of exosomes may rely on as many as three different mechanisms and that all three could be involved in parallel in reticulocytes (6, 7). Those putative pathways are represented schematically in Fig. 8 and are classified following the proposal of Blanc and Vidal (6) according to the location of the agents responsible for the specific recruitment into the invaginations that bud into the lumen of the endosomes to form exosomes. Various players of each pathway were identified in our analyses of the protein and lipid content of the exosomes, suggesting that those three distinct pathways are indeed present in reticulocytes. Their respective abundance, however, changed over time, and we can draw the following hypothetical sketch of how the prominence of these various pathways is altered over the course of the maturation process of the reticulocytes (Fig. 8).
FIGURE 8.
Scheme of different pathways for biogenesis of exosomes in reticulocytes and hypothetical changes in their respective importance in generation of D2, D4, and D7 exosomes.
The Alix-associated pathway involves partners present at the cytoplasmic face of the endosomes, either ESCRT complexes and/or a V-H+-ATPase and lysobisphosphatidic acid. ESCRT complexes recruit mono-ubiquitinylated proteins, in a collaborative network between the four ESCRT complexes and various chaperones/heat shock proteins such as Hsc70. V-H+-ATPases establish a pH gradient across the outer membrane of the endosomes driving a conformational change of lysobisphosphatidic acid and consequently the budding of exosomes (15, 16). Our results suggest that this pathway would be very effective during the early phase of the reticulocyte maturation process for eliminating many proteins associated with the plasma membrane such as TfR (20) and MHC molecules (44, 45). The predominance of V-H+-ATPase in early exosomes suggests that pH gradients could play an important role in this process. This pathway would also eliminate other proteins expected to be obsolete after enucleation such as histones and other nuclear proteins and ribonucleoproteins. Concerning heat shock proteins, Hsp40 and Hsc70 appear to be particularly involved during the early phases, whereas Hsp90 and TCP1 are found to predominate at later time points.
The ceramide pathway, initially described in oligodendrocytes (18), is thought to involve the recruitment of proteins such as flotillin or stomatin into raft-like structures within the MVB outer membrane. This pathway would remain relatively constant during maturation. In this regard, although the ceramide/sphingolipids ratio was found to go up over time, this 2-fold increase could correspond either to an up-regulation of this pathway or to a down-regulation of the other pathways. Accordingly, proteins that are classically considered as associated with rafts, such as flotillins, stomatin, and CD59, were all found to have intermediate D2/D7 ratios, between 1.1 and 1.7. Regarding tetraspanins, the observations reported on the RBL cell line (45) would tend to suggest that they would also be eliminated via this pathway. Their predominance at early time points (Fig. 6) may thus correspond to their very efficient recruitment in this pathway, leading to their very prompt overall disappearance once synthesis of new ones has stopped.
The Galectin 5 pathway, in which this β-galactoside-binding lectin, present in the lumen of MVBs, is presumed to recruit various glycosylated proteins (19). In light of our results, this pathway would remain relatively constant during the different stages of maturation of the reticulocytes and would be responsible for the elimination of proteins that may well still have been needed at earlier stages, such as those involved in cell structure and motility.
ESCRT proteins have been proposed as major players in the biogenesis of exosomes in various cell types (46). Somewhat surprisingly, we found that not all ESCRT complexes followed the same patterns of changes over the maturation course of reticulocytes. The increase of ESCRT-0 and -II in later stage exosomes correlated with their disappearance from reticulocytes. This elimination may be related to the earlier elimination of recycling proteins such as IST1 and VPS4 (see Table 2). For ESCRT-III, however, much more protein was retained in reticulocytes, and no particular changes over time were noted in the small amounts that were found in exosomes, both by MS and Western blot. Those observations suggest that the mechanisms responsible for eliminating the components of the ESCRT-0 and -II complexes probably do not apply to those of ESCRT-III. For ESCRT-I, the situation remains to be clarified because of technical difficulties with the antibody used, but our results may be pointing to a situation similar to that of ESCRT-III.
One of the burning questions that is raised by these observations is whether those different mechanisms of recruitment into exosomes all converge into one production line, which would then result in the production of one relatively homogeneous population of exosomes for which the contents would change over time. Alternatively, several parallel but separate pathways may exist, which would then result in the production of clearly separate populations of exosomes of distinct composition. In support of the latter picture, Laulagnier et al. (45) reported that the RBL cell line may produce at least two subpopulations of exosomes with different proteolipidic composition as follows: some contain different tetraspanins (CD81 or CD63) and enriched in ceramides, and the others are enriched in MHC-II. To show this, they made use of a protocol where the RBL cell line was pre-loaded with fluorescent lipids that targeted preferentially the Golgi apparatus or the plasma membrane and showed preferential association of certain lipids in exosomes immunopurified with antibodies against CD63, CD81, or MHC class II. To obtain a more definite picture regarding exosomes produced by reticulocytes, we are currently attempting to obtain immunopurified populations of exosomes derived from those various pathways with a view to submit them to proteomic analyses, but the difficult problems of yields, aggregation of the exosomes, and contamination by antibodies still need to be overcome.
It has previously been suggested that exosomes could play a role in the pathogenesis of sickle cell anemia (6), Indeed, the erythrocytes of patients suffering from drepanocytosis have been found to harbor excessive amounts of various components such as the α4β1 integrin (47, 48), as well as the chaperones Hsc70 and TCP1, whereas those of flotillin-1 and stomatin were found to be reduced (49). In line with this hypothesis, our results suggest a scheme that could explain differences in degrees of elimination of those components via exosomes. Indeed, one could envisage that large amounts of misfolded hemoglobin could act as a trap for the Hsc70 and TCP1 chaperones, thus resulting in their retention inside the cell and causing an inhibition of the ESCRT-dependent exosomal pathway. Because reticulocytes cannot call upon additional transcription, their response capacity must be extremely limited. Their incapacity to produce more chaperones such as Hsc70 would thus explain the reduced elimination of membrane proteins such as the α4β1 integrin (47) or the TfR (20). Conversely, the decrease of the raft proteins flotillin-1 and stomatin in drepanocytes (50, 51) could be due to compensation via the ceramide pathway. Previous reports have suggested that red blood cells from drepanocytic patients harbor increased activity of ionic channels (52). Interestingly, in our results, the following three major players in the ionic balance of erythrocytes were identified predominantly in D2 exosomes: Gardos channel, KCC1, and Na+/K+-ATPase pump (KCNN4, Slc12A4, and subunits AT1XX in supplemental Table S1, respectively) (48, 52). The inefficient elimination of ion channels in exosomes via the ESCRT pathway could thus be related to the reported ionic imbalance and increased sensitivity of TfR-positive sickle cells to dehydration.
Because they result from a natural process of selective proteolipidic sorting in biological membranes, exosomes are a model of choice for investigating the rules that govern the formation of membrane microdomains. In contrast to rafts, exosomes are remarkably stable structures that can be purified without the intervention of invasive techniques such as detergents or ultrasounds. We thus hope that deciphering the mechanisms involved in exosome biogenesis will not only be of interest for understanding erythrocytic diseases such as sickle cell anemia but also shed light on the thermodynamics of membrane organization and trafficking in living cells (53–58).
Supplementary Material
Acknowledgments
We are thankful to S. Powis, A. Saoudi, and I. Bernard-Cadenat for the gifts of antibody reagents and to members of the O. Cuvillier and M-L. Maddelein teams for providing us with materials and reagents for the Western blot analyses. We gratefully acknowledge L. Kaddoum, B. Monsarrat, M. Record, C. Théry, and M. Vidal for helpful discussions and technical advice, as well as the anonymous referees for their helpful suggestions. This work benefited from the assistance of the electron microscopy facility of the IFR 109 and of the TRI Imaging platform of Toulouse. The IPBS proteomic platform was supported by grants from the Agence Nationale de la Recherche (Agence Nationale de la Recherche/INCa Programme Plates-Formes Technologiques du Vivant), the Fondation pour la Recherche Médicale (Programme Grands Equipements), and the Région Midi-Pyrénées. Institut de Pharmacologie et de Biologie Structurale belongs to the CNRS Consortium CellTiss.
This work was supported in part by the CNRS (PEPS 2009 Program).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
- TfR
- transferrin receptor
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- chol
- cholesterol
- DPPC
- 1,2-palmitoyl-sn-glycero-3-phosphocholine
- SM
- sphingomyelin
- PL
- phospholipid
- FC
- free cholesterol
- CE
- cholesterol ester
- DPH
- 1,6-diphenyl-1,3,5-hexatriene
- FSC
- forward size scatter
- MVB
- multivesicular body.
REFERENCES
- 1. Chasis J. A., Mohandas N. (2008) Blood 112, 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mel H. C., Prenant M., Mohandas N. (1977) Blood 49, 1001–1009 [PubMed] [Google Scholar]
- 3. Gronowicz G., Swift H., Steck T. L. (1984) J. Cell Sci. 71, 177–197 [DOI] [PubMed] [Google Scholar]
- 4. Koury M. J., Koury S. T., Kopsombut P., Bondurant M. C. (2005) Blood 105, 2168–2174 [DOI] [PubMed] [Google Scholar]
- 5. Harding C., Heuser J., Stahl P. (1983) J. Cell Biol. 97, 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blanc L., Vidal M. (2010) Curr. Opin. Hematol. 17, 177–183 [DOI] [PubMed] [Google Scholar]
- 7. Vidal M. (2010) Transfus. Clin. Biol. 17, 131–137 [DOI] [PubMed] [Google Scholar]
- 8. Pan B. T., Johnstone R. M. (1983) Cell 33, 967–978 [DOI] [PubMed] [Google Scholar]
- 9. Théry C., Zitvogel L., Amigorena S. (2002) Nat. Rev. Immunol. 2, 569–579 [DOI] [PubMed] [Google Scholar]
- 10. Zitvogel L., Regnault A., Lozier A., Wolfers J., Flament C., Tenza D., Ricciardi-Castagnoli P., Raposo G., Amigorena S. (1998) Nat. Med. 4, 594–600 [DOI] [PubMed] [Google Scholar]
- 11. Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., Geuze H. J. (1996) J. Exp. Med. 183, 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 13. Keller S., Sanderson M. P., Stoeck A., Altevogt P. (2006) Immunol. Lett. 107, 102–108 [DOI] [PubMed] [Google Scholar]
- 14. Amigorena S. (2000) Medicina 60, Suppl. 2, 51–54 [PubMed] [Google Scholar]
- 15. Raiborg C., Stenmark H. (2009) Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 16. Matsuo H., Chevallier J., Mayran N., Le Blanc I., Ferguson C., Fauré J., Blanc N. S., Matile S., Dubochet J., Sadoul R., Parton R. G., Vilbois F., Gruenberg J. (2004) Science 303, 531–534 [DOI] [PubMed] [Google Scholar]
- 17. Wubbolts R., Leckie R. S., Veenhuizen P. T., Schwarzmann G., Möbius W., Hoernschemeyer J., Slot J. W., Geuze H. J., Stoorvogel W. (2003) J. Biol. Chem. 278, 10963–10972 [DOI] [PubMed] [Google Scholar]
- 18. Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. (2008) Science 319, 1244–1247 [DOI] [PubMed] [Google Scholar]
- 19. Barrès C., Blanc L., Bette-Bobillo P., André S., Mamoun R., Gabius H. J., Vidal M. (2010) Blood 115, 696–705 [DOI] [PubMed] [Google Scholar]
- 20. Géminard C., De Gassart A., Blanc L., Vidal M. (2004) Traffic 5, 181–193 [DOI] [PubMed] [Google Scholar]
- 21. Rouser G., Fkeischer S., Yamamoto A. (1970) Lipids 5, 494–496 [DOI] [PubMed] [Google Scholar]
- 22. Mouton-Barbosa E., Roux-Dalvai F., Bouyssié D., Berger F., Schmidt E., Righetti P. G., Guerrier L., Boschetti E., Burlet-Schiltz O., Monsarrat B., de Peredo A. G. (2010) Mol. Cell. Proteomics 9, 1006–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bouyssié D., Gonzalez de Peredo A., Mouton E., Albigot R., Roussel L., Ortega N., Cayrol C., Burlet-Schiltz O., Girard J. P., Monsarrat B. (2007) Mol. Cell. Proteomics 6, 1621–1637 [DOI] [PubMed] [Google Scholar]
- 24. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 25. Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 26. Vieu C., Tercé F., Chevy F., Rolland C., Barbaras R., Chap H., Wolf C., Perret B., Collet X. (2002) J. Lipid Res. 43, 510–522 [PubMed] [Google Scholar]
- 27. Barrans A., Collet X., Barbaras R., Jaspard B., Manent J., Vieu C., Chap H., Perret B. (1994) J. Biol. Chem. 269, 11572–11577 [PubMed] [Google Scholar]
- 28. Yao J. K., Rastetter G. M. (1985) Anal. Biochem. 150, 111–116 [DOI] [PubMed] [Google Scholar]
- 29. Leborgne N., Dupou-Cézanne L., Teulières C., Canut H., Tocanne J. F., Boudet A. M. (1992) Plant Physiol. 100, 246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lakowicz J. R. (2006) Principe of Fluorescence Spectroscopy, pp. 353–369, Springer Science, Heidelberg, Germany [Google Scholar]
- 31. Flanagan J. P., Lessler M. A. (1970) Ohio J. Sci. 70, 300 [Google Scholar]
- 32. Criswell K. A., Sulkanen A. P., Hochbaum A. F., Bleavins M. R. (2000) J. Appl. Toxicol. 20, 25–34 [DOI] [PubMed] [Google Scholar]
- 33. Thery C., Amigorena S., Raposo G., Clayton A. (2006) Curr. Protoc. Cell Biol. Chapter 3, Unit 3.22 [DOI] [PubMed] [Google Scholar]
- 34. Vidal M., Sainte-Marie J., Philippot J. R., Bienvenue A. (1989) J. Cell. Physiol. 140, 455–462 [DOI] [PubMed] [Google Scholar]
- 35. Laulagnier K., Motta C., Hamdi S., Roy S., Fauvelle F., Pageaux J. F., Kobayashi T., Salles J. P., Perret B., Bonnerot C., Record M. (2004) Biochem. J. 380, 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mathivanan S., Ji H., Simpson R. J. (2010) J. Proteomics 73, 1907–1920 [DOI] [PubMed] [Google Scholar]
- 37. Woodman P. G., Futter C. E. (2008) Curr. Opin. Cell Biol. 20, 408–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hanson P. I., Shim S., Merrill S. A. (2009) Curr. Opin. Cell Biol. 21, 568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schroeder F., Gallegos A. M., Atshaves B. P., Storey S. M., McIntosh A. L., Petrescu A. D., Huang H., Starodub O., Chao H., Yang H., Frolov A., Kier A. B. (2001) Exp. Biol. Med. 226, 873–890 [DOI] [PubMed] [Google Scholar]
- 40. de Gassart A., Geminard C., Fevrier B., Raposo G., Vidal M. (2003) Blood 102, 4336–4344 [DOI] [PubMed] [Google Scholar]
- 41. Subra C., Laulagnier K., Perret B., Record M. (2007) Biochimie 89, 205–212 [DOI] [PubMed] [Google Scholar]
- 42. Théry C., Ostrowski M., Segura E. (2009) Nat. Rev. Immunol. 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 43. Mathivanan S., Simpson R. J. (2009) Proteomics 9, 4997–5000 [DOI] [PubMed] [Google Scholar]
- 44. Lynch S., Santos S. G., Campbell E. C., Nimmo A. M., Botting C., Prescott A., Antoniou A. N., Powis S. J. (2009) J. Immunol. 183, 1884–1891 [DOI] [PubMed] [Google Scholar]
- 45. Laulagnier K., Vincent-Schneider H., Hamdi S., Subra C., Lankar D., Record M. (2005) Blood Cells Mol. Dis. 35, 116–121 [DOI] [PubMed] [Google Scholar]
- 46. van Niel G., Porto-Carreiro I., Simoes S., Raposo G. (2006) J. Biochem. 140, 13–21 [DOI] [PubMed] [Google Scholar]
- 47. Brittain J. E., Parise L. V. (2008) Transfus. Clin. Biol. 15, 19–22 [DOI] [PubMed] [Google Scholar]
- 48. De Franceschi L. (2009) Mediter. J. Hematol. Infect. Dis. 1 [Google Scholar]
- 49. Kakhniashvili D. G., Griko N. B., Bulla L. A., Jr., Goodman S. R. (2005) Exp. Biol. Med. 230, 787–792 [DOI] [PubMed] [Google Scholar]
- 50. Bickel P. E., Scherer P. E., Schnitzer J. E., Oh P., Lisanti M. P., Lodish H. F. (1997) J. Biol. Chem. 272, 13793–13802 [DOI] [PubMed] [Google Scholar]
- 51. Mairhofer M., Steiner M., Mosgoeller W., Prohaska R., Salzer U. (2002) Blood 100, 897–904 [DOI] [PubMed] [Google Scholar]
- 52. Apovo M., Beuzard Y., Galacteros F., Bachir D., Giraud F. (1994) Biochim. Biophys. Acta 1225, 255–258 [DOI] [PubMed] [Google Scholar]
- 53. van Meer G. (2010) EMBO Rep. 11, 331–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. van Meer G., de Kroon A. I. (2011) J. Cell Sci. 124, 5–8 [DOI] [PubMed] [Google Scholar]
- 55. Simons K., Gerl M. J. (2010) Nat. Rev. Mol. Cell Biol. 11, 688–699 [DOI] [PubMed] [Google Scholar]
- 56. Bagatolli L. A., Ipsen J. H., Simonsen A. C., Mouritsen O. G. (2010) Prog. Lipid Res. 49, 378–389 [DOI] [PubMed] [Google Scholar]
- 57. Joly E. (2004) BMC Cell Biol. 5, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Destainville N., Dumas F., Salomé L. (2008) J. Chem. Biol. 1, 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zak B., Dickenman R. C., White E. G., Burnett H., Cherney P. J. (1954) Am. J. Clin. Pathol. 24, 1307–1315 [DOI] [PubMed] [Google Scholar]
- 60. Folch J., Ascoli I., Lees M., Meath J. A., LeBaron N. (1951) J. Biol. Chem. 191, 833–841 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.