Background: In G2, the transcription factor FoxM1 is activated by phosphorylation to induce the expression of G2/M genes.
Results: Protein phosphatase 2A (PP2A), together with its regulatory subunit B55α, dephosphorylates FoxM1 and inhibits its transcriptional activity.
Conclusion: PP2A/B55α dephosphorylation prevents premature activation of FoxM1 in G1.
Significance: This is a new control mechanism to regulate temporally the expression of G2/M genes.
Keywords: CDK (Cyclin-dependent Kinase), Cell Cycle, Cyclins, Phosphatase, Transcription, B55, FoxM1, PP2A
Abstract
The forkhead transcription factor FoxM1 controls expression of a large number of genes that are specifically expressed during the G2 phase of the cell cycle. Throughout most of the cell cycle, FoxM1 activity is restrained by an autoinhibitory mechanism, involving a repressor domain present in the N-terminal part of the protein. Activation of FoxM1 in G2 is achieved by Cyclin A/Cyclin-dependent kinase (Cdk)-mediated phosphorylation, which alleviates autoinhibition by the N-terminal repressor domain. Here, we show that FoxM1 interacts with B55α, a regulatory subunit of protein phosphatase 2A (PP2A). B55α binds the catalytic subunit of PP2A, and this promotes dephosphorylation and inactivation of FoxM1. Indeed, we find that overexpression of B55α results in decreased FoxM1 activity. Inversely, depletion of B55α results in premature activation of FoxM1. The activation of FoxM1 that is observed upon depletion of B55α is fully dependent on Cyclin A/Cdk-mediated phosphorylation of FoxM1. Taken together, these data demonstrate that B55α acts to antagonize Cyclin A/Cdk-dependent activation of FoxM1, to ensure that FoxM1 activity is restricted to the G2 phase of the cell cycle.
Introduction
Orderly progression through the cell cycle depends on sequential activation of distinct gene clusters (1). For example, during the late stages of the G1 phase, phosphorylation of the retinoblastoma protein by Cyclin D/Cyclin-dependent kinase (Cdk)2 4–6 complexes relieves repression of E2F transcription factors that control expression of a cluster of genes required for DNA replication (2). Similarly, in the G2 phase, a number of transcription factors have been shown to be activated by Cyclin A/Cdk2 to promote expression of a cluster of genes that controls progression through G2 and mitosis (3–5). One of these transcription factors is the forkhead factor FoxM1 (6). Expression of FoxM1 itself is restricted to proliferating cells and is first activated in late G1 (7). Despite its production in late G1 and early S phase, the FoxM1 protein is not transcriptionally active before cells reach G2 phase. During G1/S, FoxM1 activity is kept low through its N-terminal autoinhibitory domain, which interacts with the C-terminal transactivation domain. In G2, phosphorylation of its transactivation domain by Cyclin A/Cdk inhibits its interaction with the N-terminal autorepressor domain, allowing FoxM1-mediated gene transcription (4, 8). However, CyclinA/Cdk2 activity is already present in cells as early as late G1 phase, and it is unclear how phosphorylation and activation of FoxM1 are prevented at this stage and restricted to G2.
Here, we identify B55α, a regulatory subunit of the protein phosphatase 2A (PP2A), as a novel interactor of FoxM1. PP2A is the major serine-threonine phosphatase in mammalian cells and plays critical roles in cell cycle, cell proliferation, development, and regulation of multiple signaling pathways (9). PP2A holoenzymes consist of a catalytic subunit C, a structural subunit A, and a variable regulatory subunit B that determines the substrate specificity and spatial and temporal functions of PP2A. There are at least four families of the regulatory subunit: B (also known as B55 or PR55), B′ (B56 or PR61), B″ (PR72), and B‴ (PR93/PR110), with at least 16 members in these families (10). We show here that FoxM1 interacts with the B55α subunit, and this acts to prevent FoxM1 activation at the early stages of the cell cycle. We find that depletion of B55α from cells synchronized at the G1/S transition results in premature phosphorylation and activation of FoxM1. Importantly, this activation is fully dependent on the presence of Cyclin A, indicating that B55α acts to antagonize Cyclin A/Cdk-dependent activation of FoxM1 during G1 and S phase, to limit the transcriptional activation of FoxM1 to G2 phase.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfections, and Drugs
U2OS and 293-T cells were maintained in DMEM with 6% fetal calf serum and antibiotics. Thymidine and nocodazole were purchased from Sigma and used at 2.5 mm and 250 ng/ml, respectively. Okadaic acid (OA) was purchased from Calbiochem and used at 100–200 nm. Cells were transfected with plasmid DNA using the standard calcium phosphate transfection protocol. Small inhibitory RNA (siRNA) oligonucleotides were transfected with HiPerFect (Qiagen) following the manufacturer's protocol.
Plasmids and Oligonucleotides
The 6XDBE luciferase reporter has been described previously (11). Plasmids encoding FoxM1wt and FoxM1 ΔN/ΔKEN have also been described (12). FoxM1 T611E was generated by site-directed mutagenesis. FLAG-B55α was obtained from Addgene (plasmid 13804) (13), and GFP-B55α was generated by PCR amplification of the full-length B55α from FLAG-B55α and subsequent subcloning into pEGFP-C2 (Clontech). pGEX-B55α was generated by subcloning full-length B55α from GFP-B55α into pGEX6P-1 vector (Amersham Biosciences). The siRNA targeting Cyclin A2 (GTAGCAGAGTTTGTGTACA) was purchased from Ambion, and the siRNA against B55α is an On-target SMART pool from Dharmacon.
Antibodies
FoxM1 (C-20), Cyclin A2 (H-432), CDK4 (C-22), and Actin (I-19) antibodies were from Santa Cruz. Anti-FLAG (M2) and anti-GFP were from Sigma and Roche Applied Science, respectively. Anti-B55 (PP2A B subunit) and anti-PP2A/C (Y119) were purchased from Cell Signaling and Abcam, respectively. Plk1 and pS10-histone H3 antibody were from Upstate. Phosphothreonine MAPK/CDK substrate antibody was purchased from Cell Signaling.
Reporter Assays
Luciferase activity was determined using the dual luciferase kit (Promega) according to the manufacturer's instructions. Relative luciferase was expressed as the ratio of firefly luciferase activity to control Renilla luciferase activity.
RT-PCR
Total RNA was isolated with the Qiagen RNeasy kit, according to the manufacturer's instructions. cDNA was synthesized from 1 μg of total RNA by using Superscript II reverse transcriptase (Invitrogen) and oligo(dT) primers. The resultant cDNA was used as a template for PCR amplification with specific primers.
Automated Image Analysis
Cells were grown in 96-well plates (Viewplate-96; PerkinElmer Life Sciences). At the indicated time points, cells were fixed with 4% formaldehyde and methanol, and were stained with DAPI and the indicated antibodies. Image acquisition was performed using a Cellomics ArrayScan VTI (Thermo Scientific) using a 20 × 0.40 NA objective. Image analysis was performed using Cellomics ArrayScan HCS Reader (Thermo Scientific).
FACS Analysis
Fluorescence-activated cell sorting (FACS) analysis was performed as described (14).
Immunoprecipitations
Cells were lysed in PLB buffer (PBS, 2 mm EDTA, 1 mm EGTA, 0.5 mm DTT, and 1% Triton X-100) supplemented with protease and phosphatase inhibitors. Cell lysates were incubated for 2 h with the appropriate antibody coupled to protein A/G-agarose beads. Immunoprecipitates were extensively washed, then denatured in sample buffer, loaded on SDS-PAGE and transferred to nitrocellulose blots. Blots were probed with the indicated antibodies.
Dephosphorylation Assays
For endogenous FoxM1 dephosphorylation, U2OS cells were lysed in phosphatase buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.25% Nonidet P-40, and protease inhibitors) and incubated at 30 °C for the indicated times, in the presence or absence of OA. For in vitro PP2A/B55 dephosphorylation assays, FoxM1 was immunoprecipitated from 293-T cells overexpressing FLAG-FoxM1wt and used as a substrate. The dephosphorylation assay was performed at 30 °C for 30 min in phosphatase buffer (20 mm Hepes, pH 7.4, 1 mm DTT, 1 mm MgCl2, 100 μg/ml BSA, and protease inhibitors), containing 0.2 unit of purified PP2A A/C heterodimer (Millipore) and 0.25 μg of recombinant GST-B55α. Where indicated, OA was added at 200 nm.
Mass Spectrometry Analysis
To identify FoxM1-interacting proteins, FoxM1 was purified from U2OS cells stably expressing YFP-S peptide-FoxM1. Ten 15-cm dishes were harvested and cells were lysed in PLB buffer, after which FoxM1 was isolated by pull-down with protein S-agarose beads. FoxM1 complexes were subsequently subjected to SDS-PAGE, and protein bands were visualized by silver staining. Each lane was cut into 16 gel pieces and subjected to in-gel digestion with trypsin. Peptides were extracted with 10% FA. The extracted peptides were subjected to nanoscale liquid chromatography tandem mass spectrometry (nanoLC-MS/MS) analysis, performed on an Agilent 1100 HPLC system (Agilent Technologies) connected to an LTQ Linear Ion Trap Mass Spectrometer combined with an Orbitrap (ThermoFisher) (15). Raw MS data were converted to peak lists using DTASuperCharge version 1.27 (16). Spectra were searched against the International Protein Index (IPI) Human data base version 3.37 (69,164 sequences; 29,064,824 residues) using Mascot software version 2.2.0. Peptides with a mascot score of 35 or higher were considered. The results were combined and compared using Scaffold 2 (Proteome software).
For phosphosites analysis, samples from an in vitro dephosphorylation assay were subjected to SDS-PAGE followed by Coomassie staining. Protein bands corresponding to FoxM1 were sliced out from the gels and subjected to in-gel digestion as described above. Using the dimethyl labeling system (17), extracted peptides from phosphatase-treated and untreated samples were labeled with intermediate and light label respectively, mixed with equal ratio, and subjected to MS analysis as described above. Identification was done against the IPI Human data base using Mascot. Peptides with a Mascot score of 25 or higher were considered. Both mascot result and raw MS data were used to quantify the relative abundance of each peptide. Quantification was performed by dimethyl version of MSQuant (16).
RESULTS
FoxM1 Interacts with B55α
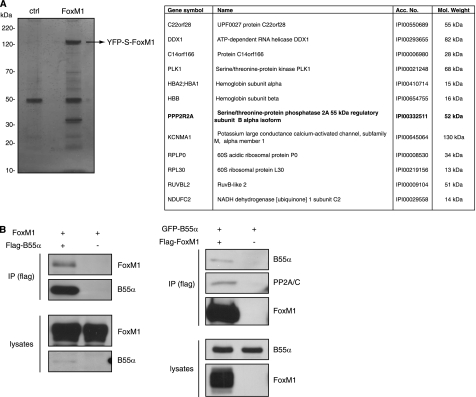
To identify proteins that interact with FoxM1, we generated cell lines that stably express a version of FoxM1 N-terminal tagged to YFP and an S peptide domain. The YFP-S-FoxM1 fusion protein was pulled-down with S-protein-agarose beads, and the complexes were analyzed by SDS-PAGE. A clear band corresponding to YFP-FoxM1 could be seen, in addition to several other additional bands (Fig. 1A). We subsequently subjected the immunocomplexes to LC-MS/MS, to try to identify the proteins that specifically co-purified with FoxM1. To this end, the control pulldown was compared with the pulldown from YFP-FoxM1-expressing cells. Two independent experiments were performed, and several unique peptides were identified in the YFP-FoxM1 complexes in both purifications, corresponding to a total of 12 known proteins (Fig. 1A). These included several peptides from Plk1, a mitotic kinase known to interact with FoxM1 and enhance its transcriptional activity (18). Among the potential novel interactors of FoxM1, we identified PPP2R2A, also known as B55α, a regulatory subunit of the PP2A phosphatase (Fig. 1A). Because FoxM1 activity is known to be restricted to specific stages in the cell cycle through Cyclin- and Plk1-dependent phosphorylation, we decided to investigate the possible contribution of B55α/PP2A to the control of FoxM1 activity.
FIGURE 1.
Identification of B55α as a FoxM1-interacting protein. A, FoxM1 was purified from U2OS cells stably expressing YFP-S peptide-FoxM1, as described under “Experimental Procedures.” FoxM1 complexes were subsequently subjected to SDS-PAGE, and protein bands were visualized by silver staining. Each lane was cut into 16 gel pieces and followed by tryptic digestion and MS analysis. The table shows the potential FoxM1-interacting proteins, for which a significant number of peptides were identified in FoxM1 samples but not in control samples from parental U2OS or U2OS cells expressing only YFP-S peptide. Two independent experiments were performed, and only the proteins identified in both of them are indicated. B, 293-T cells were co-transfected with plasmids expressing FoxM1 and FLAG-B55α. B55α immunocomplexes were purified by immunoprecipitation with an anti-FLAG antibody, and FoxM1 was detected with an anti-FoxM1 antibody (left). Cells co-transfected with plasmids expressing FLAG-FoxM1 and GFP-B55α were subjected to immunoprecipitation (IP) with an anti-FLAG antibody, and B55α and PP2A/C were detected with anti-GFP and anti-PP2A antibodies, respectively (right).
First, we validated the interaction between FoxM1 and B55α by performing co-immunoprecipitation experiments in 293-T cells using differentially tagged variants of FoxM1 and B55α. When we expressed FLAG-tagged B55α with nontagged FoxM1, we could clearly detect FoxM1 in the B55α immunoprecipitates (Fig. 1B). Inversely, if FLAG-tagged FoxM1 was co-expressed with GFP-tagged B55α we could detect GFP-B55α in the FLAG-FoxM1 immunocomplexes (Fig. 1B). In addition to B55α, we also found that the catalytic subunit of PP2A (PP2A/C) was present in complex with FoxM1 (Fig. 1B), suggesting that B55α forms a bridge between PP2A/C and FoxM1.
B55α Controls FoxM1 Activity
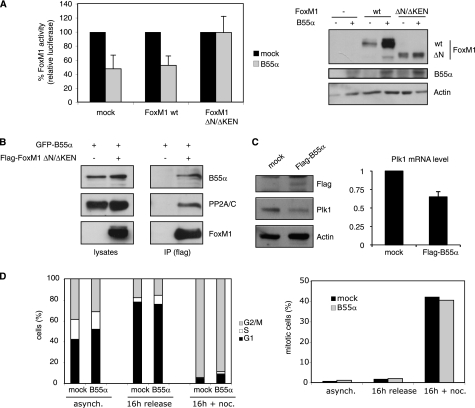
If the function of B55α is to bring FoxM1 in contact with the core enzyme of the PP2A phosphatase, this is likely to contribute to regulation of FoxM1 activity. Transcriptional activation by FoxM1 is highly cell cycle-dependent, and this is primarily controlled through phosphorylation by various cell cycle-dependent kinases, such as Cyclin A/Cdk2 and Plk1. Therefore, we reasoned that B55α could antagonize the cell cycle-dependent phosphorylation of FoxM1, causing a reduction in FoxM1 activity. To test this, we analyzed the effect of B55α overexpression on FoxM1 activity. For this purpose, we introduced a plasmid carrying FoxM1 consensus sites in front of a luciferase reporter gene into cells to assay FoxM1 transcriptional activity. We have previously shown that the basal activity of this promoter is due to its transactivation by endogenous FoxM1 (19). Interestingly, activity of this reporter was clearly reduced upon overexpression of B55α, suggesting that B55α inhibits the transcriptional activity of endogenous FoxM1 (Fig. 2A). Similarly, B55α also suppressed the activity of overexpressed FoxM1 toward this reporter (Fig. 2A). Interestingly, whereas activity of the overexpressed FoxM1 was reduced, FoxM1 levels were increased by overexpression of B55α (Fig. 2A), suggesting that B55α can positively affect FoxM1 stability while negatively influencing FoxM1 activity.
FIGURE 2.
Overexpression of B55α inhibits FoxM1 transcriptional activity. A, transactivation of the 6XDBE luciferase reporter, which contains six forkhead response elements, was measured in U2OS cells transfected with an empty vector (mock) or plasmids encoding FoxM1wt and FoxM1 ΔN/ΔKEN, in the absence or presence of an exogenously expressed FLAG-B55α. The graph shows the average of three independent experiments, and error bars represent S.D. Activity of FoxM1 in the absence of B55α overexpression was set as 100%. Expression levels of FoxM1 and B55α were monitored by Western blotting. B, 293-T cells co-expressing FLAG FoxM1 ΔN/ΔKEN and GFP-B55α were subjected to immunoprecipitation (IP) with an anti-FLAG antibody, and B55α and PP2A/C were detected with anti-GFP and anti-PP2A antibodies, respectively. C, U2OS cells transfected with an empty vector (mock) or with FLAG-B55α were collected 48 h after transfection and analyzed by Western blotting with the indicated antibodies (left), or by RT-PCR (right). D, U2OS cells transfected with an empty vector (mock) or with a plasmid expressing FLAG-B55α were synchronized with thymidine for 24 h and subsequently released in fresh medium for 16 h in the presence or absence of nocodazole. Cell cycle distribution was determined by staining with propidium iodide and FACS analysis. The graph shows the percentage of cells in each phase of the cell cycle at the indicated conditions (left). The percentage of mitotic cells was determined by phosphohistone H3 staining and FACS analysis (right).
In previous studies, we have shown that cell cycle-dependent phosphorylation of FoxM1 alleviates intramolecular repression of FoxM1 activity by the N-terminal domain (4). In those studies we described an N-terminal deletion mutant of FoxM1, lacking both the N-terminal repressor domain, as well as the KEN motif that targets FoxM1 for degradation (12). This mutant was shown to be constitutively active and did not display any cell cycle-regulated activation (4, 8). Importantly, the activity of the ΔN/ΔKEN mutant of FoxM1 was unaffected by overexpression of B55α (Fig. 2A). This was not due to a lack of interaction of this mutant with B55α or PP2A/C, as we could easily detect both proteins in co-immunoprecipitates with the ΔN/ΔKEN mutant of FoxM1 (Fig. 2B). In agreement with the luciferase results, overexpression of B55α also reduced the level of the FoxM1 target gene Plk1. We detected a significant reduction in endogenous Plk1, both at protein and mRNA levels, upon overexpression of B55α (Fig. 2C). Importantly, the effects of B55α overexpression on FoxM1 activity were not due to a cell cycle arrest, as overexpression of B55α did not result in any dramatic changes in the cell cycle distribution (Fig. 2D). Taken together, these results indicate that B55α directly affects the cell cycle-dependent activation of FoxM1.
Phosphorylation of FoxM1 Is Regulated by B55α
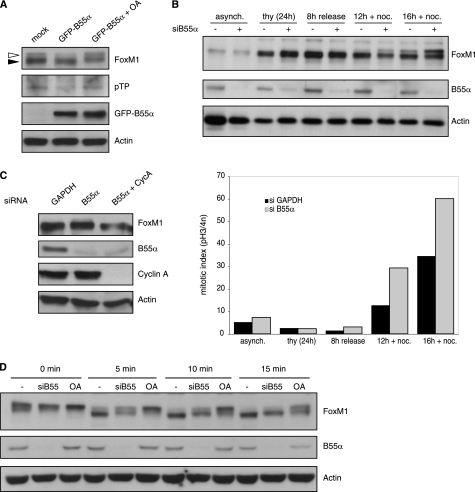
During the early stages of the cell cycle, FoxM1 is kept inactive by an autoinhibitory mechanism involving the N terminus of FoxM1. Upon phosphorylation of FoxM1 by Cyclin A/Cdk complexes, autoinhibition is relieved (4, 8). Because B55α seems to inhibit FoxM1 activity, we reasoned that it acts to antagonize Cyclin A/Cdk-dependent phosphorylation. To test this, we examined the effect of overexpression and depletion of B55α on cell cycle-dependent phosphorylation of FoxM1. As cells progress through G2 and enter mitosis, FoxM1 becomes hyperphosphorylated, leading to a shift in its electrophoretic mobility (4). Overexpression of B55α led to a reduction in that phosphoshift in cells synchronized in G2/M, which was reverted by treatment with the phosphatase inhibitor okadaic acid (Fig. 3A, white arrowhead). This effect was specific to B55α, because overexpression of another B subunit for PP2A, B56γ, did not affect phosphorylation of FoxM1 (supplemental Fig. S1). The effect of B55α overexpression on the phosphorylation of endogenous FoxM1 was also confirmed with a phosphoantibody that recognizes phosphorylated threonine residues only when followed by proline (Fig. 3A). Next, we analyzed the effect of B55α depletion on FoxM1 phosphorylation. We synchronized U2OS cells at the G1/S transition using a thymidine block and subsequently released these cells using fresh medium containing nocodazole to trap cells in mitosis. Under these conditions, a slower migrating form of FoxM1 became visible at 12 h after release, and its abundance increased at the 16-h time point (Fig. 3B). Importantly, the abundance of hyperphosphorylated FoxM1 was clearly enhanced at these time points when B55α was depleted by RNA interference (Fig. 3B). Coincident with the higher levels of hyperphosphorylated FoxM1, we observed a clear increase in the amount of mitotic cells in the cultures depleted of B55α, which fits with the recently reported function of B55α in mitotic exit (20). This result indicates that depletion of B55α enhances phosphorylation of FoxM1, although from these data we cannot conclude whether this effect is direct or whether it is achieved indirectly via a more general effect of B55α inactivation on cell cycle progression. To address this issue, we analyzed the status of FoxM1 phosphorylation upon depletion of B55α in cells synchronized in G1/S with thymidine, when there are no differences in cell cycle profile between control and B55α-depleted cells. In this case, we could also detect the appearance of a slower migrating band of endogenous FoxM1, which was not observed if we co-depleted Cyclin A (Fig. 3C). Thus, depletion of B55α does increase Cyclin A-dependent phosphorylation of FoxM1.
FIGURE 3.
Effect of B55α on FoxM1 phosphorylation. A, U2OS cells transfected with empty vector (mock) or GFP-B55α were synchronized with nocodazole for 16 h. Where indicated, OA was added for 1 h at 200 nm before collecting the cells. Lysates were subjected to Western blot analysis with the indicated antibodies. Phosphorylation of endogenous FoxM1 was detected by Western blotting with a phosphothreonine-proline antibody on FoxM1 immunoprecipitates. B, U2OS cells transfected with siRNA control (GAPDH) or a siRNA targeting B55α were synchronized with thymidine in G1/S and subsequently released from the thymidine block in fresh medium with nocodazole for the indicated times. Whole cell lysates were analyzed by Western blotting with antibodies against FoxM1 and B55α, and actin was used as a loading control (upper). The mitotic index was determined by pH 3 staining and FACS analysis (lower). C, U2OS cells transfected with siRNA control (GAPDH) or a siRNA targeting B55α, alone or in combination with a siRNA targeting Cyclin A, were synchronized with thymidine in G1/S. The expression pattern of endogenous FoxM1 was analyzed by Western blotting. Efficiency of B55α and Cyclin A depletion was also determined by Western blot analysis. D, cell extracts from control or B55α-depleted cells were subjected to in vitro dephosphorylation for the indicated time points at 30 °C, in the absence or presence of OA (100 nm). Phosphoshifts of endogenous FoxM1 and B55α depletion were determined by Western blotting with the indicated antibodies. Actin was used as a loading control.
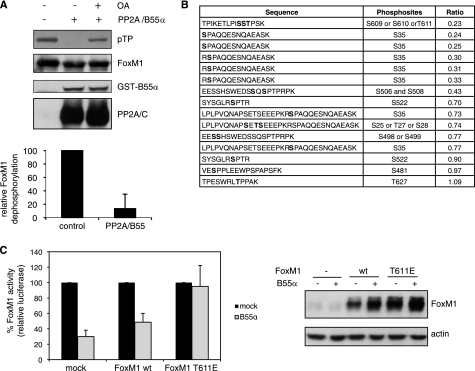
To provide more evidence for a direct link between FoxM1 dephosphorylation and B55α, we monitored the rate of in vitro dephosphorylation of endogenous FoxM1 by incubating mitotic cell extracts, in which FoxM1 is hyperphosphorylated, at 30 °C for different periods of time. FoxM1 dephosphorylation was clearly delayed in B55α-depleted extracts compared with control extracts, as demonstrated by longer persistence of slower migrating bands (Fig. 3D). This result indicates that B55α is indeed directly involved in FoxM1 dephosphorylation. Moreover, we found that FoxM1 can be efficiently dephosphorylated in vitro by incubation with purified PP2A, containing both A and C subunits and recombinant B55α (Fig. 4A). Importantly, PP2A/C can directly dephosphorylate FoxM1 in vitro (supplemental Fig. S2), indicating that B55α is not required for the catalytic activity of PP2A/C. However, the activity of PP2A/C toward FoxM1 is enhanced by the addition of B55α (supplemental Fig. S2), consistent with increased substrate recognition. Phosphorylation of FoxM1 detected with the phospho-threonine-proline antibody was dramatically reduced upon addition of recombinant PP2A/B55α, and this was reverted by preincubation with OA (Fig. 4A). This result clearly demonstrates that PP2A can directly dephosphorylate FoxM1.
FIGURE 4.
Identification of FoxM1 phosphosites targeted by PP2A/B55α. A, in vitro dephosphorylation assay of FoxM1 with purified PP2A A/C and GST-B55α. Where indicated, OA was added at 200 nm. Phosphorylation of FoxM1 was detected by Western blotting with a phosphothreonine-proline antibody. Quantification of the level of phosphorylation was performed with ImageJ. The graph shows the average of FoxM1 dephosphorylation of three independent replicates, and error bars represent S.D. Phosphorylation of the untreated control was set as 100%. B, samples from an in vitro phosphatase assay, performed as in A, subjected to quantitative MS analysis. The table shows all FoxM1 phosphopeptides identified. Ratio values indicate the relative abundance of the corresponding phosphopeptide in the phosphatase-treated sample versus the untreated control. Phosphosites are marked in bold. C, transactivation of the 6XDBE luciferase reporter measured in U2OS cells transfected with an empty vector (mock) or plasmids encoding FoxM1wt and FoxM1 T611E, in the absence or presence of an exogenously expressed FLAG-B55α. The graph shows the average of three independent experiments, and error bars represent S.D. Activity of FoxM1 in the absence of B55α overexpression was set as 100%. Expression levels of FoxM1 were monitored by Western blotting.
To find out which sites in FoxM1 are targets of PP2A/B55α, we analyzed PP2A dephosphorylation of FoxM1 by MS. FoxM1 bands from an in vitro phosphatase assay were isolated from the gel and subjected to tryptic digestion. Peptides from phosphatase-treated and untreated control were dimethyl-labeled with intermediate and light isotopes, respectively, mixed in equal ratio, and subjected to LC-MS/MS. Quantification was performed by comparing the relative abundance of intermediate versus light peptides. We identified 79 peptides of FoxM1 present in both samples, of which 15 were phosphopeptides (supplemental Table 1 and Fig. 4B). Those phosphopeptides contained 10 unique phosphosites, most of which we previously found to be phosphorylated in vivo (4). Six of those phosphopeptides showed a ratio lower than 0.4, of which 5 contained phosphorylated Ser35 and one contained a phosphorylated residue in position 609, 610, or 611 (Fig. 4B). Importantly, the ratio of their unmodified counterparts was equal to or higher than 1 (supplemental Table 1), indicating that the low abundance of the phosphopeptide in the phosphatase-treated sample is indeed due to a reduction in phosphorylation. We previously showed that mutation of Ser35 does not have any significant effect on the transcriptional activity of FoxM1 in G2 (4). In the same study we found that Thr611 was phosphorylated in vitro and in vivo by Cyclin A/Cdk complexes, and this phosphorylation was required to relieve the autoinhibition of FoxM1 by the N-terminal repressor domain (4). Therefore, we generated a phosphomimetic mutant of FoxM1 in which the T611 residue was substituted by a glutamic acid (FoxM1 T611E) to test the effect of B55α overexpression on FoxM1 transcriptional activity in G2. Interestingly, we found that the activity of FoxM1 T611E was not inhibited upon B55α overexpression, indicating that the effect of PP2A/B55 on FoxM1 transactivation is mediated via dephosphorylation of Thr611 (Fig. 4C).
B55α Suppresses Premature Activation of FoxM1
Because depletion of B55α increases the phosphorylation of FoxM1, we decided to analyze whether it also affects its transcriptional activity. In line with the inhibition of FoxM1 activity we observed upon overexpression of B55α (Fig. 2A), we found that depletion of B55α leads to enhanced FoxM1 activity in cells arrested at the G1/S transition (Fig. 5A). Again, the activity of the ΔN/ΔKEN mutant of FoxM1 was unaffected by perturbation of B55α expression (Fig. 5A). In addition, the activating effect of B55α depletion was no longer seen in cells released from the thymidine block and synchronized in G2/M (Fig. 5A), indicating that inhibition of FoxM1 by B55α occurs at the early stages of the cell cycle.
FIGURE 5.
Effect of B55α depletion on FoxM1 activity. A, U2OS cells expressing the 6XDBE reporter together with FoxM1 wt or FoxM1 ΔN/ΔKEN were co-transfected with siRNA control (GAPDH) or siRNA targeting B55α. Luciferase activity was determined in cells synchronized at the G1/S transition by thymidine treatment and in G2/M by releasing them from the thymidine block in fresh medium for 14 h. FoxM1 transcriptional activity in control (siGAPDH) was set as 1. FoxM1 protein levels and efficiency of B55α depletion were assessed by Western blotting. CDK4 was used as a loading control. B, cells were transfected with siRNA control (GAPDH) or siRNA targeting B55α and synchronized in G1/S with thymidine for 24 h. mRNA levels of FoxM1 targets were analyzed by RT-PCR (left). Cyclin A protein levels were determined by immunofluorescence and automated image analysis. At least 500 cells were counted per well. Measurements were performed in triplicate, and error bars represent S.D. (right). C, cells transfected with the 6XDBE reporter and FoxM1wt were co-transfected with siRNA control (GAPDH) and siRNAs targeting either B55α or Cyclin A. FoxM1 transcriptional activity was determined in G1/S, 24 h after treatment with thymidine. Protein levels were determined by Western blot analysis with the indicated antibodies. Actin was used as a loading control. Graphs represent the average of three independent experiments, and error bars indicate S.D.
In agreement with the luciferase results, depletion of B55α at the G1/S transition resulted in higher mRNA levels of Plk1 and Cyclin A, two well known transcriptional targets of FoxM1 (Fig. 5B). This effect correlated with an increase in the protein level of Cyclin A in B55α-depleted cells, as detected by immunofluorescence at the single cell level (Fig. 5B).
Cyclin A/Cdk activity was shown to peak in G2, coincident with the timing of activation of FoxM1-dependent target genes (4). However, the Cyclin A/Cdk2 complex is first activated at the G1/S transition, but is somehow prevented from activating FoxM1. Our findings that the effect of B55α on FoxM1 activity depends on a Cyclin A/Cdk phosphorylation site and that depletion of B55α leads to activation of FoxM1 specifically at the G1/S transition suggest that it acts to prevent premature activation of FoxM1 by low levels of Cyclin A/Cdk2. Indeed, when Cyclin A was co-depleted with B55α, activation of FoxM1 activity was no longer observed in cells synchronized at the G1/S transition (Fig. 5C), demonstrating that the effect of B55α depletion on FoxM1 activity is mediated through Cyclin A/Cdk-dependent phosphorylation.
DISCUSSION
In this study we aimed to identify new FoxM1-interacting proteins that contribute to FoxM1 function. We have found that B55α, a regulatory subunit of the PP2A phosphatase, interacts with FoxM1 and modulates its transcriptional activity. We have previously shown that phosphorylation by Cyclin A/Cdk complexes is required to activate FoxM1 in the G2 phase of the cell cycle (4). Here, we provide evidence that the phosphatase B55α/PP2A counteracts that phosphorylation and negatively regulates FoxM1 activity. Whereas overexpression of B55α inhibits FoxM-dependent transactivation in G2, depletion of B55α leads to premature activation of FoxM1 in G1/S, in a Cyclin A-dependent manner. We also provide evidence that Thr611 in FoxM1 is the main phosphoresidue through which PP2A/B55α influences FoxM1 transcriptional activity in G2. We and others have previously shown that phosphorylation of this residue by Cyclin/Cdk complexes is important for FoxM1 transactivation, allowing recruitment of the transcriptional co-activator CREB-binding protein and mediating the release of the repression of the N-terminal domain of FoxM1 (4, 21). Moreover, Thr611 has also been shown to be one of the two key sites of FoxM1 involved in its phosphorylation-dependent interaction with Plk1 (18). It is therefore likely that PP2A/B55α might counteract indirectly the activation of FoxM1 by Plk1.
It is unclear how the PP2A complexes are regulated and whether their activity changes throughout the cell cycle. Our results suggest that B55α/PP2A complexes are active during G1/S, but they could be somehow inactivated in G2, because depletion of B55α in this phase does not affect FoxM1 transactivation. However, we find that overexpression of B55α reduces FoxM1 activity in G2 and that FoxM1 also interacts with B55α in G2 (data not shown). This indicates that, rather than the phosphatase being inactivated in G2, it is more likely that the increase in Cyclin A/Cdk2 kinase activity in this phase results in a shift in the balance between phosphorylation/dephosphorylation toward a hyperphosphorylated active form of FoxM1, in the presence of active B55α/PP2A. All together, our results suggest that B55α/PP2A-dependent regulation of FoxM1 is more important during the early stages of the cell cycle, when Cyclin A/Cdk2 activity is low.
In addition to FoxM1, other transcription factors, such as B-Myb or NF-Y, also play an important role in the expression of genes required for G2/M progression (22). Similar to FoxM1, their transcriptional activity is regulated by Cyclin A/Cdk phosphorylation, and so far no phosphatase has been found to regulate their activity. An interesting possibility would be that B55/PP2A phosphatase exerts a more general role in regulating gene expression during the cell cycle by limiting also the activity of these other transcription factors to G2.
In summary, our data show that the combined action of Cyclin A/Cdk and B55α/PP2A fine tunes FoxM1 transcriptional activity to ensure that FoxM1 is only active in G2. At the G1/S transition, the phosphatase PP2A, through the subunit B55α, prevents premature activation of FoxM1 when there is low level of Cyclin A/Cdk activity. Later in G2, when Cyclin A/Cdk reaches its maximum activity, phosphorylation of FoxM1 releases its autorepressor domain, allowing FoxM1 activation and expression of the genes required for G2/M progression.
Supplementary Material
Acknowledgments
We thank the other members of the laboratory for helpful discussions. We also thank W. C. Hahn for the pMIG-FLAG B55α plasmid.
This work was supported by Dutch Cancer Society Grant UU-2007-3826) and The Netherlands Genomic Initiative of Netherlands Organization for Scientific Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1 and S2.
- Cdk
- Cyclin-dependent kinase
- OA
- okadaic acid
- PP2A
- protein phosphatase 2A.
REFERENCES
- 1. Futcher B. (2002) Curr. Opin. Cell Biol. 14, 676–683 [DOI] [PubMed] [Google Scholar]
- 2. Weinberg R. A. (1995) Cell 81, 323–330 [DOI] [PubMed] [Google Scholar]
- 3. Chae H. D., Yun J., Bang Y. J., Shin D. Y. (2004) Oncogene 23, 4084–4088 [DOI] [PubMed] [Google Scholar]
- 4. Laoukili J., Alvarez M., Meijer L. A., Stahl M., Mohammed S., Kleij L., Heck A. J., Medema R. H. (2008) Mol. Cell. Biol. 28, 3076–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saville M. K., Watson R. J. (1998) Oncogene 17, 2679–2689 [DOI] [PubMed] [Google Scholar]
- 6. Laoukili J., Kooistra M. R., Brás A., Kauw J., Kerkhoven R. M., Morrison A., Clevers H., Medema R. H. (2005) Nat. Cell Biol. 7, 126–136 [DOI] [PubMed] [Google Scholar]
- 7. Korver W., Roose J., Clevers H. (1997) Nucleic Acids Res. 25, 1715–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Park H. J., Wang Z., Costa R. H., Tyner A., Lau L. F., Raychaudhuri P. (2008) Oncogene 27, 1696–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Janssens V., Goris J. (2001) Biochem. J. 353, 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Janssens V., Longin S., Goris J. (2008) Trends Biochem. Sci. 33, 113–121 [DOI] [PubMed] [Google Scholar]
- 11. Furuyama T., Nakazawa T., Nakano I., Mori N. (2000) Biochem. J. 349, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laoukili J., Alvarez-Fernandez M., Stahl M., Medema R. H. (2008) Cell Cycle 7, 2720–2726 [DOI] [PubMed] [Google Scholar]
- 13. Chen W., Possemato R., Campbell K. T., Plattner C. A., Pallas D. C., Hahn W. C. (2004) Cancer Cell 5, 127–136 [DOI] [PubMed] [Google Scholar]
- 14. Smits V. A., Klompmaker R., Arnaud L., Rijksen G., Nigg E. A., Medema R. H. (2000) Nat. Cell Biol. 2, 672–676 [DOI] [PubMed] [Google Scholar]
- 15. Pinkse M. W., Mohammed S., Gouw J. W., van Breukelen B., Vos H. R., Heck A. J. (2008) J. Proteome Res. 7, 687–697 [DOI] [PubMed] [Google Scholar]
- 16. Mortensen P., Gouw J. W., Olsen J. V., Ong S. E., Rigbolt K. T., Bunkenborg J., Cox J., Foster L. J., Heck A. J., Blagoev B., Andersen J. S., Mann M. (2010) J. Proteome Res. 9, 393–403 [DOI] [PubMed] [Google Scholar]
- 17. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J. (2009) Nat. Protoc 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 18. Fu Z., Malureanu L., Huang J., Wang W., Li H., van Deursen J. M., Tindall D. J., Chen J. (2008) Nat. Cell Biol. 10, 1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alvarez-Fernández M., Halim V. A., Krenning L., Aprelia M., Mohammed S., Heck A. J., Medema R. H. (2010) EMBO Rep. 11, 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmitz M. H., Held M., Janssens V., Hutchins J. R., Hudecz O., Ivanova E., Goris J., Trinkle-Mulcahy L., Lamond A. I., Poser I., Hyman A. A., Mechtler K., Peters J. M., Gerlich D. W. (2010) Nat. Cell Biol. 12, 886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Major M. L., Lepe R., Costa R. H. (2004) Mol. Cell. Biol. 24, 2649–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lindqvist A., Rodríguez-Bravo V., Medema R. H. (2009) J. Cell Biol. 185, 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.