Abstract
Expression of the STAT3 transcription factor in the heart is cardioprotective and decreases the levels of reactive oxygen species. Recent studies indicate that a pool of STAT3 resides in the mitochondria where it is necessary for the maximal activity of complexes I and II of the electron transport chain. However, it has not been explored whether mitochondrial STAT3 modulates cardiac function under conditions of stress. Transgenic mice with cardiomyocyte-specific overexpression of mitochondria-targeted STAT3 with a mutation in the DNA-binding domain (MLS-STAT3E) were generated. We evaluated the role of mitochondrial STAT3 in the preservation of mitochondrial function during ischemia. Under conditions of ischemia heart mitochondria expressing MLS-STAT3E exhibited modest decreases in basal activities of complexes I and II of the electron transport chain. In contrast to WT hearts, complex I-dependent respiratory rates were protected against ischemic damage in MLS-STAT3E hearts. MLS-STAT3E prevented the release of cytochrome c into the cytosol during ischemia. In contrast to WT mitochondria, ischemia did not augment reactive oxygen species production in MLS-STAT3E mitochondria likely due to an MLS-STAT3E-mediated partial blockade of electron transport through complex I. Given the caveat of STAT3 overexpression, these results suggest a novel protective mechanism mediated by mitochondrial STAT3 that is independent of its canonical activity as a nuclear transcription factor.
Keywords: Cellular Regulation, Ischemia, Respiratory Chain, Signal Transduction, STAT Transcription Factor, Transcription Factors
Introduction
STAT3 was originally identified as an IL-6-induced transcriptional activator of acute phase genes (1). However, other members of the IL-6 family, which utilize gp-130 receptor, as well as leptin, IL-12, IFNα/β, IL-10, GM-CSF, several growth factors, oncogenes, and stress such as hypoxia, also activate STAT3 (1). STAT3 is vital to embryonic development and STAT3-null mice are embryonic lethal (2). Analysis of tissue-specific conditional STAT3 knock-out mice has provided strong evidence that transcriptional activity of STAT3 plays a central role in the control of cell growth and host responses to inflammation and cellular stress (1). STAT3 positively regulates expression of anti-apoptotic (Bcl-2 and Bcl-xL) (1) and anti-oxidative proteins (MnSOD and metallothionein-1 and -2) (3, 4).
Expression of STAT3 in the heart is associated with cardiac survival (5). When STAT3 is selectively deleted in cardiomyocytes, mice develop enhanced cardiac inflammation with fibrosis, dilated cardiomyopathy, and die prematurely due to congestive heart failure (5). Female mice, where STAT3 is not expressed in cardiomyocytes, develop post-partum cardiomyopathy, which is also seen in humans with reduced STAT3 expression in the myocardium (6). Ventricles from STAT3-null hearts show elevated levels of reactive oxygen species (ROS)2 (6). Ischemic and pharmacologic preconditioning protected the viability of wild type but not STAT3−/− cardiomyocytes (5). When STAT3 is overexpressed in cardiomyocytes, mice are less sensitive to the cardiotoxic effects of doxorubicin, which exerts its actions in part via the generation of ROS (7). Doxorubicin treatment decreases complex I activity in heart mitochondria (8), which in turn increases ROS production. STAT3−/− cardiomyocytes demonstrate increased sensitivity to endotoxin/LPS-mediated toxic shock (9). Thus, the expression of STAT3 in the heart protects from a variety of stresses that induce the formation of ROS.
One of the major sources of ROS in the cell are mitochondria (10). Recent studies demonstrated that STAT3 is constitutively present in the mitochondria (11, 12) and is important in regulation of the activity of the electron transport chain (ETC) independent of its action in the nucleus (12). In mice where STAT3 has been deleted from cardiomyocytes there is a decrease in the activities of complexes I and II of the ETC, leading to a decreased rate of oxidative phosphorylation (12).
Because STAT3 expression is required for normal cardiac function and STAT3 regulates mitochondrial respiration, we examined the role of mitochondrial STAT3 during stress conditions. To address this issue, a transgenic mouse was generated that overexpressed selectively in cardiomyocytes a DNA-binding mutant of STAT3 containing a mitochondrial targeting sequence. We have used these mice to both examine the actions of mitochondrial STAT3 in the response to ischemia as well as to begin define the molecular mechanisms by which this subcellular pool of STAT3 modulates respiration.
EXPERIMENTAL PROCEDURES
Reagents
Restriction enzymes and T4 ligase were purchased from New England Biolabs, Ipswich, MA. The following antibodies were used: STAT3, lamin A/C (Cell Signaling, Danvers, MA), tubulin, FLAG M2 (Sigma), porin (Calbiochem, Gibbstown, NJ), cytochrome c, MnSOD (BD Biosciences), and MT (Santa Cruz Biotechnology, Santa Cruz, CA). All chemicals were purchased from Sigma unless indicated otherwise.
Animals
Animals were treated in compliance with the Guide for the Care and Use of Laboratory Animals under the protocols approved by Virginia Commonwealth University Institutional Animal Care and Use Committee.
Generation of MLS-STAT3E Mice
A construct containing the MLS (mitochondria-localizing sequence taken from human cytochrome c oxidase subunit VIII gene) followed by mouse Stat3 cDNA harboring the DNA-binding mutation (E434A/E435A) (13), termed MLS-STAT3E, was inserted in the murine stem cell virus-internal ribosome entry site-GFP vector as previously described (12). MLS-STAT3E cDNA was amplified by PCR using a set of primers that introduced a SalI restriction site followed by a Kozak consensus sequence on the 5′-end (forward primer) and FLAG tag sequence followed by a STOP codon and HindIII restriction site on the 3′-end (reverse primer) of Stat3 cDNA. The amplified fragment was ligated into the pBSIISK(+) vector containing α-myosin heavy chain α gene promoter (a generous gift from Jeffrey Robbins from Children's Hospital Research Foundation, Cincinnati, OH) (Fig. 1A). The pBSIISK(+) vector backbone was removed from the targeting sequence prior to DNA injection into the fertilized eggs by NotI restriction digestion. Transgenic mice were obtained in the Transgenic Targeting Facility of Case Western Reserve University, Cleveland, OH. Mice expressing the transgene were mated with homozygous floxed-STAT3 (exons 12–14 flanked by loxP sequences) mice on 129X1/SvJ background (14). DNA samples obtained from tails of newborns were screened for the presence of the transgene to establish the founders. MLS-STAT3E transgenic DNA (genotyping) was detected as a 300-bp product of PCR using the primers designed to complement to the 3′-end of Stat3 (forward: 5′-GCG ACC AAC ATC CTG GTG TCT CCA C-3′) and to the FLAG sequence (reverse: 5′-CTT GTC GTC ATC GTC TTA GTA GTC C-3′), and GoTaq Hot Start Green Master Mix (Promega, Madison, WI). Amplification was performed under the following conditions: 95 °C for 3 min; 35 cycles of: 95 °C for 30 s, 54 °C for 30 s, and 73 °C for 30 s; and 73 °C for 5 min. Pups from founders were tested for the presence of transgene mRNA in the heart and other organs using standard RT-PCR methods. Founder lines positive for transgenic mRNA in heart tissue were screened for protein expression in the mitochondria by anti-FLAG tag immunoprecipitation followed by Western blot analysis using anti-STAT3 mAb. Before any experiments were conducted, mice were bred nine times with homozygous STAT3-floxed mice of the 129X1/SvJ strain (14) to establish a pure background.
FIGURE 1.
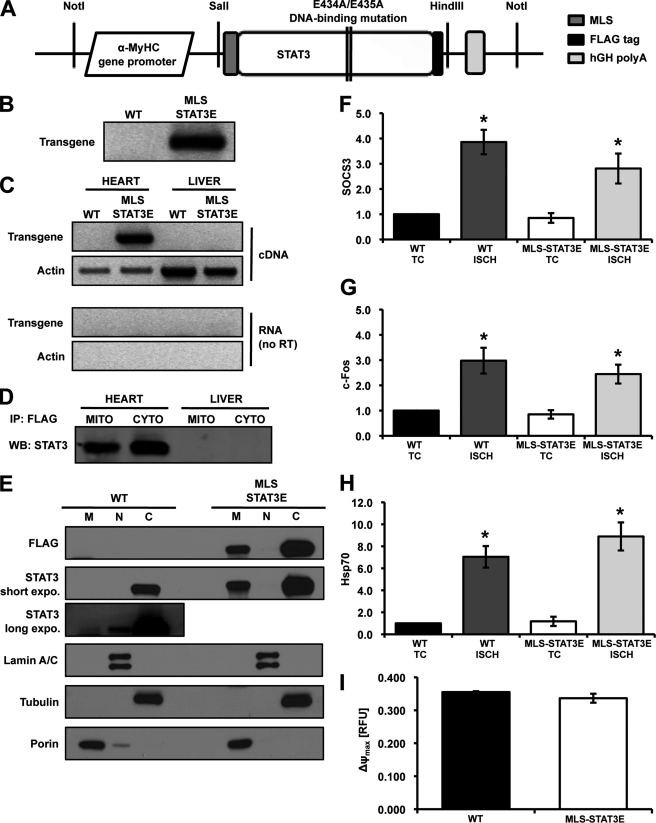
Cardiac-restricted overexpression of transcriptionally inactive MLS-STAT3E. A, schematic representation of the MLS-STAT3E transgene construct. B, isolated genomic DNA was analyzed for the presence of the transgene by PCR. C, hearts and livers from MLS-STAT3E and WT mice were screened for the presence of transgenic mRNA using qualitative RT-PCR. Primers complement to actin mRNA were used as an internal control. RNA (no reverse transcription, no RT) was used to exclude the possibility of RNA contamination with genomic DNA. D, protein expression was analyzed by anti-FLAG tag immunoprecipitation from mitochondrial (MITO) and cytosolic (CYTO) extracts isolated from hearts and livers of WT and MLS-STAT3E mice, followed by SDS-PAGE and immunoblotting for STAT3. A representative immunoblot from three independent experiments is shown. E, expression of MLS-STAT3E protein was analyzed in mitochondrial (M), nuclear (N), and cytosolic (C) fractions of the heart. Equal amounts of mitochondrial and cytosolic fractions (5 μg) were loaded on the gel. A representative immunoblot from three independent experiments is shown. F–H, MLS-STAT3E does not drive the expression of STAT3-dependent genes under basal and stress conditions. Total RNA was isolated from ischemic (ISCH) and TC hearts of MLS-STAT3E and WT mice, and mRNA levels of Socs3 (F), c-Fos (G), and Hsp70i (H) were assayed by real-time qPCR. Results are mean ± S.E. defined as a fold-induction over WT (set as 1) and normalized to β-actin, n = 5 for each group. *, p < 0.05 versus WT TC. I, the maximum potential that mitochondria were capable to generate (ΔΨmax) was measured in intact organelles using 0.1 μm tetramethylrhodamine methyl ester and was defined as the difference in membrane potential between fully polarized mitochondria in the presence of 1 nm oligomycin (state 4oligomycin) and fully uncoupled mitochondria (Ψ0) after titration with 25 μm DNP. Results are mean ± S.E., n = 4 for each group. RFU, relative fluorescence units. hGH, human growth hormone.
RNA Isolation, Reverse Transcription, Qualitative PCR, and Real Time qPCR
RNA was isolated according to the protocol previously described (15) using TRI Reagent (Molecular Research Center, Cincinnati, OH) and treated with DNase (Promega). 2 μg of total RNA was transcribed to cDNA using Tetro cDNA Synthesis Kit (Bioline, Tauton, MA). Qualitative RT-PCR was performed on samples before (RNA) and after reverse transcriptase reaction (cDNA). RNA was isolated from hearts and livers of WT and transgenic mice. DNase-treated RNA samples and RNase-treated cDNA samples were subjected to PCR to test for the presence of the transgenic mRNA in the heart and liver tissues of the screened mice. Primers for β-actin gene (Actb) (forward: 5′-GTG GGC CGC TCT AGG CAC CAA-3′, reverse, 5′-CTC TTT GAT GTC ACG CAC GAT TTC-3′) were used as an internal control. The conditions of the PCR were as described for genotyping protocol. The mRNA levels of genes of interest were analyzed by quantitative real-time PCR using SensiMix SYBR and Fluorescein Kit (Bioline). All the samples were assayed in triplicate and analyzed using the CFX96 Real-time PCR Detection System (Bio-Rad). Appropriate primers for Socs3, c-Fos, and Hsp70i were obtained from SABiosciences, Frederick, MD. The no template control (without cDNA in the reaction mixture) and the no reverse-transcribed RNA control were used as negative controls in the real-time qPCR. To calculate relative expression, all results were analyzed according to the ΔCt method (variation of Livak method (16)) using a reference gene, β-actin (Actb).
Treatment of Mice with Leukemia Inhibitory Factor (LIF)
8–10-Week-old MLS-STAT3E and WT littermate male mice were injected intravenously with either LIF (8 × 104 units/kg) (Millipore, Billerica, MA) or vehicle. After 1 h, hearts were excised and RNA was isolated.
Ex Vivo Model of Stop Flow-simulated Ischemia
Hearts were excised from 8–10-week-old MLS-STAT3E and WT littermate mice and placed in sterile 1.5-ml tubes containing 1 ml of saline (0.9% NaCl in sterile H2O). Hearts designated as time controls were kept on ice, whereas hearts that underwent simulated 45 min ischemia were incubated at 37 °C in a digitally controlled tube shaker set at 1,400 rpm (17). After the incubation, hearts were washed in CP1 buffer and mitochondria were isolated as described. For each separate experiment, two mice were used, one for ischemia and one for time control (TC).
Doppler Echocardiography
Doppler echocardiography was performed using the Vevo770TM imaging system (VisualSonics Inc., Toronto, Canada) as described previously (18). Light anesthesia was used during the exam with the injection of pentobarbital (30 mg/kg; intraperitoneally). The mice were placed in the supine position and ECG limb electrodes were attached. The chest was carefully shaved and ultrasound gel was used on the thorax to optimize visibility during the exam. A 30-MHz probe was utilized to obtain two-dimensional, M-mode, and Doppler imaging from parasternal short-axis views at the level of the papillary muscles and the apical four-chamber view. M-mode images of the left ventricle (LV) were obtained and systolic and diastolic wall thickness (anterior and posterior) and LV end-systolic and end-diastolic diameters (LVESD and LVEDD, respectively) were measured. LV fractional shortening was calculated as (LVEDD − LVESD)/LVEDD × 100. Ejection fraction was calculated using the Teichholz formula (19). The LV mass was calculated using the following formula ((LVEDD + AWDT + PWDT)3 − (LVEDD)3 × 0.8 × 1.04 + 0.6)/1000, where AWDT and PWDT are anterior and posterior wall diastolic thickness, respectively (20). The heart rate was monitored in mice under anesthesia with pentobarbital using echocardiography. No differences in heart rate were observed between MLS-STAT3E and WT mice, indicating that the effect of anesthesia had no impact on LV function in this study. The investigator performing and reading the echocardiogram was blinded to the mouse genotype.
Isolation of Mitochondrial, Cytosolic, and Nuclear Fractions
Hearts were excised and ventricles were used to isolate a single population of cardiac mitochondria. Tissue was briefly washed in a modified Chappell-Perry (CP) buffer (buffer CP1 at pH 7.4: 100 mm KCl, 50 mm MOPS, 1 mm EGTA, 5 mm MgSO4·7H2O, 1 mm ATP), dried with Whatman filter paper, weighed, then placed in glass beaker, and thoroughly minced. Ventricular tissue was homogenized in 3 ml of CP1 buffer using a Polytron tissue blender (Kinematica, Bohemia, NY) for 2.5 s at a rheostat setting of 10,000 rpm. The Polytron homogenate was centrifuged at 6,000 × g for 10 min at 4 °C and the supernatant was saved as a crude cytosol for further purification. The homogenate pellets were re-suspended in 3 ml of CP1 buffer supplemented with 5 mg/g (wet weight) trypsin (number T0303, Sigma), incubated with stirring for 15 min at 4 °C followed by addition of 3 ml of CP2 buffer (CP1 buffer containing 0.2% BSA (number A7030, Sigma) to attenuate trypsin activity). Digested tissue was further homogenized by two strokes using a digital steady-stirring tight Teflon pestle/glass tube homogenizer set at 600 rpm (Fisher Scientific). 600 μl of the homogenate was saved for RNA isolation. Undigested tissue and heavier cell fractions in the remaining volume were pelleted by centrifugation at 500 × g for 10 min at 4 °C. The mitochondria-containing supernatant was centrifuged at 3000 × g for 10 min at 4 °C. The mitochondrial pellet was washed with 2 ml of KME buffer, pH 7.4 (100 mm KCl, 50 mm MOPS, 0.5 mm EGTA). Mitochondria were re-suspended in 80–120 μl of KME and used within 4 h after isolation or frozen. Crude cytosolic fraction was supplemented with protease and phosphatase inhibitor mixtures (Roche Applied Science) and purified by ultra microcentrifugation at 100,000 × g for 1 h at 4 °C (Thermo Scientific, Waltham, MA). Nuclear fraction was isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagent Kit (Pierce/Thermo Scientific) according to the manufacturer's protocol. The protein concentration was measured by Lowry (21) using BSA as a standard and sodium deoxycholate as a detergent.
SDS-PAGE and Immunoblotting
Proteins were separated using the Novex NuPAGE electrophoresis system (Invitrogen) using 4–12% gradient BisTris gels and MES SDS running buffer according to the manufacturer's protocol. Gels were transferred to Immobilon-P PVDF membrane (Millipore) using semi-dry transfer (Bio-Rad). The blots were incubated for 1 h at room temperature in 5% (w/v) nonfat dry milk (Bio-Rad) in TBS-T buffer (10 mm Tris, pH 7.5, 150 mm NaCl, 0.1% Tween 20) followed by overnight incubation at 4 °C with primary antibody. After a 1-h incubation at room temperature with a 1:50,000 dilution of HRP-conjugated anti-mouse or anti-rabbit IgG F(ab)2 (GE Healthcare), blots were developed using Amersham Biosciences ECL Plus Western blotting Detection Reagents (GE Healthcare). An anti-FLAG immunoprecipitation protocol was applied to 1 mg/ml of protein extracts solubilized in modified RIPA buffer, pH 7.4 (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, protease and phosphatase inhibitor mixtures) using 40 μl of 1:1 slurry of EZview Red Anti-FLAG M2-agarose affinity beads (Sigma). After overnight incubation, samples were washed in RIPA buffer and re-suspended in 20 μl of 2× lithium dodecyl sulfate sample loading buffer (Invitrogen), incubated for 5 min at room temperature, and transferred to the new tube. After addition of 50 mm DTT, samples were incubated for 10 min at 70 °C and subjected to SDS-PAGE electrophoresis using the Novex NuPAGE system followed by already described immunoblotting protocol.
Mitochondrial Oxidative Phosphorylation
Oxygen consumption by intact mitochondria was measured using a Clark-type oxygen electrode (Strathkelvin Instruments, North Lanarkshire, UK) at 30 °C in respiration buffer at pH 7.4 (80 mm KCl, 50 mm MOPS, 1 mm EGTA, 5 mm KH2PO4, 1 mg/ml of defatted BSA) as previously described (22, 23). Substrates for complex I (20 mm glutamate + 5 mm malate), complex II (20 mm succinate with 7.5 μm rotenone), and complex IV (1 mm TMPD, 20 mm l-ascorbate with 7.5 μm rotenone) were used and state 3 (0.2 mm ADP-stimulated), state 4 (ADP-limited) respiration, respiratory control ratio (RCR), maximal rate of state 3 respiration (2 mm ADP), and rate of uncoupled respiration (0.04 mm dinitrophenol, DNP) were determined. In the case of TMPD/ascorbate, only the 2 mm ADP-dependent maximum state 3 rate was measured and 2 mm azide was added to determine the specificity of complex IV-dependent oxygen consumption. RCR was determined as a ratio of 0.2 mm stimulated state 3 to ADP-limited state 4 respiration.
Detection of H2O2 Production
The enzymatic reduction of mitochondria-generated H2O2 by horseradish peroxidase (HRP) was coupled with the oxidation of the fluorogenic indicator Amplex Red (Invitrogen) to the fluorescent resorufin as described previously (24). Intact mitochondria (30 μg) were incubated with 25 μm Amplex Red and 0.25 units/ml of HRP in Chelex-treated buffer, pH 7.4 (120 mm KCl, 5 mm KH2PO4, 1 mm EGTA), in the presence or absence of complex I- and/or complex III-specific inhibitors: 2.5 μm rotenone and 3.33 μg/ml of antimycin A.
ETC and Citrate Synthase Enzyme Activities
The following enzyme activities were measured spectrophotometrically in detergent-solubilized mitochondria using previously described methods (22–24): NADH-ubiquinone oxidoreductase, rotenone-sensitive (complex I), NADH-ferricyanide reductase (NFR); succinate-ubiquinone oxidoreductase, theonyltrifluoroacetone sensitive (complex II); ubiquinol-cytochrome c oxidoreductase, antimycin A sensitive (complex III); cytochrome c oxidase (complex IV) and citrate synthase. Frozen-thawed mitochondria were solubilized at a final protein concentration of 1 μg/μl in 0.5% cholate in MSM/EDTA buffer, pH 7.4 (220 mm mannitol, 70 mm sucrose, 5 mm MOPS, 2 mm EDTA). Calculated enzyme activities were expressed as nanomole/min/mg of mitochondrial protein or 1/min/mg in the case of complex IV.
Mitochondrial Membrane Potential
Membrane potential of intact mitochondria (ΔΨm) was measured using tetramethylrhodamine methyl ester dye (Invitrogen) as described previously (25). Mitochondria (200 μg) were added to 2 ml of respiration buffer at pH 7.4 supplemented with 0.1 μm tetramethylrhodamine methyl ester and stirred at 30 °C. Mitochondria were polarized by 5 mm glutamate + 1.25 mm malate followed by depolarization with 0.5 mm ADP. Membrane potential was re-generated by 1 nm oligomycin (state 4oligomycin) followed by full uncoupling (ΔΨ0) achieved by titration with 25 μm DNP. The maximal membrane potential (ΔΨmax) was determined as a difference between state 4oligomycin and DNP-uncoupled mitochondria (ΔΨ0).
Statistical Analysis
The data consisting of two groups were analyzed by two-tailed Student's t test. In the case of more than two groups (WT versus MLS-STAT3E in context of time control and ischemia), data were subjected to two-way analysis of variance with Holm-Sidak post test (pairwise multiple comparison procedures) for multiple groups. All analysis was executed using SigmaPlot software (Systat Software Inc., Chicago, IL). p < 0.05 was considered statistically significant.
RESULTS
Generation of MLS-STAT3E Transgenic Mice
Transgenic mice were generated with cardiomyocyte-restricted expression of STAT3 that was targeted to the mitochondria with a mitochondrial localization sequence (MLS) (Fig. 1A). The STAT3 transgene also contained mutations in the DNA-binding domain (E434A/E435A) and a carboxyl-terminal FLAG epitope tag. The transgenic animals expressing this modified version of STAT3 were termed MLS-STAT3E. Founders were back-crossed nine times into homozygous floxed-STAT3 mice to obtain a pure genotypic background (129X1/SvJ). The mice defined in this report as wild type (WT) were STAT3flx/flx and were previously shown not to exhibit any differences in phenotype when compared with STAT3+/+ animals (14). The MLS-STAT3E mice also expressed endogenous STAT3 on a floxed STAT3 background.
Pups were genotyped for the presence of transgenic DNA (Fig. 1B). MLS-STAT3E mice were phenotypically normal and fertile. Genotypes of offspring occurred in accordance with Mendelian predictions. There were no gross differences in mortality of transgenic animals (data not shown). As depicted in Fig. 1C, the expression of the MLS-STAT3E mRNA was limited to heart tissue. Hearts were further screened for the presence of MLS-STAT3E protein. The transgenic protein was found in the hearts but not in the livers, kidneys, or spleens of MLS-STAT3E mice (Fig. 1D and data not shown). Fractionation of heart homogenates revealed that the transgenic protein was present in the mitochondria and cytosol (Fig. 1, D and E), but very little if any of MLS-STAT3E was detected in the nuclear fraction (Fig. 1E). Under basal conditions there was a small amount of endogenous STAT3 in the heart nuclei.
Trypsin was used to increase mitochondrial protein yield and to remove potential contamination with other cellular fractions. The purity of isolated fractions was tested using anti-tubulin (cytosolic marker), anti-porin (mitochondrial marker), anti-lamin A/C (nuclear marker), and anti-SERCA2 (sarco/endoplasmic reticulum and Ca2+-ATPase) (endoplasmic marker) antibodies (Fig. 1E and data not shown). Densitometric analysis revealed that the amount of STAT3 in transgenic heart homogenates was increased 20-fold compared with WT littermates (data not shown). There was approximately a 100-fold increase of STAT3 protein in transgene mitochondria compared with WT endogenous levels (supplemental Fig. S1B). Densitometric analysis showed 2.5 times more STAT3 protein in the cytosol compared with mitochondrial fraction of MLS-STAT3E hearts (data not shown).
Expression of MLS-STAT3E Does Not Alter Nuclear-encoded Genes Activated by STAT3
The STAT3 transgene contained two point mutations in the DNA-binding domain to block its function as a transcription factor (13). To verify if MLS-STAT3E overexpression did not alter induction of STAT3-dependent genes, mRNA levels of Socs3 (26) and c-Fos (27) genes were analyzed by real-time qPCR. No changes in concentration of these RNAs were observed under baseline conditions (Fig. 1, F and G).
Oxidative stress affects transcriptional activity leading to changes in mRNA levels of a variety of different genes (28) including those regulated by STAT3 (29–31). We examined the expression of STAT3-dependent genes (Socs3 and c-Fos) in hearts from MLS-STAT3E and WT mice subjected to 45 min of global ischemia versus TC. mRNA levels of both genes were increased ∼3-fold by ischemia in both WT and MLS-STAT3E hearts (Fig. 1, F and G), indicating that the presence of MLS-STAT3 in cardiomyocytes did not alter the expression of these well described STAT3-regulated genes. Analysis of heat-shock protein 70 (Hsp70i) expression was used as a control of responsiveness of heart tissue to the stress conditions (28). Hsp70i mRNA was up-regulated over 7-fold by ischemia when compared with time controls in both transgenic and WT hearts (Fig. 1H).
We also examined LIF-induced expression of STAT3-dependent RNAs. LIF binds to the gp130 receptor (30). WT and MLS-STAT3E mice were injected with LIF and RNA from heart tissue was assayed for the expression of c-Fos and Socs3 (supplemental Fig. S1A). Both of these RNAs were induced to similar levels in WT and MLS-STAT3E mice, which confirms that the expression of the transgene has no effects on classical activation of STAT3-dependent genes.
Cardiac Function Is Unchanged in MLS-STAT3E Mice
Although MLS-STAT3E did not alter baseline or ischemia-induced expression of the RNAs, whose expression was dependent on the nuclear actions of STAT3, we wanted to examine whether there were any physiological consequences of overexpressing MLS-STAT3E. It is known that overexpression of STAT3 in the heart leads to the up-regulation of hypertrophic genes and development of cardiac dysfunction with age (7). We examined cardiac function in hearts of 1-year-old MLS-STAT3E and WT mice using echocardiography. No differences in LVEDD, LVESD, LVFS, and LVEF were observed between MLS-STAT3E and wild type littermates (Table 1), indicating normal heart function in the transgenic mice. Moreover, the LV mass was not different in the two groups (Table 1), illustrating that the MLS-STAT3E mice did not promote the development of cardiac hypertrophy. These results confirmed that the overexpressed MLS-STAT3E did not display any of the cardiac abnormalities reported in mice that expressed a transcriptionally active transgene (7) and reinforced the data that MLS-STAT3E expression did not alter STAT3-dependent gene expression.
TABLE 1.
Echocardiography of 1-year-old WT and MLS-STAT3E mouse hearts
Results are mean ± S.E., n = 5 for WT and n = 7 for MLS-STAT3E group.
| LVEDD | LVESD | FS | EF | HR | LV mass | |
|---|---|---|---|---|---|---|
| mm | % | bpm | mg | |||
| WT | 4.18 ± 0.2 | 2.57 ± 0.3 | 40 ± 6 | 70 ± 7 | 398 ± 55 | 158 ± 5 |
| MLS-STAT3E | 3.98 ± 0.1 | 2.25 ± 0.2 | 44 ± 4 | 74 ± 5 | 313 ± 33 | 166 ± 10 |
Mitochondrial Respiration Is Modestly Decreased in MLS-STAT3E Hearts
There was a marginally lower protein yield of heart mitochondria from MLS-STAT3E compared with WT (30.8 ± 0.5 versus 29.0 ± 0.5 mg of mitochondrial protein/g of wet heart tissue; p < 0.05, Student's t test; n = 11 for each group). DNA levels of three mitochondria-encoded genes (Nd2 from complex I, Cytb from complex III, and Cox1 from complex IV) were also examined as a reflection of mitochondrial content in hearts and livers (control) of transgenic and WT mice. No differences in mitochondrial DNA content were observed between MLS-STAT3E mice and WT (data not shown). Activity of citrate synthase, exclusively localized to mitochondrial matrix, was also similar in both MLS-STAT3 and WT mitochondria (Table 3).
TABLE 3.
Effect of ischemia on activity of electron transport chain complexes
Activities are expressed in nmol/min/mg, except for complex IV in 1/min/mg. Results are mean ± S.E., n = 6 for each group, except n = 4 for complex IV data.
| Enzymatic activity |
||||
|---|---|---|---|---|
| WT |
MLS-STAT3E |
|||
| TC | Ischemia | TC | Ischemia | |
| Complex I | 937 ± 56 | 669 ± 48a | 750 ± 45b | 649 ± 36 |
| NFR | 1904 ± 58 | 1749 ± 76 | 2103 ± 120 | 1897 ± 68 |
| Complex II | 248 ± 9 | 238 ± 16 | 197 ± 15b | 200 ± 11 |
| Complex III | 8471 ± 274 | 8500 ± 99 | 8392 ± 410 | 8167 ± 333 |
| Complex IV | 414 ± 16 | 299 ± 28a | 401 ± 21 | 291 ± 21a |
| CS | 2770 ± 41 | 2679 ± 47 | 2691 ± 162 | 2821 ± 62 |
a p < 0.05 Ischemia versus corresponding TC.
b p < 0.05 MLS-STAT3E versus corresponding WT.
To investigate if expression of STAT3 in cardiac mitochondria influenced oxidative function, oxidative phosphorylation was analyzed in mitochondria isolated from hearts of MLS-STAT3E and WT mice. Respiration was tightly coupled in both experimental groups with similar rates of state 4 respiration and RCR (defined as state 3/state 4) (Table 2). The maximal rates of oxidative phosphorylation measured after addition of saturating 2 mm ADP were modestly decreased in MLS-STAT3E hearts when glutamate + malate or succinate were used as substrates (Table 2). DNP-uncoupled respiration was reduced in MLS-STAT3E mitochondria to a similar extent (Table 2), localizing the defects in MLS-STAT3E mitochondria to the ETC. In contrast to glutamate + malate and succinate rates, TMPD/ascorbate-dependent respiration was similar in both MLS-STAT3E and WT hearts (Table 2). These data indicated that the site of the defect in MLS-STAT3E mitochondria was located in the ETC proximal to complex IV.
TABLE 2.
Ischemia affects mitochondrial oxidative phosphorylation
Results are mean ± S.E., n = 6 for each group.
| Respiration |
||||
|---|---|---|---|---|
| WT |
MLS-STAT3E |
|||
| TC | Ischemia | TC | Ischemia | |
| nAO/min/mg | ||||
| Glutamate + malate | ||||
| State 3 | 294 ± 12 | 197 ± 11a | 277 ± 6 | 217 ± 12a |
| State 4 | 46 ± 2 | 59 ± 3a | 42 ± 2 | 66 ± 5a |
| RCR | 6.30 ± 0.10 | 3.42 ± 0.16a | 6.90 ± 0.30 | 3.30 ± 0.40a |
| 2 mm ADP | 377 ± 13 | 231 ± 22a | 316 ± 14b | 260 ± 13a |
| DNP | 368 ± 13 | 224 ± 24a | 309 ± 14b | 264 ± 14 |
| Succinate | ||||
| State 3 | 516 ± 19 | 218 ± 13a | 456 ± 17b | 202 ± 18a |
| State 4 | 184 ± 7 | 139 ± 8a | 171 ± 11 | 143 ± 14 |
| RCR | 2.80 ± 0.10 | 1.56 ± 0.05a | 2.70 ± 0.10 | 1.40 ± 0.10a |
| 2 mm ADP | 542 ± 18 | 297 ± 21a | 467 ± 20b | 266 ± 21a |
| DNP | 512 ± 19 | 297 ± 18a | 444 ± 16b | 262 ± 16a |
| TMPD/ascorbate | ||||
| 2 mm ADP | 1478 ± 53 | 1042 ± 70a | 1469 ± 71 | 1028 ± 73a |
a p < 0.05 ischemia versus corresponding TC.
b p < 0.05 MLS-STAT3E versus corresponding WT.
Mitochondrial membrane potential (ΔΨ) is a key indicator of inner mitochondrial membrane intactness (32). The maximum membrane potential that mitochondria could generate (ΔΨmax), expressed as a difference between fully polarized mitochondria in the presence of 1 nm oligomycin (state 4oligo) and fully uncoupled mitochondria after addition of DNP (Ψ0), was similar in MLS-STAT3E mitochondria compared with WT (Fig. 1I). This result corroborated the similar state 4 respiratory rates and RCR values, indicating that the inner membrane was intact in both MLS-STAT3E and WT mitochondria.
Complex I and II Activities Are Decreased in MLS-STAT3E Mitochondria
Activities of complex I (NADH-ubiquinone oxidoreductase) and complex II (succinate-ubiquinone oxidoreductase) were decreased in MLS-STAT3E mitochondria compared with WT, whereas complex III (ubiquinol-cytochrome c oxidoreductase) and complex IV (cytochrome c oxidase) activities were unchanged (Table 3). These results localized the defects in MLS-STAT3E mitochondria to the major entry points of electron transport, i.e. complexes I and II. These data also corroborated the decrease in integrated respiration observed with complex I and II substrates (Table 2).
Despite the decreased activity of NADH-ubiquinone oxidoreductase, the activity of NADH-ferricyanide reductase, i.e. the proximal part of complex I containing flavin mononucleotide center (FMN) and responsible for NADH reduction, was unchanged in MLS-STAT3E mitochondria when compared with WT (NRF data in Table 3). These results localized the defect to the distal portion of complex I, i.e. the chain of iron-sulfur clusters or the quinone-binding site.
Because complex I and II activities were impaired in MLS-STAT3E mitochondria, the mRNA and protein expression of representative subunits of these complexes were tested: from complex I, two subunits of NADH dehydrogenase (NDUFV1 and NDUFV2), two subunits of the iron-sulfur protein fraction (NDUFS1 and NDUFS2), and two proteins responsible for proper assembly of the complex (NDUFAF1 and Grim-19 (33)); from complex II, two catalytic subunits (70-kDa SDHa and 30-kDa SDHb). mRNA and protein contents of these subunits were similar in MLS-STAT3E and WT mitochondria (data not shown). In addition, the abundance and mobility of single ETC complexes, and the composition of supercomplexes (“respirasomes”) (34), were unaltered in MLS-STAT3E (data not shown).
Complex I-dependent Respiration Is Protected against Ischemic Damage in MLS-STAT3E Hearts
Hearts isolated from MLS-STAT3E mice and WT littermates were subjected to 45 min of ex vivo-simulated stop flow ischemia (17) and ischemia-induced changes in mitochondria function, ETC activities, and ETC-driven ROS production were investigated.
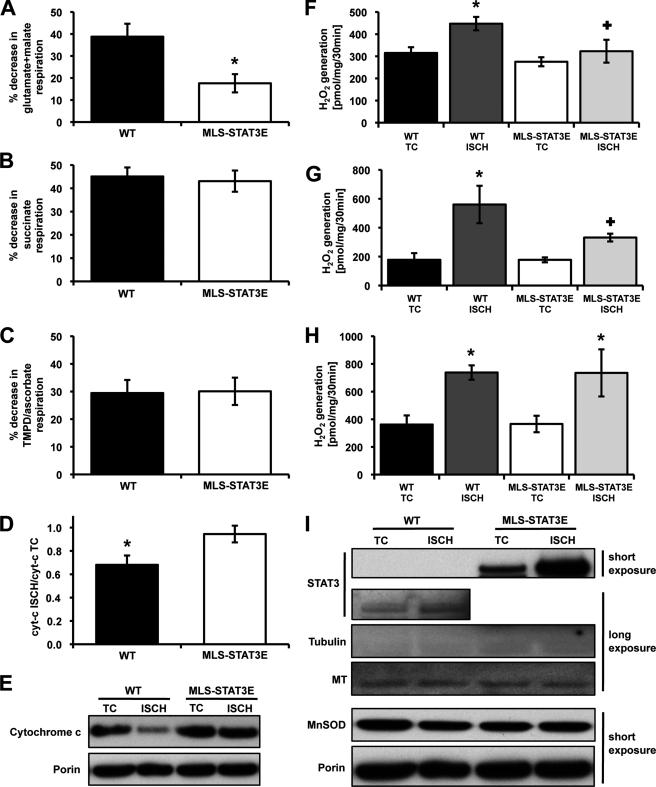
Ischemia induced a moderate decrease of 2 mm ADP-stimulated state 3 respiration using glutamate + malate as substrate in WT hearts (Fig. 2A and Table 2). In contrast, in mitochondria expressing MLS-STAT3E there was only a minimal decrease in glutamate + malate rates, indicating a protective role of MLS-STAT3E during ischemia. Furthermore, MLS-STAT3E and WT mitochondria demonstrated equally reduced succinate and TMPD/ascorbate rates (Fig. 2, B and C, and Table 2). Thus, MLS-STAT3E protected complex I-dependent respiration against ischemic damage.
FIGURE 2.
MLS-STAT3-mediated protection of glutamate + malate respiration, preserved cytochrome c retention, and attenuation of ROS production in mitochondria subjected to ischemia. A–C, WT and MLS-STAT3E mitochondria were isolated from hearts subjected to 45 min of ex vivo stop flow-simulated ischemia, and 2 mm ADP-stimulated state 3 respiration on glutamate + malate (A), succinate (B), and TMPD/ascorbate (C) was measured. Results are expressed as % decrease compared with time control rates set as 100%. The actual rates are shown in Table 2. Results are mean ± S.E., n = 6 for each group. *, p < 0.05 versus WT. D and E, equal amounts of mitochondrial extracts (10 μg) from ischemic (ISCH) and TC hearts from WT and MLS-STAT3E mice were resolved by SDS-PAGE, transferred to PVDF membrane, and immunoblotted against cytochrome c and porin (mitochondrial loading control). D, densitometry was measured using ImageJ (NIH) and expressed as ratio of cytochrome c signal in ischemia sample to TC sample, normalized to the ratio of porin signal from ischemia to TC. Results are mean ± S.E., n = 4 for each group. *, p < 0.05 versus MLS-STAT3E. E, one representative immunoblot for cytochrome c is shown. F–H, hearts of WT and MLS-STAT3E mice were subjected to 45 min of ex vivo stop flow-simulated ischemia. WT and MLS-STAT3E mitochondria from ischemia and TC hearts were subjected to H2O2 net release using HRP/Amplex Red protocol. F, mitochondria were incubated with glutamate + malate to measure ROS production dependent on electron flow through complex I and complex III. G, rotenone was used in addition to glutamate + malate to establish the maximal capacity of complex I to generate ROS. H, mitochondria were supplied with succinate and rotenone (to block reverse electron transfer into complex I) to measure complex III-dependent ROS production. Results are mean ± S.E., n = 4 for each group. *, p < 0.05 ischemia versus corresponding TC and +, p < 0.05 MLS-STAT3E versus corresponding WT. I, mitochondrial protein extracts (10 μg) from TC and ischemia hearts were resolved by SDS-PAGE and immunoblotted against STAT3, tubulin (cytosolic loading control), and porin (mitochondrial loading control). A representative blot from three independent experiments is shown.
Consistent with previously published data (24), ischemia increased uncoupling, indicated by augmented state 4 rates and decreased RCRs, to the same extent in both MLS-STAT3E and WT mitochondria (Table 2). DNP-uncoupled respiration localized the protective effect of MLS-STAT3E to the ETC.
MLS-STAT3E Expression Prevents Ischemia-induced Decrease in Complex I Activity and Cytochrome c Release
The initial site of ischemic damage to the ETC is complex I (22, 35) and with increasing duration of the insult, complexes III (36) and IV (37, 38) are affected. The enzymatic activities of complexes I–IV were measured in mitochondria from MLS-STAT3E and WT hearts subjected to ischemia. As depicted in Table 3, ischemia decreased the activity of complex I in mitochondria from WT hearts compared with TC. In contrast, complex I activity after ischemia was unchanged in mitochondria expressing MLS-STAT3E. Ischemia did not alter the activity of NADH dehydrogenase (NFR data in Table 3), in either WT or MLS-STAT3E hearts, consistent with previous findings showing the absence of ischemic damage to NFR (22). These results indicated that MLS-STAT3E expression prevented ischemia-induced damage to the distal part of complex I.
Complex II activity was not decreased by ischemia in mitochondria from WT or MLS-STAT3E hearts (Table 3). The basal enzymatic rates of complex II in TC samples from the MLS-STAT3E group were lower when compared with WT TC and did not decrease further due to the insult. Thus, the reduced rates of respiration with succinate after ischemia were likely a result of damage to the distal part of ETC. Results showing ubiquinone-cytochrome c oxidoreductase (complex III) activity unchanged by ischemia in both WT and MLS-STAT3E mitochondria (Table 3) indicated that the ischemia-induced blockade of ETC was localized to cytochrome c oxidase (complex IV). It has been shown that ischemic damage to TMPD/ascorbate respiration was related to cytochrome c release from mitochondria (23). Western blot analysis revealed over 30% reduced amounts of cytochrome c in WT mitochondria after ischemia (Fig. 2, D and E). In contrast, MLS-STAT3E expression blocked cytochrome c release from mitochondria during ischemia. Because cytochrome c content after ischemia was preserved in MLS-STAT3E mitochondria, the first-order enzymatic rates of complex IV were also measured. As shown in Table 3, ischemia equally decreased cytochrome c oxidase activity in both WT and MLS-STAT3E mitochondria, confirming the presence of ischemic damage to complex IV (38).
Ischemia Does Not Augment ROS Generation from Complex I in MLS-STAT3E Hearts
Ischemia enhances production of ROS from complexes I and III (22), the major mitochondrial sources of ROS (10). Mitochondria-driven ROS generation was measured as the net release of hydrogen peroxide (H2O2) from mitochondria isolated from ischemic and time control hearts. Basal production of H2O2 did not differ between MLS-STAT3E and WT mitochondria (compare TC groups in Fig. 2, F and H). Ischemia increased the release of H2O2 in WT mitochondria respiring on glutamate + malate (Fig. 2F). In contrast, no such effect was observed in mitochondria expressing MLS-STAT3E. The maximal capacity of complex I to produce H2O2 after ischemia, established by inhibiting complex I with rotenone and supplying glutamate + malate as a substrate, was decreased in MLS-STAT3E compared with WT mitochondria (Fig. 2G). However, mitochondria incubated with succinate and rotenone, to target complex III, generated equally higher amounts of H2O2 after ischemia in both WT and MLS-STAT3E (Fig. 2H).
It has been reported that the expression of MnSOD depended on the transcriptional activity of STAT3 (3). Constitutively active STAT3 was expressed in heart induced metallothionein-1 and -2 (Mt-1 and Mt-2) mRNA and protein levels, which in turn decreased ROS during ischemia and reperfusion (4). Therefore, the expression levels of these scavengers were analyzed. Neither the mRNA nor protein expression of MnSOD in cardiac mitochondria exhibited any differences between MLS-STAT3E and WT (Fig. 2I and data not shown). Ischemia did not alter MnSOD protein content in the mitochondria. These results indicated that up-regulation of MnSOD did not account for the decreases in ROS production from MLS-STAT3E mitochondria. Similarly, there was no increase in Mt-1 mRNA or protein levels in MLS-STAT3E hearts under both basal and ischemic conditions (Fig. 2I and data not shown). Thus, the observed inhibition of ROS generation in mitochondria with MLS-STAT3E was a result of decreased production rather than enhanced scavenging capability.
Ischemia Increases Mitochondrial STAT3
The Western blot analysis of the cardiac mitochondria subjected to ischemia showed an enhanced signal from STAT3 in the mitochondria when compared with time control hearts (Fig. 2I). This increase was likely to be a result of a de novo import of STAT3 into the mitochondria because other proteins examined (MnSOD, MT-1, Porin, complex I subunits: NDUFV1 and NDUFA19) exhibited no such increase (Fig. 2I and data not shown). This enrichment was much more apparent in MLS-STAT3E transgenic hearts compared with WT hearts. A similar trend was observed in WT samples; however, the increase in mitochondrial STAT3 was less apparent.
DISCUSSION
Using a variety of experimental approaches numerous reports have highlighted the importance of STAT3 in cardiac function (5). Mice and humans that express decreased amounts or lack expression of STAT3 in the heart develop premature cardiomyopathy and heart failure. Coincident with heart failure, STAT3−/− mice also have elevated levels of ROS. In contrast to a lack of STAT3 being detrimental in the heart, overexpression of STAT3 in the heart can also be injurious (7). These observations suggest that maintaining the “correct” concentration or activity of STAT3 is essential for its cardioprotective effects.
Our previous studies indicate that a small pool of STAT3 is located in the mitochondria where it modulates the activities of complexes I and II of the ETC (12). The activities of these complexes are decreased in cardiomyocytes from mice that did not express STAT3. The current studies were initiated to begin to determine the mechanisms by which mitochondrial-localized STAT3 modulates respiration. It should be emphasized that the experiments in this report were performed on hearts where the expression of endogenous STAT3 was intact. Experiments examining the role of mitochondrial-targeted STAT3 in the absence of endogenous protein address a different issue and the experimental design of such experiments must take into consideration the possible actions of the ablation of STAT3 on heart development as well as the multiple effects in fully developed adult animals. It is unlikely that STAT3 in the mitochondria is present in stoichiometric amounts to other subunits of complexes I or II (39). Rather, STAT3 likely acts as a dynamic modulator of the ETC through protein-protein interactions or post-translational modifications.
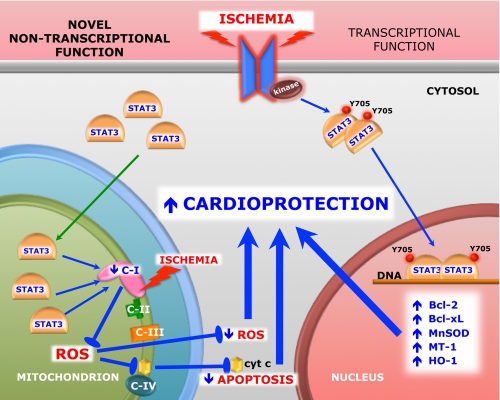
Given the caveat that the MLS-STAT3E mice used for these experiments overexpress the protein on a background of endogenous STAT3 expression, there are a number of important observations that suggest the targets for the actions of MLS-STAT3E are in the mitochondria and not the nucleus (see Fig. 3). 1) We observe very little if any of the MLS-STAT3E in isolated nuclei (Fig. 1E). 2) Although the mutant STAT3 used in these experiments has been reported to translocate to the nucleus (13), it does not bind DNA or activate transcription. 3) Metallothionein-1, which is increased in transgenic mice expressing a constitutively active form of STAT3 (4), is not altered in expression in MLS-STAT3E mice (Fig. 2I). We would expect this protein to be increased in MLS-STAT3E mice, as it is in mice that express constitutively active STAT3 if MLS-STAT3E acted as a transcription factor. 4) STAT3-dependent genes in both WT and MLS-STAT3E mice exposed to LIF or in hearts subjected to ischemia were equally induced (Fig. 1, F and G, and supplemental Fig. S1A). 5) In contrast to mice that overexpress a transcriptionally active STAT3 transgene in the heart and develop LV hypertrophy at 3 months (7), the MLS-STAT3E transgenic mice display normal cardiac function and the absence of ventricular hypertrophy at 12 months (Table 1). These observations are consistent with the notion that MLS-STAT3E functions in the mitochondria to protect complex I against ischemic damage (Fig. 2A and Table 3), to decrease ROS production from complex I (Fig. 2, F and G), and to block cytochrome c release into the cytosol (Fig. 2, D and E) during ischemia. 6) The effects of the MLS-STAT3E transgene are observed with 45 min of ischemia, which is probably not sufficient time for isolated hearts to transcribe and translate STAT3-activated RNAs for the protective effects of the transgene to be due to its actions in the nucleus. In previous reports, where it has been shown that STAT3 protects against ischemia/reperfusion injury (4, 40, 41), it is likely that the actions of STAT3 are at least partially a result of its activation of nuclear genes. These protocols subject the hearts to 2–3 h of combinations of ischemia and reperfusion, which is sufficient time for STAT3-mediated induction and translation of nuclear encoded RNAs. It should be emphasized that we recognize that STAT3-dependent gene expression does play a major role in the cardioprotective effects of STAT3. However, our results indicate that its actions in the mitochondria are also significant, especially during the very early phases of ischemic injury when STAT3-induced genes have not yet been translated.
FIGURE 3.
Model of the role of STAT3 in protection against cellular stress. During stress conditions, such as cardiac ischemia, STAT3 works both as a signaling molecule involved in regulation of cardioprotective gene expression and as a direct modulator of complex I of the mitochondrial electron transport chain. Jak, Janus kinase; Y705, tyrosine 705; C-I, -II, -III and -IV, respiratory complex I, II, III, and IV; cyt c, cytochrome c; Bcl-2, B-cell lymphoma-2; Bcl-xL, B-cell lymphoma-extra large.
Previous experiments in our lab indicated that expression of mitochondria-localized STAT3 containing a mutated DNA-binding domain restored respiration deficits observed in STAT3 null cells (12). Therefore, we expected that the expression of the same STAT3 construct in cardiac tissue, which already had endogenous STAT3, would not change the rates of oxidative phosphorylation using complex I- and II-dependent substrates. Surprisingly, heart mitochondria expressing MLS-STAT3E exhibited a small but significant decrease in the basal activities of complexes I and II (Table 3). At first glance our results appear to contradict those of Wegrzyn et al. (12) who demonstrated that in the absence of STAT3 there was an inhibition of complexes I and II activities. However, the complete absence of STAT3 in mitochondria results in a profound inhibition of complexes I and II (12) accompanied by a substantially decreased membrane potential (42), decreased ATP production (42), and enhanced ROS generation (6, 42). In contrast, overexpression of the MLS-STAT3E transgene does not increase basal ROS production (Fig. 2, F–H) and the ability to generate and maintain membrane potential in MLS-STAT3E mitochondria is preserved (Fig. 1I). It is not surprising, then, the hearts with mitochondrial-targeted STAT3 do not exhibit any signs of cardiomyopathy at 1 year of age (Table 1), in contrast to STAT3−/− mice (9).
In response to ischemia there is an increase in the amount of endogenous STAT3 as well as MLS-STAT3E in mitochondria (Fig. 2I). Ischemia also decreases complex I activity in mitochondria from WT hearts (Table 3) (22). In the MLS-STAT3E transgenic mice there is already an excess of MLS-STAT3E in the mitochondria and the basal complex I activity is suppressed even in the absence of ischemia. These results suggest that under basal conditions there is sufficient transgenic protein to maximally protect the heart from an ischemic insult. The fact that complex I is already suppressed in MLS-STAT3E hearts and provides protection against ischemic damage to mitochondria is likely a result of a STAT3-dependent partial blockade of electron flow through complex I. Consistent with this possibility, inhibitors of complex I, such as rotenone or amobarbital, protect mitochondria and heart tissue from ischemic injury (43–45). Studies are underway to examine the relationship between expression levels of STAT3 in the mitochondria and decreases in complex I and II activities with ischemia.
Results from these studies lead us to propose an initial model of how STAT3 regulates mitochondrial respiration. We postulate that during cell stress STAT3 is an anti-apoptotic factor that works both as a signaling molecule involved in regulation of gene expression (1, 3, 4) and as a direct immediate modulator of the mitochondrial electron transport chain (Fig. 3). STAT3-dependent partial blockade of electron flow through complex I results in lower ROS production in the mitochondria and decreased release of cytochrome c during ischemia and possibly other forms of cellular stress. MLS-STAT3E expression did not influence basal ROS production (Fig. 2, F–H).
The two tightly bound ubiquinones are a likely site of ischemic damage that results in augmented ROS production, probably from the iron-sulfur cluster N2, which is proximal to the Q-binding site within complex I (22). In line with this concept, the lack of the ischemia-mediated increase in ROS release from complex I, observed in MLS-STAT3E mitochondria, suggests that the site of STAT3 interaction with this complex may be located proximal to the N2 cluster. The fact that NFR activity, which measures complex I function proximal to the N2 cluster, is the same in WT and MLS-STAT3E hearts (Table 3) is consistent with the proposal of STAT3 functioning near or in this iron-sulfur cluster.
In WT hearts, ischemia-induced increase in mitochondrial STAT3 is either not sufficient in amount or not rapid enough to completely protect mitochondria. We speculate that overexpression of MLS-STAT3E augments the potential of STAT3 to protect mitochondria during stress. Future work is needed to develop approaches to enhance activation of STAT3 in the mitochondria during critical times of ischemia and reperfusion.
Supplementary Material
Acknowledgments
We thank Ying Hu and Elizabeth Bowler from the Virginia Commonwealth University Department of Internal Medicine for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA098924 (to A. C. L.) and 2PO1AG15885 (to E. J. L.) and a grant from the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (to E. J. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- ROS
- reactive oxygen species
- DNP
- 2,4-dinitrophenol
- ETC
- electron transport chain
- Hsp70i
- inducible heat shock protein 70
- LIF
- leukemia inhibitory factor
- LV
- left ventricle
- LVESD
- left ventricle end-systolic diameter
- LVEDD
- left ventricle end-diastolic diameter
- MLS
- mitochondria-localizing sequence
- MnSOD
- manganese superoxide dismutase
- MT
- metallothionein
- NFR
- NADH-ferricyanide reductase
- RCR
- respiratory control ratio
- SOCS
- suppressor of cytokine signaling
- TC
- time control
- TMPD
- N,N,N′,N′-tetramethyl-p-phenylenediamine
- qPCR
- quantitative PCR
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Levy D. E., Lee C. K. (2002) J. Clin. Invest. 109, 1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takeda K., Noguchi K., Shi W., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T., Akira S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3801–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Negoro S., Kunisada K., Fujio Y., Funamoto M., Darville M. I., Eizirik D. L., Osugi T., Izumi M., Oshima Y., Nakaoka Y., Hirota H., Kishimoto T., Yamauchi-Takihara K. (2001) Circulation 104, 979–981 [DOI] [PubMed] [Google Scholar]
- 4. Oshima Y., Fujio Y., Nakanishi T., Itoh N., Yamamoto Y., Negoro S., Tanaka K., Kishimoto T., Kawase I., Azuma J. (2005) Cardiovasc. Res. 65, 428–435 [DOI] [PubMed] [Google Scholar]
- 5. Hilfiker-Kleiner D., Hilfiker A., Drexler H. (2005) Pharmacol. Ther. 107, 131–137 [DOI] [PubMed] [Google Scholar]
- 6. Hilfiker-Kleiner D., Kaminski K., Podewski E., Bonda T., Schaefer A., Sliwa K., Forster O., Quint A., Landmesser U., Doerries C., Luchtefeld M., Poli V., Schneider M. D., Balligand J. L., Desjardins F., Ansari A., Struman I., Nguyen N. Q., Zschemisch N. H., Klein G., Heusch G., Schulz R., Hilfiker A., Drexler H. (2007) Cell 128, 589–600 [DOI] [PubMed] [Google Scholar]
- 7. Kunisada K., Negoro S., Tone E., Funamoto M., Osugi T., Yamada S., Okabe M., Kishimoto T., Yamauchi-Takihara K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 315–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiong Y., Liu X., Lee C. P., Chua B. H., Ho Y. S. (2006) Free Radic. Biol. Med. 41, 46–55 [DOI] [PubMed] [Google Scholar]
- 9. Jacoby J. J., Kalinowski A., Liu M. G., Zhang S. S., Gao Q., Chai G. X., Ji L., Iwamoto Y., Li E., Schneider M., Russell K. S., Fu X. Y. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12929–12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Turrens J. F. (2003) J. Physiol. 552, 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boengler K., Hilfiker-Kleiner D., Heusch G., Schulz R. (2010) Basic Res. Cardiol 105, 771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wegrzyn J., Potla R., Chwae Y. J., Sepuri N. B., Zhang Q., Koeck T., Derecka M., Szczepanek K., Szelag M., Gornicka A., Moh A., Moghaddas S., Chen Q., Bobbili S., Cichy J., Dulak J., Baker D. P., Wolfman A., Stuehr D., Hassan M. O., Fu X. Y., Avadhani N., Drake J. I., Fawcett P., Lesnefsky E. J., Larner A. C. (2009) Science 323, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horvath C. M., Wen Z., Darnell J. E., Jr. (1995) Genes Dev. 9, 984–994 [DOI] [PubMed] [Google Scholar]
- 14. Alonzi T., Maritano D., Gorgoni B., Rizzuto G., Libert C., Poli V. (2001) Mol. Cell. Biol. 21, 1621–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 16. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 17. Chen Q., Lesnefsky E. (2009) J. Investig. Med. 57, 558 [Google Scholar]
- 18. Salloum F. N., Abbate A., Das A., Houser J. E., Mudrick C. A., Qureshi I. Z., Hoke N. N., Roy S. K., Brown W. R., Prabhakar S., Kukreja R. C. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H1398–1406 [DOI] [PubMed] [Google Scholar]
- 19. Teichholz L. E., Kreulen T., Herman M. V., Gorlin R. (1976) Am. J. Cardiol. 37, 7–11 [DOI] [PubMed] [Google Scholar]
- 20. Devereux R. B., Alonso D. R., Lutas E. M., Gottlieb G. J., Campo E., Sachs I., Reichek N. (1986) Am. J. Cardiol. 57, 450–458 [DOI] [PubMed] [Google Scholar]
- 21. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 22. Chen Q., Moghaddas S., Hoppel C. L., Lesnefsky E. J. (2008) Am. J. Physiol. Cell Physiol. 294, C460–466 [DOI] [PubMed] [Google Scholar]
- 23. Lesnefsky E. J., Tandler B., Ye J., Slabe T. J., Turkaly J., Hoppel C. L. (1997) Am. J. Physiol. Heart Circ. Physiol. 273, H1544–1554 [DOI] [PubMed] [Google Scholar]
- 24. Chen Q., Vazquez E. J., Moghaddas S., Hoppel C. L., Lesnefsky E. J. (2003) J. Biol. Chem. 278, 36027–36031 [DOI] [PubMed] [Google Scholar]
- 25. Scaduto R. C., Jr., Grotyohann L. W. (1999) Biophys. J. 76, 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang L., Badgwell D. B., Bevers J. J., 3rd, Schlessinger K., Murray P. J., Levy D. E., Watowich S. S. (2006) Mol. Cell. Biochem. 288, 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang E., Lerner L., Besser D., Darnell J. E., Jr. (2003) J. Biol. Chem. 278, 15794–15799 [DOI] [PubMed] [Google Scholar]
- 28. Das D. K., Maulik N., Moraru I. I. (1995) J. Mol. Cell Cardiol. 27, 181–193 [DOI] [PubMed] [Google Scholar]
- 29. Hattori R., Maulik N., Otani H., Zhu L., Cordis G., Engelman R. M., Siddiqui M. A., Das D. K. (2001) J. Mol. Cell Cardiol. 33, 1929–1936 [DOI] [PubMed] [Google Scholar]
- 30. Darnell J. E., Jr. (1997) Science 277, 1630–1635 [DOI] [PubMed] [Google Scholar]
- 31. Booz G. W., Day J. N., Baker K. M. (2002) J. Mol. Cell Cardiol. 34, 1443–1453 [DOI] [PubMed] [Google Scholar]
- 32. Chen L. B. (1988) Annu. Rev. Cell Biol. 4, 155–181 [DOI] [PubMed] [Google Scholar]
- 33. Huang G., Lu H., Hao A., Ng D. C., Ponniah S., Guo K., Lufei C., Zeng Q., Cao X. (2004) Mol. Cell. Biol. 24, 8447–8456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schägger H., Pfeiffer K. (2000) EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rouslin W. (1983) Am. J. Physiol. Heart Circ. Physiol. 244, H743–748 [DOI] [PubMed] [Google Scholar]
- 36. Lesnefsky E. J., Gudz T. I., Migita C. T., Ikeda-Saito M., Hassan M. O., Turkaly P. J., Hoppel C. L. (2001) Arch. Biochem. Biophys. 385, 117–128 [DOI] [PubMed] [Google Scholar]
- 37. Lesnefsky E. J., Slabe T. J., Stoll M. S., Minkler P. E., Hoppel C. L. (2001) Am. J. Physiol. Heart Circ. Physiol. 280, H2770–2778 [DOI] [PubMed] [Google Scholar]
- 38. Prabu S. K., Anandatheerthavarada H. K., Raza H., Srinivasan S., Spear J. F., Avadhani N. G. (2006) J. Biol. Chem. 281, 2061–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Phillips D., Reilley M. J., Aponte A. M., Wang G., Boja E., Gucek M., Balaban R. S. (2010) J. Biol. Chem. 285, 23532–23536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Butler K. L., Huffman L. C., Koch S. E., Hahn H. S., Gwathmey J. K. (2006) Am. J. Physiol. Heart Circ. Physiol. 291, H797–803 [DOI] [PubMed] [Google Scholar]
- 41. Smith R. M., Suleman N., Lacerda L., Opie L. H., Akira S., Chien K. R., Sack M. N. (2004) Cardiovasc. Res. 63, 611–616 [DOI] [PubMed] [Google Scholar]
- 42. Sarafian T. A., Montes C., Imura T., Qi J., Coppola G., Geschwind D. H., Sofroniew M. V. (2010) PLoS One 5, e9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen Q., Moghaddas S., Hoppel C. L., Lesnefsky E. J. (2006) J. Pharmacol. Exp. Ther. 319, 1405–1412 [DOI] [PubMed] [Google Scholar]
- 44. Ambrosio G., Zweier J. L., Duilio C., Kuppusamy P., Santoro G., Elia P. P., Tritto I., Cirillo P., Condorelli M., Chiariello M. (1993) J. Biol. Chem. 268, 18532–18541 [PubMed] [Google Scholar]
- 45. Lesnefsky E. J., Chen Q., Moghaddas S., Hassan M. O., Tandler B., Hoppel C. L. (2004) J. Biol. Chem. 279, 47961–47967 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.