Abstract
Substrate-specific protein degradation mediated by the ubiquitin proteasome system (UPS) is crucial for the proper function of the cell. Proteins are specifically recognized and ubiquitinated by the ubiquitin ligases (E3s) and are then degraded by the proteasome. BTB proteins act as the substrate recognition subunit that recruits their cognate substrates to the Cullin 3-based multisubunit E3s. Recently, it was reported that missense mutations in KLHL7, a BTB-Kelch protein, are related to autosomal dominant retinitis pigmentosa (adRP). However, the involvement of KLHL7 in the UPS and the outcome of the adRP causative mutations were unknown. In this study, we show that KLHL7 forms a dimer, assembles with Cul3 through its BTB and BACK domains, and exerts E3 activity. Lys-48-linked but not Lys-63-linked polyubiquitin chain co-localized with KLHL7, which increased upon proteasome inhibition suggesting that KLHL7 mediates protein degradation via UPS. An adRP-causative missense mutation in the BACK domain of KLHL7 attenuated only the Cul3 interaction but not dimerization. Nevertheless, the incorporation of the mutant as a heterodimer in the Cul3-KLHL7 complex diminished the E3 ligase activity. Together, our results suggest that KLHL7 constitutes a Cul3-based E3 and that the disease-causing mutation inhibits ligase activity in a dominant negative manner, which may lead to the inappropriate accumulation of the substrates targeted for proteasomal degradation.
Keywords: Autophagy, Proteasome, Ubiquitin, Ubiquitin Ligase, Ubiquitylation, Degenerative Disease, Retinitis Pigmentosa
Introduction
Covalent attachment of ubiquitin (Ub)2 onto proteins is involved in the regulation of a wide array of cellular phenomena, ranging from the cell cycle to stress response and DNA damage repair (1, 2). Proteins targeted for ubiquitination are attached with either a single Ub or a poly-Ub chain, formed by the coupling of Ub molecules via one of the seven Lys residues or the N terminus of the first Ub molecule and the C terminus of the second. The poly-Ub chain via the Lys-48 is known to act as a degradation signal, which is recognized by the proteasome and consequently leads to the degradation of the ubiquitinated protein. Lys-63 linkage of the poly-Ub chains is involved in nonproteolytic regulation such as signal transduction, damaged DNA repair (3) and autophagy-mediated protein degradation (4). Other modes of the Ub chain are present in eukaryotic cells (5), but the functions of each form are yet to be elucidated.
In the ubiquitination reaction, Ub is first activated by the Ub-activating enzyme E1 and then carried by the Ub-conjugating enzyme E2 to the Ub ligase E3, where the Ub is attached onto a lysine residue of the specific substrate of the E3s (1, 2). The substrate specificity of this pathway is determined by the E3s of which the Cullin-RING ligases (CRLs) constitute the largest subclass. CRLs are multisubunit complexes where the enzymatic core Rbx1/Roc1 and substrate recognition modules each assemble onto the C and N terminus of a Cullin (Cul1, Cul2, Cul3, Cul4A, Cul4B, Cul5, and Cul7) (6–10). The most well characterized CRL is the SCF Ub ligase, comprised of Cul1, Roc1, an F-box family protein, and Skp1 (Skp1, Cullin, and F-box protein). The F-box protein, which recruits the substrate, binds to the adaptor protein Skp1, which in turn binds onto Cul1 (11–14). In Cul3-based CRLs, instead of F-box proteins, BTB (Bric-à-brac, Tramtrack, and Broad complex)/POZ (poxviruses and zinc finger) family proteins are employed as the substrate adaptor (15–18). There are more than 180 BTB proteins encoded in the human genome (19), and some of them have been related to substrate targeting in Cul3 complexes. BTB proteins bind to Cul3 through the BTB domain and the adjacent “3-box (Cul3-interacting box)” region, an element involved in facilitating this interaction (20). While interacting with Cul3, BTB proteins recruit substrates with their additional protein-protein interaction domain, such as the Kelch and MATH domains.
Of the BTBs involved in ubiquitination processes, the BTB-Kelch protein Keap1 is the best characterized. By regulating the turnover of the transcription factor Nrf2, Keap1 has an important role in the cellular defense mechanism for oxidative and xenobiotic stresses (21, 22). Another two BTB-Kelch proteins, KLHL9 and KLHL13, are involved in the cell cycle by targeting Aurora B, thereby regulating cytokinesis (23). The emerging roles of BTB family proteins and the discovery of mutations in BTB proteins that are responsible for diseases, such as gigaxonin for giant axonal neuropathy patients (24) and KLHL9 in autosomal dominant distal myopathy (25), have set forth the importance of BTB proteins.
Recently, three disease-causing missense mutations in the BACK domain of KLHL7, a BTB-Kelch protein, were found in patients with autosomal dominant retinitis pigmentosa (adRP) (26, 27). Retinitis pigmentosa is a genetically heterogeneous group of progressive retinal dystrophies, resulting in degeneration of rod and cone photoreceptors. Patients experience night blindness and visual field loss, often leading to complete blindness. Retinitis pigmentosa can be inherited in an autosomal dominant, autosomal recessive, and X-linked manner (28, 29).
Here, we report that KLHL7 forms Cul3-KLHL7 Ub E3 ligase in cells and that an adRP causative mutation, A153V, leads to the attenuation of the E3 ligase activity of Cul3-KLHL7 in a dominant negative manner. Our results provide molecular insights into the function of KLHL7 and the outcome of an adRP-causative mutation.
EXPERIMENTAL PROCEDURES
Expression Plasmids
Epitope-tagged full-length KLHL7 was amplified from human placenta cDNA library using Phusion DNA polymerase (Finnzymes) and cloned into EcoRI and NotI sites of pcDNA3 or pEFBOS (−) vectors. Cul3 and Ub were generated likewise and cloned into EcoRI and XhoI sites. Deletion mutants of KLHL7 were generated by a PCR-based method. The S150N, A153V, and A153T amino acid substitutions were introduced by QuikChange site-directed mutagenesis method using overlapping primers. The PCR-amplified coding regions of Cul3, KLHL7 (wild-type and mutants), and Ub were verified by sequence analyses.
Cell Culture, Transfection, and Treatment
HCT116 cells were cultured in Dulbecco's modified Eagle's medium (low glucose) (Wako) supplemented with 10% fetal bovine serum, 1% nonessential amino acid solution (Invitrogen), and 1% penicillin/streptomycin (Invitrogen) in a 37 °C incubator with 5% CO2. HeLa cells were cultured as above without nonessential amino acid solution. ATG5+/+:GFP-LC3 and ATG5−/−:GFP-LC3 mouse embryo fibroblasts (30) were cultured in medium without nonessential amino acid solution, supplemented with 0.8% puromycin. Transfections were carried out using FuGENE 6 transfection reagent (Roche Applied Science), Lipofectamine 2000 (Invitrogen), or Multifectam (Promega) according to the manufacturers' specifications. For proteasome inhibition, cells were treated with MG132 (benzyloxycarbonyl-Leu-Leu-Leu-H) (Peptide Institute) for 1 h at 20 μm. For lysosome inhibition, cells were treated with NH4Cl (Wako) for 1 h at 10 mm.
Antibodies
The following antibodies were used for immunoblot analyses: anti-FLAG monoclonal (Sigma, M2), rabbit polyclonal (Rockland), anti-HA monoclonal (Covance, HA. 11), polyclonal (Bethyl), anti-c-Myc monoclonal (Santa Cruz Biotechnology, 9E10), and anti-Cul3 rabbit polyclonal antibodies. Anti-Cul3 antibody was raised using recombinant His-tagged Cul3. Anti-Roc1 rabbit polyclonal antibody has been described previously (31). Peroxidase-conjugated anti-rabbit and anti-mouse antibodies (Jackson ImmunoResearch) were used as secondary antibodies. For immunocytochemical analyses, anti-FLAG (Sigma, M2), anti-HA rabbit polyclonal (Bethyl), anti-multi-Ub (MBL, FK2), anti-Ub Lys-48-specific (Millipore, Apu2), anti-Ub Lys63-specific (Millipore, Apu3), and anti-GFP rabbit polyclonal (MBL) were used with Alexa Fluor 488- and Alexa Fluor 568-conjugated anti-rabbit and anti-mouse antibodies (Invitrogen).
Immunoprecipitation and Immunoblot Analyses
Cells were lysed with ice-cold lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.5% CHAPS (Sigma), 1 mm dithiothreitol (DTT)). The supernatant was subjected to immunoprecipitation with anti-FLAG M2-agarose or anti-HA-agarose beads (Sigma) and incubated at 4 °C overnight. The beads were then washed with lysis buffer containing 750 mm NaCl and 0.2% CHAPS. The protein samples were added to SDS sample buffer, boiled for 5 min, and immunoblotted using Western Lightning Plus-EDL reagent (PerkinElmer Life Sciences).
Immunofluorescence Analyses
HeLa cells were cultured on coverslips and were transfected with various expression plasmids. The cells were fixed in 4% paraformaldehyde in PBS for 10 min, permeabilized and blocked in 0.5% Triton X-100, 5% BSA in PBS for 30 min. The coverslips were then immersed in primary antibodies diluted in 5% BSA in PBS followed by secondary immunofluorescent antibodies diluted in 5% BSA in PBS. Nuclei were stained with Hoechst 33342 (Invitrogen). The coverslips were mounted onto slides using Fluoromount/Plus mounting solution (Diagnostic BioSystems), and images were obtained using a fluorescent microscope (Keyence model BZ-9000).
In Vitro Ubiquitination Assays
Cul3-KLHL7 complex was immunoprecipitated from cells expressing FLAG-KLHL7 and eluted in 1× FLAG peptide (Sigma) in TBS. The purified protein was incubated at 30 °C for 1 or 2 h with 10 μg of Ub (Sigma), 0.2 μl of Ub-activating enzyme UBE1 (Boston Biochem), 0.2 μg of Ub-conjugating enzyme His-UBC4 purified from Escherichia coli, in reaction buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 0.5 mm DTT) either with or without 5 mm ATP at the final volume of 50 μl. The reaction was stopped by the addition of SDS sample buffer, followed by boiling for 5 min at 100 °C. The reaction mixtures were then subjected to immunoblot analyses with anti-multi-Ub (MBL, FK2) antibody to assess poly-Ub chain formation.
RESULTS
KLHL7 Forms Cullin3-based Complexes
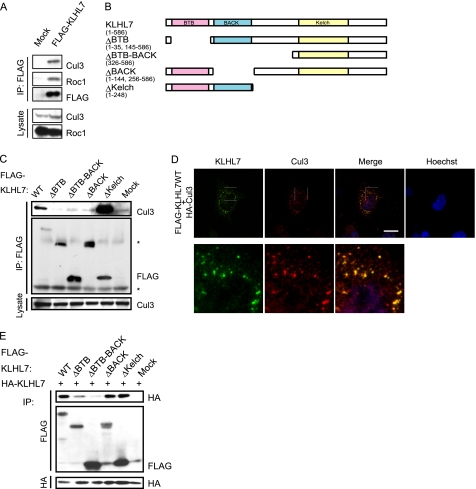
Some BTB-BACK-Kelch proteins have been reported to function as the substrate recognition subunits of Cul3-based CRLs. However, whether KLHL7 associates with Cul3 or functions as an E3 ligase is unknown. To investigate whether KLHL7 forms a Cul3-based complex, we expressed FLAG-tagged KLHL7 in HCT116 cells and immunoprecipitated the cell lysates using anti-FLAG antibody. As shown in Fig. 1A, Cul3 and Roc1 were co-immunoprecipitated with FLAG-KLHL7, indicating that KLHL7 forms a Cul3 complex.
FIGURE 1.
KLHL7 forms a Cul3-based complex. A, HCT116 cells transfected with FLAG-KLHL7 or mock vectors were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted with antibodies to Cul3, Roc1, and FLAG as indicated. B, schematic representations of KLHL7 (wild-type (WT) and deletion mutants). C, deletion mutants of FLAG-tagged KLHL7 were expressed in HCT116 cells, immunoprecipitated with anti-FLAG antibody, and immunoblotted with anti-Cul3 and anti-FLAG antibodies. Nonspecific bands are indicated with an asterisk. D, HeLa cells cultured on coverslips were transfected with HA-Cul3 and FLAG-KLHL7. The cells were fixed and stained with anti-HA and anti-FLAG antibodies and Hoechst 33342. Scale bar, 20 μm. E, deletion mutants of FLAG-tagged KLHL7 were co-expressed with HA-tagged full-length KLHL7 in HCT116 cells and immunoprecipitated with anti-FLAG and anti-HA antibodies. The immunoprecipitates were blotted with anti-HA and anti-FLAG antibodies.
To characterize the domain of KLHL7 involved in the interaction with Cul3, KLHL7 mutants lacking each domain were constructed (Fig. 1B). Wild-type KLHL7 and the mutant lacking the Kelch domain, which still has the BTB and BACK domains, were able to bind to Cul3. Conversely, the ΔBTB, ΔBTB-BACK, and ΔBACK mutants lacking either or both the BTB and the BACK domains could not (Fig. 1C). These results suggest that KLHL7 associates with Cul3 via its BTB and the BACK domains, and both are required for Cul3 binding.
Next, to examine the subcellular localization of KLHL7 and Cul3, HeLa cells transfected with FLAG-KLHL7 and HA-Cul3 were subjected to immunocytochemical analyses. As shown in Fig. 1D, KLHL7 was mostly present as punctate structures that were also positive for Cul3 staining. Although most of KLHL7 was associated with Cul3, not all of the Cul3-positive signals were stained with KLHL7, suggesting that only a subset of Cul3 is associated with KLHL7. Furthermore, the punctate structures were not formed by any of the deletion mutants, including the ΔKelch mutant (supplemental Fig. 1), indicating that Cul3 interaction is not sufficient for punctate formation. These results indicate that KLHL7 co-localizes with Cul3, and all of the domains are necessary for the punctate subcellular localization.
KLHL7 Forms a Homodimer through Its BTB Domains
Several members of the BTB family proteins, such as Keap1 and SPOP, form homodimers (20, 32). To investigate whether KLHL7 dimerize, HA-tagged wild-type KLHL7 was co-expressed with FLAG-tagged KLHL7 wild-type or deletion mutants into HCT116 cells, and their interactions were assessed by immunoprecipitation experiments, as shown in Fig. 1E. As expected, wild-type KLHL7 could homodimerize. The ΔBACK and ΔKelch mutants could still interact with wild-type KLHL7, whereas deletion of the BTB domain led to weak interaction, which was further weakened by the additional deletion of the BACK domain. These results suggest that the BTB domain is the primary domain necessary for KLHL7 dimerization, whereas the BACK domain, which is not essential for dimerization, is important for efficient dimerization. Because the ΔBACK mutant that could not interact with Cul3 (Fig. 1C) was still able to dimerize (Fig. 1E), the dimer formation of KLHL7 is not mediated indirectly by Cul3 interaction.
Cul3-KLHL7 Has Ub Ligase Activity
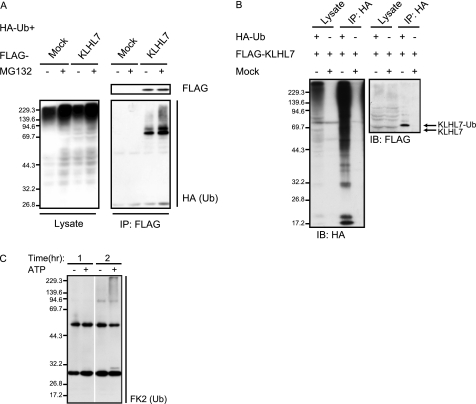
Because KLHL7 forms Cul3-based complexes, we next investigated whether Cul3-KLHL7 has Ub ligase activity. If KLHL7 is indeed a part of a Ub ligase complex, the substrates of KLHL7 should accumulate in the ubiquitinated form when the proteasome is inhibited. To test this, HCT116 cells were transfected with FLAG-KLHL7 and HA-Ub and treated with the proteasome inhibitor MG132 prior to immunoprecipitation. Immunoblot analyses with anti-HA antibody showed that ubiquitinated proteins interact with FLAG-KLHL7, and its amount increased drastically with proteasome inhibition (Fig. 2A). These data suggest that the ubiquitinated proteins that interacted with KLHL7 are targeted for proteasome degradation. There are several possibilities as to what the ubiquitinated proteins that accumulated with proteasome inhibition are. As strong bands were observed at the predicted molecular weight of ubiquitinated KLHL7 (∼70 kDa), it may be possible that KLHL7 itself is a target. However, ubiquitin positive bands were also observed below the predicted molecular weight of ubiquitinated KLHL7, suggesting that those ubiquitinated proteins represent the authentic substrates of KLHL7.
FIGURE 2.
Cul3-KLHL7 is an E3 ligase. A, FLAG-KLHL7 or mock vectors were co-expressed with HA-Ub in HCT116 cells and treated with 20 μm MG132 (+) or DMSO (−) prior to harvesting. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody and blotted with anti-FLAG and anti-HA antibodies. B, FLAG-KLHL7 was co-expressed with HA-Ub in HCT116 cells. After 20 μm MG132 treatment, the cell lysates were immunoprecipitated with anti-HA antibody and subjected to immunoblotting (IB) with anti-HA and anti-FLAG antibodies. C, in vitro ubiquitination assay using Cul3-KLHL7 complex immunopurified from HCT116 cells. Cul3-KLHL7 was incubated with recombinant E1 (UBE1), E2 (UBC4), and Ub, with or without ATP at 30 °C. The reaction was stopped at the indicated times and then immunoblotted with anti-FK2 Ub antibody.
To test whether KLHL7 is indeed self-ubiquitinated, we examined the ubiquitinated state of KLHL7 by detecting FLAG-KLHL7 after HA-Ub immunoprecipitation. After proteasome inhibition, cell lysates of HCT116 cells expressing HA-Ub and FLAG-KLHL7 were immunoprecipitated with anti-HA antibody and subsequently immunoblotted with anti-FLAG antibody to detect ubiquitinated KLHL7 (Fig. 2B). A single band of the estimated molecular weight of mono-ubiquitinated KLHL7 was strongly detected, whereas no ladder or smear was observed, indicating that the majority of KLHL7 is only present as the mono-ubiquitinated form. These results suggest that the increase of Ub moiety associated with KLHL7 upon proteasome inhibition is mostly due to the accumulation of polyubiquitinated authentic substrates.
We next examined the ligase activity of Cul3-KLHL7, in a substrate-independent in vitro ubiquitination assay (33, 34). Cul3-KLHL7, purified from HCT116 cells by immunoprecipitation, was incubated with Ub-activating enzyme E1, Ub-conjugating enzyme E2, and Ub in buffers with or without ATP (Fig. 2C). The amount of poly-Ub chain increased in a time-dependent manner, indicating that Cul3-KLHL7 has E3 ligase activity.
Ub-positive KLHL7 Puncta Increase with Proteasome Inhibition
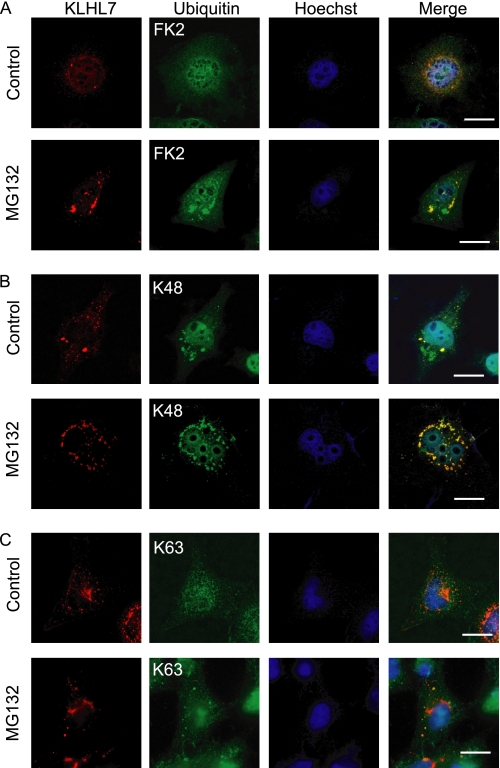
KLHL7 and Cul3 co-localized in punctate structures in the cytoplasm (Fig. 1D). KLHL7 also interacted with Ub, which increased with proteasome inhibition (Fig. 2A). Therefore, we next sought to find out whether the KLHL7 puncta co-localize with Ub upon proteasome inhibition. Immunocytochemical analyses using the FK2 anti-multi-Ub antibody, which recognizes poly-Ub chains and polyubiquitinated substrates, indicated that KLHL7 co-localized with ubiquitinated proteins, and the co-localizing structures increased with proteasome inhibition (Fig. 3A). Similar analyses were done with Lys-48-linked Ub chain-specific antibody, and the co-localization of KLHL7 with Lys-48-linked Ub chain increased with proteasome inhibition (Fig. 3B). However, no co-localization of KLHL7 with Lys-63-linked Ub was seen, regardless of proteasome inhibition (Fig. 3C). These results show that KLHL7 co-localizes with Lys-48-linked polyubiquitinated proteins but not with those with Lys-63-linked polyubiquitin, suggesting that KLHL7 mediates the linkage of Lys-48-linked but not Lys-63-linked polyubiquitin chain onto target proteins. The increase in poly-Ub and Lys-48-linked Ub-positive KLHL7 puncta with proteasome inhibition indicate that the substrates ubiquitinated by KLHL7 are targeted for proteasomal degradation.
FIGURE 3.
Ub-positive KLHL7 puncta increase with proteasome inhibition. A–C, HeLa cells cultured on coverslips were transfected with FLAG-KLHL7 and treated with either 20 μm of MG132 or DMSO and stained with antibodies to FLAG (red) and Ub (green) with anti-FK2 (A), anti-K48 (B), and anti-K63 (C). The nucleus was stained with Hoechst 33342. Scale bars, 20 μm.
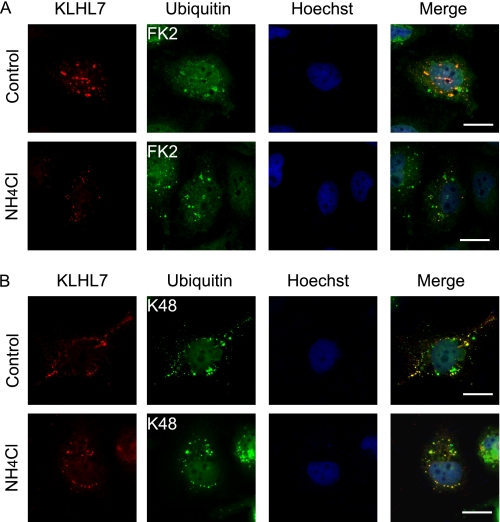
Ubiquitinated proteins can also be processed by the lysosome through the autophagic pathway. To investigate if the substrate of KLHL7 is also degraded by the lysosome, the co-localization of KLHL7 and Ub was examined under NH4Cl treatment (Fig. 4, A and B). In contrast to proteasome inhibition, no accumulation in Ub-positive KLHL7 puncta was seen with lysosome inhibition with NH4Cl. Furthermore, the involvement of the autophagic pathway was assessed by the co-localization of KLHL7 with GFP-tagged autophagosome marker LC3. To test this, we used ATG5+/+:GFP-LC3 and autophagy-deficient ATG5−/−:GFP-LC3 mouse embryo fibroblasts as negative control. No increase in the co-localization of KLHL7 with GFP-LC3 could be seen with MG132 or NH4Cl treatment, regardless of the increase in GFP-LC3 dots with lysosome inhibition (supplemental Fig. 2). Taken together, these data suggest that KLHL7-associated ubiquitinated proteins are processed mostly, if not all, by the proteasome and not the lysosome or LC3-dependent autophagy.
FIGURE 4.
Ub-positive KLHL7 does not increase by lysosome inhibition. A and B, HeLa cells cultured on coverslips were transfected with FLAG-KLHL7 and treated with or without NH4Cl for lysosome inhibition and stained with antibodies to FLAG and Ub with anti-FK2 (A) and anti-K48 (B). The nucleus was detected with Hoechst 33342. Scale bars, 20 μm.
AdRP Causing A153V and A153T Mutations Disrupt Cul3 Interaction but Not Dimerization
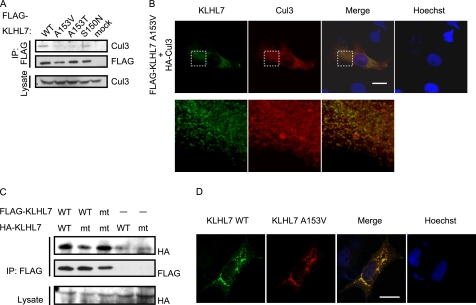
Recently, three missense mutations, S150N, A153T, and A153V in KLHL7 were found in patients with adRP. All of these mutations are in the BACK domain, and although both Ser-150 and Ala-153 are conserved within vertebrate KLHL7 (supplemental Fig. 3A), only Ala-153 is conserved within other human BTB-BACK-Kelch proteins (supplemental Fig. 3B). As the BACK domain is necessary for the interaction with Cul3 but not for KLHL7 dimerization (Fig. 1, B, C, and E), we examined if these mutants interact with Cul3 in HCT116 cells. KLHL7 S150N could still interact with Cul3 (Fig. 5A, 4th lane), whereas the A153V and A153T mutants could not (Fig. 5A, 2nd and 3rd lanes). We also investigated the subcellular localization of these missense mutations and their co-localization with Cul3. Immunocytochemical analyses in HeLa cells showed that KLHL7 S150N has similar subcellular localization with wild-type KLHL7 and co-localizes with Cul3 (supplemental Fig. 4, top). On the contrary, the subcellular localization of A153V and A153T was more peripheral than that of wild-type KLHL7, and co-localization with Cul3 was not as prominent (Figs. 1D, 5B, and supplemental Fig. 4, bottom). Taken together, these data suggest that Ala-153 mutations in KLHL7 attenuate its interaction and co-localization with Cul3.
FIGURE 5.
Ala-153 mutations of KLHL7 inhibit Cul3 interaction but not dimerization. A, HCT116 cells were transfected with either FLAG-KLHL7 wild-type or missense mutants. The cell lysates were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted with anti-Cul3 and anti-FLAG antibodies. B, HeLa cells cultured on coverslips were transfected with HA-Cul3 and FLAG-KLHL7 A153V. The cells were fixed and stained with anti-HA and anti-FLAG antibodies and Hoechst 33342. C, FLAG-KLHL7 (wild-type (WT) and A153V (mt)) and HA-KLHL7 (wild-type (WT) and A153V (mt)) were expressed in HCT116 cells as indicated. FLAG-KLHL7 was immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-HA and anti-FLAG antibodies. D, HeLa cells on coverslips were transfected with FLAG-KLHL7 and HA-KLHL7 A153V. After fixation, cells were stained with anti-FLAG and anti-HA antibodies and Hoechst 33342. Scale bars, 20 μm.
We next examined if the mutation in the Ala-153 position affects the dimerization ability of KLHL7 by using the A153V mutant, as the BACK domain was important for efficient dimerization (Fig. 1E). We expressed FLAG-KLHL7 wild-type or A153V with HA-KLHL7 wild-type or A153V in HCT116 cells and immunoprecipitated the cell lysates with anti-FLAG antibody. KLHL7 A153V was able to form both a homodimer and a heterodimer with wild-type KLHL7 (Fig. 5C). The S150N and A153T mutants could also bind to wild-type KLHL7 (supplemental Fig. 5A). These data suggest that none of the adRP-causative mutations interfere with KLHL7 dimerization. Immunocytochemical analyses showed that when both wild-type KLHL7 and A153V were expressed in HeLa cells, co-localization was observed in perinuclear punctate structures, resembling the subcellular localization of wild-type KLHL7 (Fig. 5D). These data indicate that the subcellular localization of KLHL7 A153V seems to be restored to that of wild-type KLHL7 by forming a heterodimer.
KLHL7 A153V Does Not Inhibit Cul3-Wild-type KLHL7 Interaction
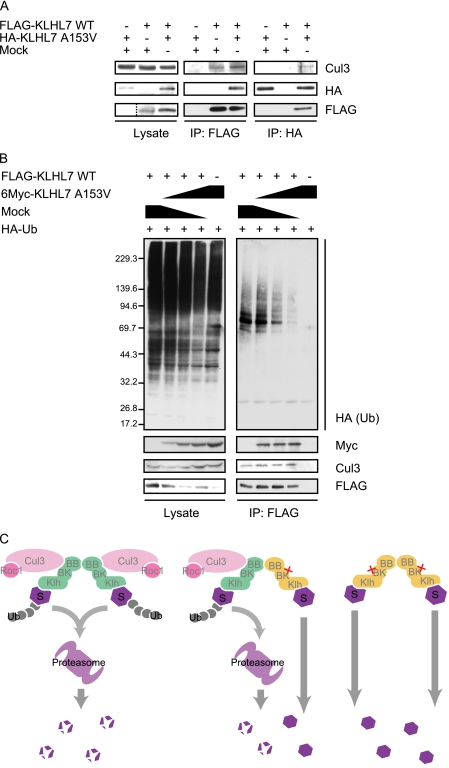
As KLHL7 A153V associates with wild-type KLHL7 but not directly with Cul3, we next examined the possibility that the heterodimerization of KLHL7 A153V with wild-type KLHL7 inhibits the formation of the Cul3-KLHL7 complex. To examine this, we co-expressed KLHL7 A153V with wild-type KLHL7 and investigated the amount of Cul3 that co-immunoprecipitate with wild-type KLHL7 (Fig. 6A). Contrary to our expectations, the co-immunoprecipitation of Cul3 did not decrease even when KLHL7 A153V was co-expressed (Fig. 6A, IP: FLAG, 2nd and 3rd lanes). Moreover, a slight amount of Cul3 could be co-immunoprecipitated by KLHL7 A153V when the wild-type KLHL7 was co-expressed (Fig. 6A, IP: HA, 1st and 3rd lanes). Similarly, A153T was also able to associate with Cul3 by the co-expression of wild-type KLHL7 (supplemental Fig. 5B). Taken together, it seems that although KLHL7 A153V cannot directly interact with Cul3, the mutation in one molecule of the KLHL7 dimer does not inhibit the assembly of Cul3-KLHL7 in a dominant negative manner.
FIGURE 6.
KLHL7 A153V attenuates Cul3-KLHL7 ligase activity. A, HCT116 cells were transfected with FLAG-KLHL7 and HA-KLHL7 A153V as indicated. The cell lysates were immunoprecipitated (IP) with anti-FLAG or anti-HA antibodies and immunoblotted with anti-Cul3, anti-HA, and anti-FLAG antibodies. B, HCT116 cells were transfected with FLAG-KLHL7, HA-Ub, and increasing amounts of 6Myc-KLHL7 A153V. The cells were treated with 20 μm MG132 for 1 h prior to harvesting and immunoprecipitated with anti-FLAG antibodies. Immunoblot analyses were done using anti-HA, anti-Myc, anti-Cul3, and anti-FLAG antibodies. C, molecular models of KLHL7-mediated ubiquitination. KLHL7 is indicated in green, the Ala-153 mutant in yellow, and the substrate as S. The domains of KLHL7 are as indicated: BB, BTB; BK, BACK; Klh, Kelch. The wild-type KLHL7 homodimer binds to two sets of Cul3/Roc and targets the substrates for ubiquitination (left panel). The heterodimer of wild-type and Ala-153 mutant can only bind to one set of Cul3/Roc1 and may ubiquitinate one of the two substrates, or not at all (middle panel). The Ala-153 mutant homodimer cannot bind to Cul3/Roc1 and fails to target the substrates for ubiquitination (right panel).
KLHL7 A153V Attenuates the E3 Ligase Activity of Cul3-KLHL7
To understand the effect of KLHL7 with mutations in Ala-153 in the Cul3-KLHL7 complex, we next assessed whether the incorporation of KLHL7 A153V affects the E3 ligase activity of Cul3-KLHL7. FLAG-KLHL7 and HA-Ub were transfected with increasing amounts of 6Myc-KLHL7 A153V, and ubiquitinated proteins associated with FLAG-KLHL7 were examined. Under proteasome inhibition, the amount of Ub that associated with wild-type KLHL7 decreased in proportion to the co-expressed KLHL7 A153V, even though there was no change in the amount of co-immunoprecipitated Cul3 and FLAG-KLHL7 (Fig. 6B). These data indicate that the A153V mutant decreases the E3 activity of Cul3-KLHL7.
DISCUSSION
In this study, we have shown that KLHL7 dimerizes and forms a novel Cul3-based E3 ligase. Although the RP-causative KLHL7 S150N mutant did not have obvious effects, and remains to be further investigated, our study provides insights into how the Ala-153 mutations affect the ligase activity of Cul3-KLHL7. The A153V and A153T mutants have the putative substrate-binding Kelch, and the dimerization BTB domains intact. Therefore, it is conceivable that the homodimers would be able to bind onto its authentic substrate. As the Ala-153 mutants could not directly associate with Cul3, the substrate bound to the mutant homodimer would not be recruited to the Cul3-Roc1 complex for ubiquitination (Fig. 6C, right). The subcellular localization of A153V and A153T mutants differed from that of wild type, suggesting that Cul3 binding and subsequent ubiquitination of the substrate may regulate the subcellular localization of KLHL7. Consistent with this notion, none of the deletion mutants formed such punctate structures (supplemental Fig. 1), including the ΔKelch mutant that associates with Cul3.
Although KLHL7 A153V and A153T mutants did not interact with Cul3, co-expression of wild-type KLHL7 enabled the mutants to associate with Cul3. Furthermore, the subcellular localization of KLHL7 A153V returned to normal appearance with the co-expression of wild-type KLHL7. These results suggest that the mutants can be incorporated into the Cul3 complex through the dimerization with wild-type KLHL7. However, the amount of ubiquitinated proteins associated with Cul3-KLHL7 greatly diminished when the A153V mutant was present in the complex. This decrease in the ligase activity of Cul3-KLHL7 can be attributed to the change in the conformation of the complex. As the heterodimer would only be able to recruit one molecule of Cul3 with the wild-type KLHL7, the overall number of the catalytic subunit Roc1 incorporated in the Cul3-KLHL7 complex will decrease (Fig. 6C, left and middle panels). Several previous studies have also shown that CRLs take dynamic conformations (35, 36), the flexibility of which may be rate-limiting for ligase activity. Dimerizations of substrate recognition proteins may influence the positioning of the substrate within the complex, as suggested by the analysis of the F-box protein Cdc4. The dimerization of Cdc4 does not influence the affinity with its substrate Sic1, but it seems to influence the positioning of the substrate within the ligase complex (37). KLHL7 may function likewise, and the incorporation of the Ala-153 mutant into the Cul3-KLHL7 complex may result in a change in the flexibility or the correct positioning of the substrate, necessary for efficient substrate ubiquitination. The attenuation of Cul3-KLHL7 ligase activity by homo- and heterodimers of mutant KLHL7 will lead to the accumulation of the substrates, which could result in cellular toxicity.
Cul3-KLHL7 formed punctate structures that resemble vesicles or inclusion bodies. Inclusion bodies, which seclude misfolded or unnecessary proteins, are degraded by the proteasome and autophagic pathways (38). In our study, Lys-48-linked poly-Ub-positive KLHL7 puncta increased with proteasome inhibition, suggesting that proteins ubiquitinated by Cul3-KLHL7 are degraded by the proteasome. However, the puncta did not increase with NH4Cl treatment (Fig. 4) nor co-localize with the autophagosome marker LC3 (supplemental Fig. 2) and the lysosome marker LAMP1 (data not shown), making it unlikely that these puncta are processed by the lysosomal pathways.
RP is a heterogeneous disease, which is caused by defects in different biochemical cascades (28, 29). Mutations in membrane proteins such as RDS and the photon receptor RHO are known to cause RP. Several genes related to protein trafficking and the cytoskeletal structures, such as FSCN2, that bind to actin (39, 40) and microtubule-associated RP1 (41), as well as MYO7A (42–45) involved in intracellular trafficking, have also been reported as RP-causative genes. Ub modification is known to be important for the quality control, internalization, and multivesicular body sorting of membrane proteins. It is also involved in the regulation of vesicle sorting and transport along the cytoskeletal structures (46, 47). KLHL7 may be involved in the regulation of any of these processes, however, how it is involved remains to be elucidated.
In summary, we present KLHL7 as a novel substrate recognition subunit of a Cul3-based Ub ligase complex. Moreover, we have shown that the adRP causative A153V mutation inhibits the ligase activity of Cul3-KLHL7 in a dominant negative manner. Future studies should focus on determining the substrate and the components of the Cul3-KLHL7 puncta, which will undoubtedly become the key to understanding the physiological role of KLHL7.
Supplementary Material
Acknowledgments
We thank Dr. N. Mizushima (Tokyo Medical and Dental University) for providing ATG5+/+:GFP-LC3 and ATG5−/−:GFP-LC3 mouse embryo fibroblasts, Professor M. Wood (University of Tsukuba) for critical reading of the manuscript, and members of the Chiba laboratory for helpful discussions and technical support.
This work was supported by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan (to T. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- Ub
- ubiquitin
- CRL
- Cullin-RING ligase
- adRP
- autosomal dominant retinitis pigmentosa.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 3. Pickart C. M., Fushman D. (2004) Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 4. Olzmann J. A., Chin L. S. (2008) Autophagy 4, 85–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dias D. C., Dolios G., Wang R., Pan Z. Q. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16601–16606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jackson P. K., Eldridge A. G. (2002) Mol. Cell 9, 923–925 [DOI] [PubMed] [Google Scholar]
- 8. Cardozo T., Pagano M. (2004) Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 9. Willems A. R., Schwab M., Tyers M. (2004) Biochim. Biophys. Acta 1695, 133–170 [DOI] [PubMed] [Google Scholar]
- 10. Petroski M. D., Deshaies R. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 11. Willems A. R., Lanker S., Patton E. E., Craig K. L., Nason T. F., Mathias N., Kobayashi R., Wittenberg C., Tyers M. (1996) Cell 86, 453–463 [DOI] [PubMed] [Google Scholar]
- 12. Feldman R. M., Correll C. C., Kaplan K. B., Deshaies R. J. (1997) Cell 91, 221–230 [DOI] [PubMed] [Google Scholar]
- 13. Patton E. E., Willems A. R., Sa D., Kuras L., Thomas D., Craig K. L., Tyers M. (1998) Genes Dev. 12, 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 15. Geyer R., Wee S., Anderson S., Yates J., Wolf D. A. (2003) Mol. Cell 12, 783–790 [DOI] [PubMed] [Google Scholar]
- 16. Xu L., Wei Y., Reboul J., Vaglio P., Shin T. H., Vidal M., Elledge S. J., Harper J. W. (2003) Nature 425, 316–321 [DOI] [PubMed] [Google Scholar]
- 17. Pintard L., Willis J. H., Willems A., Johnson J. L., Srayko M., Kurz T., Glaser S., Mains P. E., Tyers M., Bowerman B., Peter M. (2003) Nature 425, 311–316 [DOI] [PubMed] [Google Scholar]
- 18. Furukawa M., He Y. J., Borchers C., Xiong Y. (2003) Nat. Cell Biol. 5, 1001–1007 [DOI] [PubMed] [Google Scholar]
- 19. Stogios P. J., Downs G. S., Jauhal J. J., Nandra S. K., Privé G. G. (2005) Genome Biol. 6, R82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhuang M., Calabrese M. F., Liu J., Waddell M. B., Nourse A., Hammel M., Miller D. J., Walden H., Duda D. M., Seyedin S. N., Hoggard T., Harper J. W., White K. P., Schulman B. A. (2009) Mol. Cell 36, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., Yamamoto M. (1999) Genes Dev. 13, 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kobayashi A., Kang M. I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. (2004) Mol. Cell. Biol. 24, 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sumara I., Quadroni M., Frei C., Olma M. H., Sumara G., Ricci R., Peter M. (2007) Dev. Cell 12, 887–900 [DOI] [PubMed] [Google Scholar]
- 24. Bomont P., Cavalier L., Blondeau F., Ben Hamida C., Belal S., Tazir M., Demir E., Topaloglu H., Korinthenberg R., Tüysüz B., Landrieu P., Hentati F., Koenig M. (2000) Nat. Genet. 26, 370–374 [DOI] [PubMed] [Google Scholar]
- 25. Cirak S., von Deimling F., Sachdev S., Errington W. J., Herrmann R., Bönnemann C., Brockmann K., Hinderlich S., Lindner T. H., Steinbrecher A., Hoffmann K., Privé G. G., Hannink M., Nürnberg P., Voit T. (2010) Brain 133, 2123–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Friedman J. S., Ray J. W., Waseem N., Johnson K., Brooks M. J., Hugosson T., Breuer D., Branham K. E., Krauth D. S., Bowne S. J., Sullivan L. S., Ponjavic V., Gränse L., Khanna R., Trager E. H., Gieser L. M., Hughbanks-Wheaton D., Cojocaru R. I., Ghiasvand N. M., Chakarova C. F., Abrahamson M., Göring H. H., Webster A. R., Birch D. G., Abecasis G. R., Fann Y., Bhattacharya S. S., Daiger S. P., Heckenlively J. R., Andréasson S., Swaroop A. (2009) Am. J. Hum. Genet. 84, 792–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hugosson T., Friedman J. S., Ponjavic V., Abrahamson M., Swaroop A., Andréasson S. (2010) Arch. Ophthalmol. 128, 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hartong D. T., Berson E. L., Dryja T. P. (2006) Lancet 368, 1795–1809 [DOI] [PubMed] [Google Scholar]
- 29. Daiger S. P., Bowne S. J., Sullivan L. S. (2007) Arch. Ophthalmol. 125, 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuma A., Matsui M., Mizushima N. (2007) Autophagy 3, 323–328 [DOI] [PubMed] [Google Scholar]
- 31. Kawakami T., Chiba T., Suzuki T., Iwai K., Yamanaka K., Minato N., Suzuki H., Shimbara N., Hidaka Y., Osaka F., Omata M., Tanaka K. (2001) EMBO J. 20, 4003–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ogura T., Tong K. I., Mio K., Maruyama Y., Kurokawa H., Sato C., Yamamoto M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2842–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ohta T., Michel J. J., Schottelius A. J., Xiong Y. (1999) Mol. Cell 3, 535–541 [DOI] [PubMed] [Google Scholar]
- 34. Tan P., Fuchs S. Y., Chen A., Wu K., Gomez C., Ronai Z., Pan Z. Q. (1999) Mol. Cell 3, 527–533 [DOI] [PubMed] [Google Scholar]
- 35. Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., Schulman B. A. (2008) Cell 134, 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Merlet J., Burger J., Gomes J. E., Pintard L. (2009) Cell. Mol. Life Sci. 66, 1924–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang X., Orlicky S., Lin Z., Willems A., Neculai D., Ceccarelli D., Mercurio F., Shilton B. H., Sicheri F., Tyers M. (2007) Cell 129, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 38. Kubota H. (2009) J. Biochem. 146, 609–616 [DOI] [PubMed] [Google Scholar]
- 39. Tubb B. E., Bardien-Kruger S., Kashork C. D., Shaffer L. G., Ramagli L. S., Xu J., Siciliano M. J., Bryan J. (2000) Genomics 65, 146–156 [DOI] [PubMed] [Google Scholar]
- 40. Saishin Y., Ishikawa R., Ugawa S., Guo W., Ueda T., Morimura H., Kohama K., Shimizu H., Tano Y., Shimada S. (2000) Invest. Ophthalmol. Vis. Sci. 41, 2087–2095 [PubMed] [Google Scholar]
- 41. Liu Q., Zuo J., Pierce E. A. (2004) J. Neurosci. 24, 6427–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. el-Amraoui A., Sahly I., Picaud S., Sahel J., Abitbol M., Petit C. (1996) Hum. Mol. Genet. 5, 1171–1178 [DOI] [PubMed] [Google Scholar]
- 43. Weil D., Levy G., Sahly I., Levi-Acobas F., Blanchard S., El-Amraoui A., Crozet F., Philippe H., Abitbol M., Petit C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Williams D. S. (2002) Vision Res. 42, 455–462 [DOI] [PubMed] [Google Scholar]
- 45. Udovichenko I. P., Gibbs D., Williams D. S. (2002) J. Cell Sci. 115, 445–450 [DOI] [PubMed] [Google Scholar]
- 46. Haglund K., Dikic I. (2005) EMBO J. 24, 3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Raiborg C., Stenmark H. (2009) Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.