Abstract
The Mef2 family of transcription factors regulates muscle differentiation, but the specific gene programs controlled by each member remain unknown. Characterization of Mef2A knock-out mice has revealed severe myofibrillar defects in cardiac muscle indicating a requirement for Mef2A in cytoarchitectural integrity. Through comprehensive expression analysis of Mef2A-deficient hearts, we identified a cohort of dysregulated genes whose products localize to the peripheral Z-disc/costamere region. Many of these genes are essential for costamere integrity and function. Here we demonstrate that these genes are directly regulated by Mef2A, establishing a mechanism by which Mef2A controls the costamere. In an independent model system, acute knockdown of Mef2A in primary neonatal cardiomyocytes resulted in profound malformations of myofibrils and focal adhesions accompanied by adhesion-dependent programmed cell death. These findings indicate a role for Mef2A in cardiomyocyte survival through regulation of costamere integrity. Finally, bioinformatics analysis identified over-represented transcription factor-binding sites in this network of costamere promoters that may provide insight into the mechanism by which costamere genes are regulated by Mef2A. The global control of costamere gene expression adds another dimension by which this essential macromolecular complex may be regulated in health and disease.
Keywords: Cardiac Muscle, Differentiation, Gene Regulation, Mouse, Transcription
Introduction
The costamere is an elaborate macromolecular complex that is necessary for the transmission of contractile force throughout a striated muscle cell. Costameres connect the outermost myofibril, at each Z-disc, to the sarcolemma of a striated muscle cell and function to laterally transmit contractile forces throughout the myocyte (1). In muscle cells, costameres can be considered specialized focal adhesions because many focal adhesion proteins, such as vinculin, talin, and integrins, are localized to the costamere (2–4). The importance of the costamere is highlighted by the observation that mutations in proteins localized to this region can destabilize the costamere and result in cardio- and/or skeletal myopathies (2, 5). Although the costamere is essential for muscle function, the vast array of mechanisms that potentially regulate this structure have not been fully explored.
Previously we reported the cardiac defects in mice lacking the Mef2A transcription factor. Mef2A knock-out mice display increased mortality within the first week of birth, and hearts from these animals exhibit severe cardiac cytoarchitectural defects and right ventricular chamber dilation (6). This phenotype has also been observed in morpholino knockdown of Mef2A in zebrafish, which resulted in myofibrillar defects and impaired cardiac contractility (7). Additionally, a cardiac-specific knock-out of the costamere-localized focal adhesion kinase (FAK)2 resulted in myofibrillar abnormalities and was associated with the down-regulation of Mef2A expression (8).
To characterize the molecular mechanisms of the abnormal cardiac phenotype in Mef2A-deficient mice, microarray analysis was previously performed to identify differentially expressed genes (6). In addition to the dysregulation in structural, mitochondrial, and stress-responsive genes, novel genes represented a large percentage of differentially expressed genes in Mef2A knock-out hearts. Two of these novel genes, myospryn and Xirp2, were subsequently determined to be direct targets of Mef2A (9, 10). Additional characterization of myospryn and Xirp2 revealed localization of their protein products to the muscle costamere.
The identification of two Mef2A-dependent genes that encode costamere-localized proteins and the pronounced cytoarchitectural abnormalities in Mef2A knock-out hearts led us to hypothesize that the structural defect in Mef2A-deficient cardiac muscle is attributable to a perturbation of the costamere. Previously, we had not considered a deficiency in this critical subcellular compartment. Using a candidate gene approach, we found that nearly one-fourth of transcripts encoding costamere proteins were significantly down-regulated in Mef2A-deficient hearts. Subsequent characterization revealed that expression of these costamere genes is dependent on Mef2A activity and is regulated directly by this factor. To corroborate our findings in Mef2A-deficient mice, we developed an independent model system to acutely knock down the expression of Mef2A, using short hairpin RNAs, in neonatal rat ventricular myocytes (NRVMs). Acute knockdown of Mef2A in NRVMs resulted in severe myofibrillar disorganization and disruption of focal adhesions. In addition, these structural defects led to widespread cardiomyocyte detachment and reduced cell viability. These results have extended our previous observations by revealing that Mef2A is indispensable for costamere/focal adhesion integrity and survival of cardiac muscle cells.
EXPERIMENTAL PROCEDURES
Cell Culture
NRVMs were isolated from 1–2-day-old Sprague-Dawley pups as previously described (11). Myocytes were preplated for 2 h to reduce nonmyocyte contamination. Enriched NRVMs were cultured on 6-cm dishes or 22 × 22-mm coverslips coated with 0.1% gelatin at a density of 1.6 × 104 cells/10 mm2. Myocytes were maintained in DMEM (Invitrogen) supplemented with 0.5× Nutridoma (Roche Applied Science) and 1% penicillin, streptomycin, and l-glutamine, and the medium was changed daily. COS1 cells were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin, streptomycin, and l-glutamine.
Adenovirus
Specific shRNA oligonucleotides for Mef2A, Mef2C, and β-galactosidase were generated using the BLOCK-iT adenoviral RNAi expression system (Invitrogen), cloned into the pENTRTM/U6 RNAi entry vector for use in transient transfections, and finally recombined into the promoter-less pAd/BLOCK-iT-DEST expression vector to allow packaging of the shRNA into adenovirions. Viral particles were expanded in 293A cells, and viral titer was determined using the end point dilution assay (Clontech). Adenoviruses used to overexpress human MEF2A and β-galactosidase were generously provided by J. Molkentin (Children's Hospital, Cincinnati, OH) and K. Walsh (Boston University Medical School), respectively. NRVMs were transduced at a multiplicity of infection of 20 and cultured for 72 h after transduction unless otherwise noted.
Antibodies and Immunofluorescence
Rabbit polyclonal antibodies for Mef2A (C-21) and GAPDH (FL-335) from Santa Cruz Biotechnology were used in a 1:1,000 dilution for Western blot analysis. Mouse anti-FLAG (F3165; Sigma), anti-FHL2 (K0055-3; MBL), anti-TCAP (612328; BD Transduction Labs), and anti-cleaved-caspase-3 (9661; Cell Signaling) were used at a 1:1,000 dilution for Western blot analysis. Secondary antibodies (PerkinElmer Life Sciences) for Western blot analysis were as follows: anti-rabbit (NEFB12001EA) and anti-mouse (NEFB22001EA) IgG (goat) HRP. Chemiluminescent detection was performed with the Western Lightening Plus-ECL detection kit (NEL103001EA; PerkinElmer Life Sciences). Antibodies specific for sarcomeric α-actinin (monoclonal, Sigma), vinculin (V4139, Sigma), FHL2, and TCAP were used for immunostaining at a 1:250 dilution. Texas Red anti-mouse (Vector Labs) and anti-mouse Alexa Fluor 568 (Invitrogen) secondary antibodies were used at a 1:5,000 and 1:200 dilution, respectively. Immunofluorescence was imaged using an Olympus spinning disk confocal microscope.
Quantitative RT-PCR
Total RNA was isolated from neonatal mouse hearts and neonatal rat cardiomyocytes by homogenization in TRIzol (Invitrogen) following the manufacturer's instructions. cDNA was synthesized from 2 μg of total RNA using reverse transcriptase (Moloney murine leukemia virus) with random hexamers according to the manufacturer's instructions (Promega). cDNA was diluted 1:2 with distilled H2O, and 2 μl was subjected to quantitative real time PCR (qRT-PCR) using the Applied Biosystems Power SYBR-Green reagent and the 7900HT Detection System (Applied Biosystems). Fold changes (FC) between samples were calculated using the equation FC = 2−ΔΔCT, where the CT values were calculated as the cycle number where the PCR was still within the linear range, by SDS 2.2.2 software (Applied Biosystems). Primers sequences are available upon request.
Cell Titer Blue Assay
NRVM cell viability was assessed with the Cell Titer Blue assay (Promega), according to the manufacturer's instructions. NRVMs were transduced with shM2A or shLacZ adenovirus at a multiplicity of infection of 20 and cultured for 3 days prior to analysis. Resazurin reduction of viable cells was measured by fluorescence, 22 h after application of cell titer blue dye, using the PerkinElmer Life Sciences Victor III fluorescent plate reader.
Gel Shift Analysis
Mef2A cDNA was in vitro translated using rabbit reticulocyte lysate according to the manufacturer's instructions (Promega). 10 μl of gel shift reaction was as follows: 1 μl of probe (50,000 cpm), 2 μl of in vitro translated protein, 2 μl of 5× gel shift buffer (125 mm HEPES, pH 7.6, 500 mm NaCl, 75% glycerol, 0.5% IGEPAL, 1× protease inhibitors (Roche Applied Science), 1 μl of unlabeled probe (for competitions), and 4–5 μl of H2O. Reactions were incubated for 10 min at room temperature. Double-stranded oligonucleotide probes harboring the core Mef2 site plus flanking sequence on either side were labeled using [32P]dCTP (Invitrogen) with Klenow. Mutant Mef2-binding sites were generated by changing the core Mef2-binding site nucleotides from four A/T nucleotides to four G nucleotides. Gel shift reactions were fractionated on a 5% nondenaturing polyacrylamide gel, dried, and exposed to a phosphorus imaging screen (Amersham Biosciences).
Chromatin Immunoprecipitation
NRVMs were transfected with 10 μg of FLAG-tagged MEF2A expression vector (6:1 of PEI transfection reagent to cDNA). NRVMs were fixed with 37% paraformaldehyde to cross-link the proteins bound to DNA for 10 min. This reaction was stopped with 380 μl of 2.5 m glycine. The cell pellets were resuspended in 800 μl of 1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1, with 1× protease inhibitors (Roche Applied Science) and sonicated to between 200 and 1,000 bases in size (25% amplitude, 10 s, four times). Chromatin was then diluted 10-fold in 0.001% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, 167 mm NaCl with 1× protease inhibitors. Ten micrograms of antibodies (MEF2 and IgG control) were used for precipitation, and antibody/chromatin precipitates were recovered with 80 μl of protein A-agarose bead 50% slurry (Upstate/Millipore). DNA isolated from the precipitated chromatin was then subjected to qRT-PCR using the Applied Biosystems Power SYBR-Green reagent and the 7900HT Detection System (Applied Biosystems). Primer sequences are available upon request.
Promoter Cloning
Costamere promoter sequences, between 2 and 3 kb, were amplified from mouse genomic DNA using promoter specific primers and high fidelity Taq polymerase (Expand Long Template Taq; Roche Applied Science). PCR products were cloned into the pGL3-Basic luciferase vector (Invitrogen). Promoters containing mutant Mef2 sites were generated by rolling circle PCR using primers containing the Mef2-binding site mutations as described for EMSA. PCR was performed using high fidelity Taq (Roche Applied Science) and the wild type promoter construct as the template DNA.
Luciferase Analysis
COS1 cells were seeded onto 24-well dishes, and each well was transfected with 150 ng of each construct (expression vector, costamere promoter in the pGL3-Basic Luciferase vector, and CMV-β-Galactosidase expression vector) using Mirus TransIT-LT1 transfection reagent at a 3:1 ratio of transfection reagent to DNA. Transfection efficiency was determined using a β-galactosidase assay with ortho-Nitrophenyl-β-galactoside as a substrate. Luciferase units were normalized to β-galactosidase activity.
Statistical Analysis
All of the numerical quantification is representative of the mean ± S.E. of at least three independently performed experiments. Student's t test was used to determine statistical significance between the two populations of data. p values of ≤0.05 were considered to be statistically significant.
RESULTS
Dysregulated Costamere Gene Expression in Mef2A-deficient Perinatal Hearts
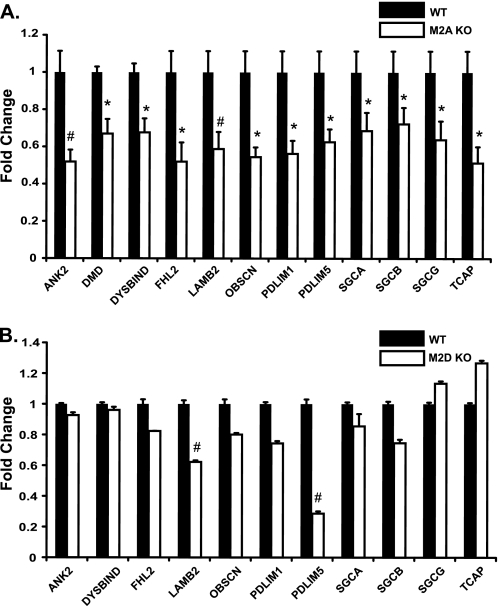
Previously, we demonstrated that Mef2A is required for myofibrillar integrity in cardiac muscle (6) and that this transcription factor directly regulates the expression of two genes, myospryn and Xirp2, whose protein products localize to the costamere (9, 10). Because the cytoskeletal abnormalities in Mef2A knock-out hearts have been incompletely characterized, we sought to further investigate the mechanisms of the structural phenotype in these mutant mice. Defects in striated muscle cytoarchitecture can originate from mutations in one or more of its various structural components. Given the aforementioned observations, we asked whether the cytoarchitectural abnormalities in Mef2A-deficient cardiac muscle may stem from impaired costameres as a result of dysregulated costamere gene expression. Toward this end, we analyzed the expression of 55 transcripts encoding known costamere-localized proteins in Mef2A-deficient mouse hearts by quantitative RT-PCR (Table 1). Through this comprehensive analysis we found 12 genes, in addition to myospryn and Xirp2, that were significantly (p ≤ 0.05) down-regulated 30% or more in knock-out hearts relative to wild type (Fig. 1A). The vast majority of these genes encode structurally unrelated proteins that play essential roles in the costamere. We also examined the expression of the atrial natriuretic factor (ANF) gene, an established marker of cardiac dysfunction that is often up-regulated in various types of cardiac disease (12, 13). The ANF gene was not up-regulated (Table 1), suggesting that this group of mutant mice had not yet progressed to heart failure as previously described (6).
TABLE 1.
List of transcripts encoding costamere proteins in Mef2A knockout hearts
A total of 55 known costamere-localized genes were analyzed by qRT-PCR. Gene names are listed according to NCBI gene nomenclature. Fold change is shown for each gene in KO relative to wild type normalized to 1. The 12 genes down-regulated by at least 30% relative to wild type and with a p value <0.05 are indicated in bold type.
| Gene | KO |
|---|---|
| AACTININ | 1.09 |
| AFIL | 1.11 |
| ALP | 0.92 |
| ANF | 1.09 |
| ANK2 | 0.52 |
| ANKRD1 | 0.84 |
| BSPECT | 1.06 |
| CALP3 | 0.60 |
| CAPZA1 | 0.73 |
| CAV3 | 0.88 |
| CYPHER | 0.90 |
| DAG-1 | 0.85 |
| DESMIN | 1.12 |
| DMD | 0.67 |
| DMN | 0.99 |
| DTNA | 1.00 |
| DYSBIND | 0.68 |
| FHL2 | 0.52 |
| G-FILAMIN | 1.14 |
| HIC-5 | 0.98 |
| IGF-1 | 0.86 |
| ILK | 0.79 |
| ITGB1 | 1.05 |
| LAMB2 | 0.59 |
| LIMS1 | 0.79 |
| MELUSIN | 0.87 |
| MLP | 0.95 |
| MYOZ1 | 0.78 |
| MYOZ2 | 0.80 |
| MYOZ3 | 1.29 |
| MYPN | 0.98 |
| NEBULIN | 0.92 |
| NNOS | 0.72 |
| OBSCN | 0.54 |
| PARB | 0.89 |
| PAXILLIN | 0.84 |
| PDLIM1 | 0.56 |
| PDLIM5 | 0.63 |
| PDLIM7 | 0.87 |
| PKP2 | 1.17 |
| PLECTIN | 0.97 |
| PTK2 | 0.99 |
| SGCA | 0.69 |
| SGCB | 0.72 |
| SGCD | 0.91 |
| SGCG | 0.64 |
| SNTA | 1.03 |
| SSPN | 0.96 |
| SYNC | 0.85 |
| TALIN | 0.81 |
| TCAP | 0.51 |
| TITIN | 0.74 |
| TTID | 1.14 |
| UTRN | 1.01 |
| VINCULIN | 0.87 |
| ZYXIN | 1.20 |
FIGURE 1.
Global down-regulation of transcripts encoding costamere proteins in perinatal Mef2A-deficient hearts. A, qRT-PCR analysis of 55 costamere transcripts in perinatal day 5 wild type and Mef2A knock-out hearts. cDNA generated from total RNA was pooled from five mice of each genotype and subjected to qRT-PCR. Twelve costamere transcripts were significantly reduced by 30% or more in Mef2A knock-out hearts: ANK2 (ankyrin 2), DMD (dystrophin), DYSB (dysbindin), FHL2 (four-and-a-half LIM domain 2), LAMB2 (laminin β2), OBSCN (obscurin), PDLIM1 (PDZ LIM domain 1), PDLIM5 (PDZ LIM domain 5), SGCA (sarcoglycan α), SGCB (sarcoglycan β), SGCG (sarcoglycan γ), and TCAP (titin-cap/telethonin). GAPDH was used as an internal control. B, costamere transcript analysis of cDNA from adult Mef2D knock-out hearts (n = 3) shows that only two of the twelve costamere target genes are significantly down-regulated. *, p ≤ 0.05; #, p ≤ 0.008 versus wild type mice.
This is the first demonstration that a cohort of costamere genes has been shown to be dependent on a Mef2 transcription factor. The dysregulation of these genes in Mef2C or Mef2D knock-out mice has not been reported, suggesting a unique function of Mef2A not shared by other family members. To further investigate this possibility, we obtained RNA from Mef2D knock-out mouse hearts (14) and analyzed the expression of the dysregulated costamere genes identified above. Only two of these genes were significantly down-regulated in Mef2D knock-out hearts (Fig. 1B), indicating that expression of a subnetwork of costamere genes is highly dependent on Mef2A activity.
Generation of Mef2A-specific Short Hairpin RNA Adenoviruses
To establish a model system that would allow us to further characterize the role of Mef2A in the regulation of the costamere, we knocked down the expression of Mef2A in primary cultures of NRVMs using an RNA interference approach. We developed a shRNA sequence targeting Mef2A (shM2A) (supplemental Fig. S1A), which was then packaged into replication-defective adenovirus to maximize the delivery of these molecules in NRVMs. In addition, we developed adenovirus expressing a β-galactosidase shRNA sequence (shLacZ) for use as a negative control. The efficacy and specificity of shM2A was determined in COS cells transiently transfected with various epitope-tagged Mef2 expression vectors (supplemental Fig. S1B). Subsequently, whole cell lysates were prepared from NRVMs transduced with either shLacZ or shM2A adenovirus and used for Western blot analysis to determine the extent of Mef2A knockdown. Western blot analysis revealed that shM2A substantially reduced Mef2A expression (supplemental Fig. S1C).
Cardiomyocyte Structural Integrity Is Dependent on Mef2A
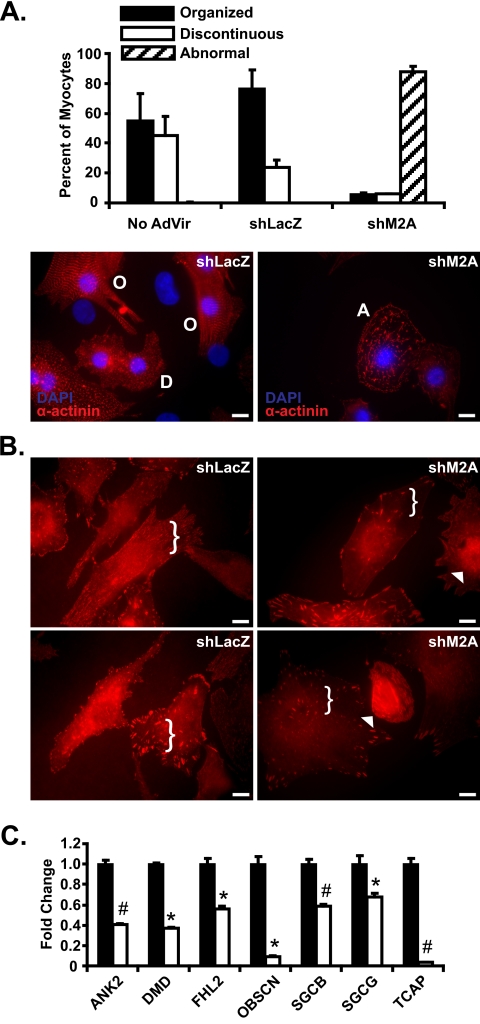
Upon determining the efficacy of shM2A adenovirus, we transduced NRVMs and examined the cytoarchitecture of shM2A NRVMs in greater detail. Primary NRVMs transduced with either LacZ or Mef2A shRNA adenovirus were stained with antibodies against α-actinin, which enabled us to identify sarcomeric structures. In shLacZ control NRVMs, we noted two distinct patterns of α-actinin immunoreactivity that were highly reproducible. As shown in Fig. 2A, nearly 75% of cardiomyocytes contained myofibrils that displayed uniform, periodic α-actinin striations arranged in a longitudinal fashion (left panel; organized, O). The remaining 25% of cardiomyocytes also exhibited abundant α-actinin staining, but the striated pattern appeared discontinuous and less well defined (Fig. 2A, bottom left panel, D). In striking contrast, cardiomyocytes transduced with Mef2A shRNA adenoviruses resulted in a greater than 90% reduction in both organized and discontinuous α-actinin staining pattern. Nearly all Mef2A-deficient cardiomyocytes displayed an abnormal structural phenotype characterized by cells devoid of well defined striations and highly irregular, fibrous α-actinin staining (Fig. 2A, bottom right panel).
FIGURE 2.
RNA interference-mediated knockdown of Mef2A in neonatal cardiomyocytes results in structural and gene expression abnormalities. A, adenoviruses harboring either Mef2A short hairpin RNA (shM2A) or lacZ shRNA (shLacZ) were transduced into NRVM cultures at a multiplicity of infection of 20 and analyzed at 72 h post transduction. Left panel, immunofluorescence analysis with α-actinin antibodies in shLacZ transduced NRVMs revealed cells with either a highly organized (O) or discontinuous (D) α-actinin staining pattern. Right panel, shM2A cultures had a dramatic loss of the characteristic organized (O) and discontinuous (D) α-actinin stain with an increase in abnormal cells with fibrous, linear α-actinin stain (A). B, immunofluorescence analysis reveals mislocalization of vinculin in Mef2A knockdown cardiomyocytes. A reduction in the amount of punctate immunoreactivity is observed throughout the cell (brackets) and accompanied by an increase in perinuclear vinculin staining (arrowheads). C, costamere transcript analysis in shM2A NRVMs. qRT-PCR reveals a significant down-regulation in seven of the 12 costamere target genes. Total RNA was collected from six individual experiments and was performed in triplicate. *, p ≤ 0.05; #, p ≤ 0.008 versus shLacZ transduced cultures. Images for immunofluorescence experiments are representative of multiple experiments. Five hundred myocytes were counted for each transduced group (n = 3). Scale bars, 10 μm.
In conjunction with the α-actinin immunofluorescence analysis, we stained transduced cardiomyocytes with vinculin antibodies as a representative costamere/focal adhesion marker. Focal adhesions are normally detected as robust, punctate immunoreactivity throughout the cardiomyocyte cytoplasm and concentrated foci along the cell membrane (Fig. 2B, left panels). Consistent with our hypothesis that Mef2A is an essential regulator of the costamere, we found a pronounced reduction in the density of vinculin immunoreactivity in most shM2A cardiomyocytes compared with shLacZ transduced cells (Fig. 2B, brackets). Moreover, we noted a reproducible increase in perinuclear/nuclear vinculin immunoreactivity in many shM2A NRVMs (Fig. 2B, arrowheads), indicating displacement of this protein from focal adhesion sites to the nucleus. The nuclear accumulation of focal adhesion proteins is known to occur under various physiological and stress conditions (15). Neonatal myocytes were also transduced with Mef2C-specific shRNA (shM2C) adenovirus at various multiplicities of infections, and in all cases this knockdown did not perturb vinculin localization (supplemental Fig. S2, A and B).
To determine whether the cytoarchitectural defects in this model system corresponded with the dysregulation of costamere target genes, we examined the expression of the 12 down-regulated costamere genes described in Fig. 1A. Quantitative expression analysis revealed that seven of these target genes were also down-regulated in shM2A NRVMs (Fig. 2C). Furthermore, qRT-PCR analysis of this same gene set in shM2C NRVMs failed to reveal a significant dysregulation of costamere gene expression (supplemental Fig. S2C). Taken together, these data highlight an important function for Mef2A in regulating the integrity of the cardiomyocyte costamere, a role not previously recognized in Mef2A knock-out hearts.
Mef2A Is Required for Cardiomyocyte Cell Survival in Vitro
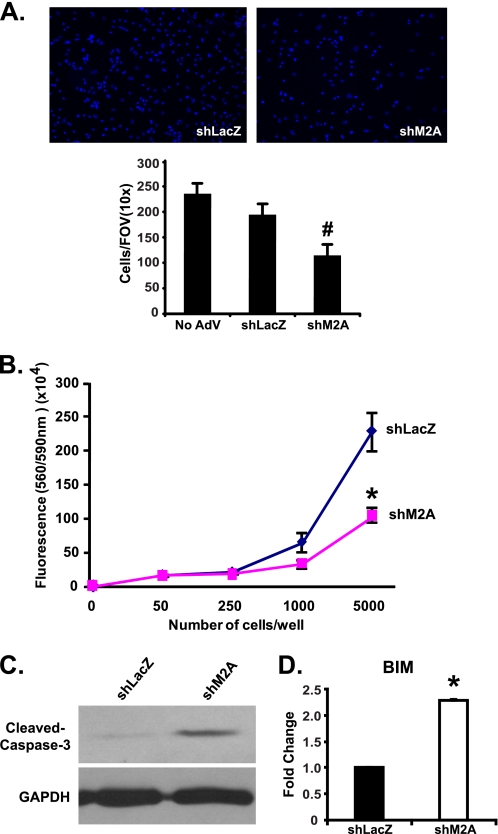
Neonatal cardiomyocytes transduced with shM2A also exhibited dramatic cell loss within 48 h compared with primary cultures transduced with shLacZ adenovirus (Fig. 3A). Quantification of the remaining attached NRVMs revealed a significant 44% decrease in the number of nuclei in shM2A-treated cells compared with shLacZ controls (Fig. 3A). To determine whether the reduction in nuclei is the result of decreased cell survival, we performed a CellTiter-Blue assay. As expected, the results of this assay revealed a significant reduction in cell viability in shM2A-transduced cells (Fig. 3B).
FIGURE 3.
Mef2A knockdown in neonatal cardiomyocytes decreases cell viability. A, upper panels, representative DAPI-stained images from each knockdown culture: shLacZ (left panel) and shM2A (right panel). Lower panel, quantification of nuclei in transduced NRVMs. DAPI-positive nuclei counted from 10 distinct areas (10× magnification) in transduced cultures from two experiments revealed a 44% reduction in nuclei number in the shM2A cultures (#, p ≤ 0.008). B, CellTiter-Blue assay of cell viability. Cell survival is significantly decreased in shM2A cultures (*, p ≤ 0.05). C, Western blot analysis of cleaved caspase-3 expression showed an increase in caspase-3 in shM2A cultures. GAPDH is shown as a loading control. D, qRT-PCR expression analysis of BIM in shM2A transduced NRVMs. BIM expression is significantly stimulated 2.3-fold (*, p ≤ 0.012) in shM2A cultures.
To support the above observations, we examined the expression of cleaved caspase-3, a marker of apoptosis. Cleaved caspase-3 expression was up-regulated in shM2A-transduced NRVMs (Fig. 3C). Furthermore, given the dysregulation of genes encoding proteins at the costamere, we reasoned that impaired integrity of focal adhesions may lead to the induction of apoptotic markers that respond to perturbations in this structure. It is established that a loss of cell anchorage can lead to a programmed cell death pathway known as anoikis (16, 17). As a marker for this process, we examined the expression of BIM, a pro-apoptotic BCL2 family member, which is activated in response to loss of cell adhesion (18, 19). The expression of BIM was significantly up-regulated (2.3-fold) in shM2A-transduced cultures, indicating activation of the anoikis apoptotic pathway (Fig. 3D). These data suggest that Mef2A knockdown in NRVMs decreases cell viability through impaired connections at focal adhesions.
Mislocalization and Reduction in Costamere Proteins in Mef2A-deficient Cardiac Myocytes
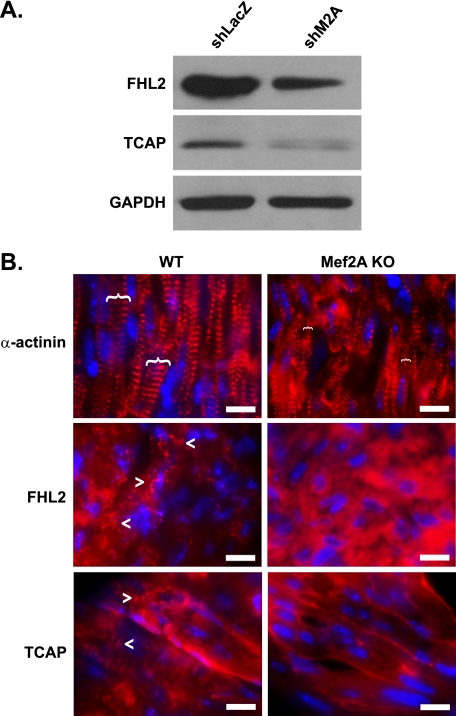
To determine whether the dysregulation of costamere genes is physiologically relevant, we examined the expression of selected costamere proteins in Mef2A-deficient cardiomyocytes. By Western blot, FHL2 (four-and-a-half LIM domain 2) and TCAP (titin-cap) protein levels were dramatically reduced in shM2A NRVM cultures compared with shLacZ controls (Fig. 4A). In vivo, immunofluorescence analysis of perinatal day 2 heart cryosections with FHL2 and TCAP antibodies revealed a loss of punctate staining in Mef2A-deficient cardiac muscle compared with wild type hearts (Fig. 4B, lower panels). Furthermore, the pattern of α-actinin immunostaining in Mef2A knock-out cardiomyocytes revealed disorganized striations with qualitatively narrower Z-discs (Fig. 4B, upper panels, brackets). Together, these data suggest that dysregulated costamere gene expression in Mef2A-deficient cardiac muscle is associated with altered costamere protein expression and structure.
FIGURE 4.
Mef2A-deficient cardiomyocytes display costamere disruption. A, Western blot analysis of costamere gene expression in NRVMs. The expression of FHL2 and TCAP proteins is decreased in shM2A-transduced cultures. GAPDH is shown as a loading control. B, immunofluorescence of α-actinin, FHL2, and TCAP in perinatal day 2 WT and mutant (Mef2A KO) hearts. Top panels, relative width of Z-discs, as determined by α-actinin, are indicated by brackets. Middle and bottom panels, WT hearts display a punctate FHL2 and TCAP staining pattern (arrowheads) that is absent in Mef2A KO hearts. Note that FHL2 and TCAP only display the characteristic, striated pattern in adult skeletal muscle (see supplemental Fig. S3). Scale bars, 10 μm.
Characterization of Mef2-binding Sites in Costamere Promoters
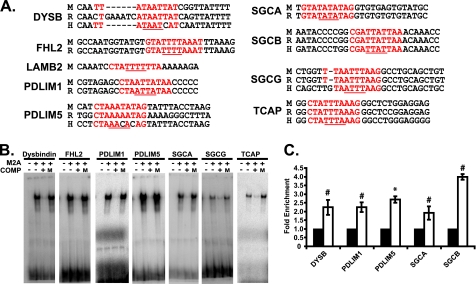
The above results prompted us to investigate whether Mef2A directly regulates the expression of this costamere gene set. For these studies we identified and analyzed nine of twelve costamere promoter sequences using the Ensembl genome browser. The three promoters that were not used for this analysis belong to the ankyrin 2 (ANK2), obcurin (OBSCN), and dystrophin (DMD) genes. The transcription start sites have not been determined for the ANK2 and OBSCN genes, and the dystrophin gene was not included because a Mef2 site was previously characterized in this promoter (20). To identify potentially important regulatory regions, we next selected 5,000 kilobases upstream of the predicted transcription start site of the nine costamere gene promoters and compared these mouse sequences to the orthologous rat and human promoters using BLAST analysis for regions of conservation. In parallel, the nine costamere promoters were analyzed for the presence of Mef2-binding sites (CTA(A/T)4TAG) using the TRED position weight matrix algorithm. Because more than one Mef2 site was found in each costamere promoter with varying position weight matrix scores, we focused our characterization on a single Mef2 site in these promoters by selecting a Mef2 site with a high position weight matrix score within a region of conservation. These Mef2 sites are indicated in Fig. 5A.
FIGURE 5.
Mef2A binds candidate Mef2 sites in costamere target gene promoters. A, conservation of Mef2 sites, from top to bottom, mouse (M), rat (R), human (H). The position of the Mef2 site relative to predicted transcription start site in the mouse gene is: DYSB (−4887), FHL2 (−1932), LAMB2 (−2767), PDLIM (−627), PDLIM5 (−304), SGCA (−497), SGCB (−3602), SGCG (−838), and TCAP (−30). Mef2 sites are highlighted in red. A/T core sequences are underlined in red. B, EMSA analysis of candidate Mef2 sites. The reactions were loaded as follows: radioactively labeled oligonucleotides with unprogrammed lysate (first lane of each gel from the left) or Mef2A protein (second lane). Unlabeled oligonucleotides containing WT (third lane) but not mutant (fourth lane) sites were able to effectively compete for binding of the labeled sites at a 50-fold molar excess. C, ChIP analysis was performed with a negative control antibody (IgG) or a Mef2A polyclonal antibody. Chromatin was subjected to quantitative PCR with primers designed to amplify ∼100 bp of promoter sequence containing the Mef2 site. Mef2A was significantly enriched on the DYSB, PDLIM1, PDLIM5, SGCA, and SGCB promoters. *, p ≤ 0.05; #, p ≤ 0.008 versus αIgG PCR values.
To determine whether the above Mef2 sites could bind Mef2A, we performed an EMSA with in vitro translated Mef2A protein and 32P-end-labeled oligonucleotides harboring the Mef2-binding site. As shown in Fig. 5B, seven of the nine Mef2-binding sites were capable of binding Mef2A. Although the Mef2 sites for LAMB2 and sarcoglycan beta (SGCB) displayed similarity to the consensus sequence and, in the case of SGCB, showed conservation, the inability to be bound by Mef2A may indicate additional constraints influenced by sequences flanking the core Mef2 site (21). We next performed competition assays using unlabeled, oligonucleotides harboring wild type and mutant ((A/T)4 core mutated to GGGG) Mef2 sites. Unlabeled, wild type oligonucleotides for each Mef2 site effectively competed with their radiolabeled Mef2 counterparts (Fig. 5B). By contrast, unlabeled, mutant Mef2 (M) oligonucleotides were unable to compete with the radiolabeled Mef2 probes (Fig. 5B). These data indicate that this cohort of costamere genes harbors bona fide Mef2 sites.
To determine whether Mef2A is associated with costamere promoters, harboring Mef2 sites, in vivo, we performed ChIP analysis on chromatin isolated from NRVMs. Sonicated, cross-linked chromatin from NRVMs was incubated with a Mef2A antibody (Santa Cruz Biotechnology). Precipitated chromatin samples were then subjected to quantitative RT-PCR using primer sets flanking the conserved Mef2 sites. As shown in Fig. 5C, quantitative PCR analysis revealed that Mef2A was significantly enriched in five of the twelve costamere promoters (Dysbindin (DYSB), PDLIM1, PDLIM5, SGCA and SGCB). Of the seven remaining costamere genes, we were unable to identify the promoter regions for ANK2 and OBSCN, and multiple primer designs for FHL2 and TCAP failed to yield amplification products from genomic DNA. Moreover, the LAMB2 and SGCG genes were subjected to ChIP, but Mef2A antibodies were unable to precipitate a Mef2-binding complex. The dystrophin gene (DMD) was not subjected to this analysis.
Mef2A Activates Costamere Promoters in Vitro and in Vivo
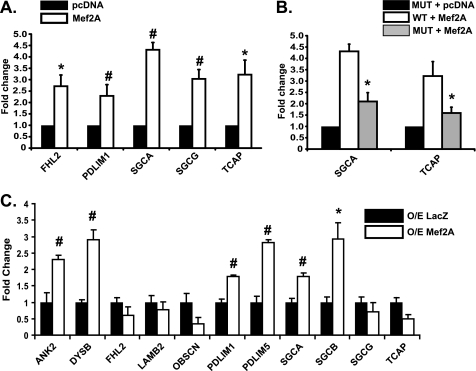
To determine whether Mef2A activates costamere genes in a heterologous system, we attempted to clone the proximal promoter region from all 12 dysregulated costamere genes into a reporter construct for use in transient transfection assays. We were able to successfully clone the upstream regions of five costamere genes: FHL2, PDLIM1, SGCA, SGCG, and TCAP, into pGL3-Basic (Invitrogen). These constructs were subsequently co-transfected along with Mef2A in COS cells. As shown in Fig. 6A, these promoters were significantly stimulated in the presence of Mef2A. To demonstrate that Mef2A is activating these promoters by binding to functional Mef2 sites, we attempted to generate point mutations in the core Mef2 site, as described in our gel shift assays, in all five cloned promoters. Mef2-binding site mutations were successfully introduced into the SGCA and TCAP promoters. Mutation of these Mef2 sites resulted in a significant decrease in transcriptional activity (Fig. 6B).
FIGURE 6.
Mef2A activates costamere target gene expression. A, luciferase reporter assays of cloned costamere promoters display significant activation with transiently transfected Mef2A in COS1 cells. Fold change was calculated using normalized luciferase readings. B, functional Mef2 sites in costamere promoters. Mutant Mef2 sites in costamere promoters reduces the ability of Mef2A to activate reporter in cells co-transfected with Mef2A. C, analysis of costamere gene expression in NRVMs overexpressing Mef2A. Total RNA was harvested from four NRVM cultures transduced with LacZ (O/E LacZ, black bars) or Mef2A (O/E Mef2A, white bars) adenovirus for overexpression. Total RNA was harvested from NRVMs 72 h after transduction and subjected to qRT-PCR analysis. A subset of costamere target genes (ANKB, DYSB, PDLIM1, PDLIM5, SGCA, and SGCB) are activated with acute expression of Mef2A (n = 4). *, p ≤ 0.05; #, p ≤ 0.008.
We next wanted to determine whether Mef2A could activate transcription of costamere genes in vivo. NRVMs were transduced with adenoviruses harboring either Mef2A or β-galactosidase (LacZ) cDNAs. Overexpression of Mef2A resulted in a significant activation of the endogenous ANK2, DYSB, PDLIM1, PDLIM5, SGCA, and SGCB genes (Fig. 6C). Interestingly, induction of five of the above six genes corresponded with the ability of these regions to be precipitated by Mef2 antibodies in ChIP assays, suggesting that the Mef2 sites exist in relatively open chromatin structure and must be readily accessible to Mef2A.
Identification of Over-represented Transcription Factor-binding Sites in Costamere Promoters
To further understand the mechanism of Mef2A-dependent costamere gene regulation, we considered the possibility that this cohort of costamere promoters harbors common cis-elements that bind transcription factor(s) that cooperate with Mef2A to regulate their expression. Toward this end, we performed a bioinformatics analysis to identify over-represented transcription factor-binding sites in the promoters of dysregulated costamere genes utilizing the Genomatix Region-Miner algorithm. Briefly, this algorithm determines over-representation statistics of transcription factor-binding sites by comparing the frequency of binding sites found within 3.0 kb of genomic sequence upstream from the predicted transcription start site of the cohort of dysregulated costamere promoters compared with the predicted frequency of each binding site found in the entire mouse genome. The results of this analysis revealed seven transcription factor-binding sites: EGR, AP2, ZF5, FOXN1, DEAF, CDEF, and NRF1, as significantly over-represented and present in five or more of this subset of costamere promoters (Table 2). Therefore, the transcription factors which bind these cis-elements may function as co-regulators of Mef2A. Given this exciting finding, we are now poised to delve deeper into the Mef2A-dependent transcriptional mechanisms of costamere gene regulation.
TABLE 2.
Overrepresented transcription factor binding sites in dysregulated costamere promoters
The 12 costamere promoters dysregulated in Mef2A knockout hearts were analyzed for overrepresentation of transcription factor (TF) binding sites using the RegionMiner algorithm. Seven TF binding sites were identified that were significantly over-represented (p < 0.05) and present in five or more of the 12 costamere genes: early growth response (EGR), activating protein 2 (AP2), zinc finger 5 (ZF5), forkhead box N1 (FOXN1), deformed epidermal autoregulatory factor (DEAF), cell cycle-dependent factor (CDEF), and nuclear respiratory factor 1 (NRF1). Multiple family members are listed for EGR: 1, 2, 3, 4, and WT1 (Wilms' tumor suppressor 1) and for AP2: A, B, and C. Also listed is the more commonly recognized role for each transcription factor (TF description).
| TF binding site identified | Number of promoters with sites | TF description |
|---|---|---|
| Early growth response (EGR) | 11 | 1: G0/G1 switch, regulates IGF-1R, TGFβ1, FN1, tumor suppressor |
| 2: Regulates late myelin genes | ||
| 3: Circadian gene, regulates muscle spindle formation | ||
| 4: Regulates cytokine gene expression, regulates LH secretion | ||
| WT1: Tumor suppressor | ||
| Activating protein 2 (AP2) | 10 | A: Important in bone/cartilage development, muscle proliferation |
| B: Expression of genes ectodermal tissue development | ||
| C: Placental expression in embryos, purine metabolism | ||
| Zinc finger 5 (ZF5) | 8 | Represses c-Myc, transcriptional activator dopamine transporter |
| Forkhead box N1 (FOXN1) | 8 | Immune response and hair follicle development |
| Deformed epidermal autoregulatory factor (DEAF) | 7 | Immune response and epithelial cell proliferation |
| Cell cycle-dependent factor (CDEF) | 6 | Regulates cell cycle progression |
| Nuclear respiratory factor 1 (NRF1) | 5 | Mitochondrial biogenesis |
DISCUSSION
Here we report that Mef2A functions as a critical regulator of the costamere in cardiac muscle and that it is essential for the maintenance of structural integrity and cell survival. These results establish for the first time a direct molecular link between Mef2A and cardiomyocyte survival through its regulation of a costamere gene program. Importantly, our results reveal a previously unrecognized transcription-based mechanism by which the costamere can be regulated on a global scale. Thus, given the importance of the costamere in muscle contraction and stability of the cytoskeleton, further investigation into the transcriptional control of this macromolecular complex is warranted.
Using a candidate approach, we identified an additional 12 costamere genes, excluding myospryn and Xirp2, whose regulation is dependent on Mef2A, and found that expression of these genes was not significantly affected in Mef2D knock-out hearts and in NRVMs transduced with Mef2C-specific shRNAs. Interestingly, Mef2C in mice (22) and both Mef2C and Mef2D in zebrafish (23) are required for cytoarchitectural integrity in skeletal muscle. These phenotypes have not been observed in cardiac muscle, suggesting that cardiac muscle cytoarchitectural gene expression, or specifically costamere genes, is more dependent on Mef2A. Furthermore, Mef2A knock-out mice do not exhibit any obvious skeletal muscle structural abnormalities (24), implying either that Mef2A lacks the ability to regulate cytoarchitectural genes in skeletal muscle or that this activity can be replaced by other Mef2 factors.
In a related study, neonatal cardiomyocytes lacking serum response factor displayed altered cytoarchitecture (25). This study also reported the dysregulated expression of a large number of cytoarchitectural genes, of which only three are down-regulated in Mef2A-deficient cardiomyocytes. Mice lacking Prox1, a Prospero-related homeobox transcription factor, also develop myofibrillar abnormalities and dysregulation of sarcomeric α-actinin and zyxin (26), genes that are unaffected in Mef2A and serum response factor conditional knock-out mice. These data suggest the existence of distinct cardiac cytoarchitectural gene programs regulated by different families of transcription factors. Subdividing the regulation of structural gene expression may be a way to modulate levels of a specific gene network to calibrate the development and remodeling of the elaborate cardiac cytoarchitecture.
It is important to note that expression of costamere genes was differentially affected in the various assays performed in this study. Initially, we identified 12 costamere genes down-regulated in perinatal Mef2A-deficient hearts, indicating that expression of these genes is dependent on Mef2A throughout development. However, when we acutely knocked down Mef2A in NRVMs, only a subset of this cohort was dysregulated. One explanation for this finding is that Mef2A-dependent costamere genes have different sensitivities to Mef2A such that genes that require a higher level or activity of Mef2A for their expression are most readily down-regulated in the acute knockdown. We also noted that the 12 costamere genes were differentially sensitive to Mef2A overexpression. We had predicted that those genes acutely sensitive to reduction in Mef2A levels would also be sensitive to increased Mef2A levels, but this was not what we observed. Of the six up-regulated costamere genes, only two were down-regulated in the acute knockdown. One possibility for this finding is that this particular subset of up-regulated genes may be regulated by Mef2A heterodimers instead of Mef2A homodimers. In other words, active Mef2 heterodimers require only one Mef2A molecule to enhance basal transcription of these genes. Alternatively, if these genes were regulated by Mef2A homodimers, twice as much Mef2A would need to be produced by overexpression to stimulate these genes above basal levels.
The anoikis phenotype in Mef2A knockdown NRVMs is reminiscent of the programmed cell death in primary cardiomyocytes resulting from overexpression of FRNK, an endogenous inhibitor of FAK (16). There is overwhelming evidence that FAK is important for mechanotransduction and that it is also required for costamere integrity (27). Therefore, a FAK-Mef2A pathway may be a critical signaling circuit for cardiac muscle cell viability through its ability to modulate the costamere. Furthermore, the anoikis phenotype is remarkable given the fact that Mef2A knock-out hearts failed to display any obvious alterations in cell survival. Perhaps this difference can be explained by the sensitivity of cardiomyocytes, grown on a two-dimensional substratum, to structural perturbations. In comparison, the abundant extracellular matrix and cell-cell contacts surrounding cardiomyocytes in the heart may help reinforce weakened attachment sites, preventing the activation of adhesion-based programmed cell death pathways. It is worth noting that conditional knock-out of serum response factor in cultured neonatal cardiomyocytes exhibited normal survival and proliferation (25). These observations suggest that although Mef2A and serum response factor both regulate the expression of cytoarchitectural genes in cardiac muscle, the unique cohort of Mef2A-dependent genes appears to be more closely associated with cell adhesion-dependent cell survival pathways.
The ability of Mef2A to regulate the costamere is conceptually, if not biologically, similar to that of the activity-dependent remodeling of neuronal synapses (28, 29). Mef2 proteins control synaptic remodeling (30, 31), in response to changes in neuronal activity, by regulating the expression of genes that function at the synapse (32). In this regard, EGR, or early growth response gene (Egr) transcription factor (33), identified in our over-representation analysis is a particularly attractive candidate co-regulator because members of this family are induced in response to neuronal activity and are involved in the regulation of synaptic plasticity genes (34, 35). Moreover, one member of this family, Egr-1, has been linked to various cardiovascular pathologies (36), and it functions in vascular smooth muscle gene expression downstream of mechanical stretch (37), a stimulus that modulates focal adhesion/costamere activity. Along these lines, mechanical stress imposed on cardiomyocytes in vitro and in vivo has been shown to result in increased Mef2 DNA binding activity (38, 39), reinforcing the notion of activity-dependent gene regulation by Mef2. Collectively, our data and other literature suggest that Mef2A is a nodal point in the reception and interpretation of mechanical signals to promote cytoarchitectural remodeling in cardiomyocytes. Thus, investigating strain-induced responses in Mef2A knock-out mice and in Mef2A-deficient cultured cardiomyocytes on an assortment of biological matrices is likely to yield physiologically important information on the role of Mef2A in mechanotransduction in the future.
Supplementary Material
Acknowledgments
Total RNA from Mef2D knock-out hearts was a kind gift from R. Bassel-Duby and E. N. Olson (University of Texas Southwestern Medical Center). Mef2A (human) and β-galactosidase adenovirus were kind gifts from J. Molkentin (Children's Hospital, Cincinnati, OH) and Ken Walsh (Boston University Medical School), respectively. We thank Nathan Waldron for excellent technical assistance and Brian Black (University of California, San Francisco) for critical reading of the manuscript.
This work was supported, in whole or in part, by a National Institutes of Health Grant (to F. J. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- FAK
- focal adhesion kinase
- NRVM
- neonatal rat ventricular myocyte
- qRT-PCR
- quantitative real time PCR.
REFERENCES
- 1. Danowski B. A., Imanaka-Yoshida K., Sanger J. M., Sanger J. W. (1992) J. Cell Biol. 118, 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ervasti J. M. (2003) J. Biol. Chem. 278, 13591–13594 [DOI] [PubMed] [Google Scholar]
- 3. Samarel A. M. (2005) Am. J. Physiol. Heart Circ. Physiol. 289, H2291–H2301 [DOI] [PubMed] [Google Scholar]
- 4. Hoshijima M. (2006) Am. J. Physiol. Heart Circ. Physiol. 290, H1313–H1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clark K. A., McElhinny A. S., Beckerle M. C., Gregorio C. C. (2002) Annu. Rev. Cell Dev. Biol. 18, 637–706 [DOI] [PubMed] [Google Scholar]
- 6. Naya F. J., Black B. L., Wu H., Bassel-Duby R., Richardson J. A., Hill J. A., Olson E. N. (2002) Nat. Med. 8, 1303–1309 [DOI] [PubMed] [Google Scholar]
- 7. Wang Y. X., Qian L. X., Yu Z., Jiang Q., Dong Y. X., Liu X. F., Yang X. Y., Zhong T. P., Song H. Y. (2005) FEBS Lett. 579, 4843–4850 [DOI] [PubMed] [Google Scholar]
- 8. Peng X., Wu X., Druso J. E., Wei H., Park A. Y., Kraus M. S., Alcaraz A., Chen J., Chien S., Cerione R. A., Guan J. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6638–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Durham J. T., Brand O. M., Arnold M., Reynolds J. G., Muthukumar L., Weiler H., Richardson J. A., Naya F. J. (2006) J. Biol. Chem. 281, 6841–6849 [DOI] [PubMed] [Google Scholar]
- 10. Huang H. T., Brand O. M., Mathew M., Ignatiou C., Ewen E. P., McCalmon S. A., Naya F. J. (2006) J. Biol. Chem. 281, 39370–39379 [DOI] [PubMed] [Google Scholar]
- 11. McCalmon S. A., Desjardins D. M., Ahmad S., Davidoff K. S., Snyder C. M., Sato K., Ohashi K., Kielbasa O. M., Mathew M., Ewen E. P., Walsh K., Gavras H., Naya F. J. (2010) Circ. Res. 106, 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boluyt M. O., O'Neill L., Meredith A. L., Bing O. H., Brooks W. W., Conrad C. H., Crow M. T., Lakatta E. G. (1994) Circ. Res. 75, 23–32 [DOI] [PubMed] [Google Scholar]
- 13. Jones W. K., Grupp I. L., Doetschman T., Grupp G., Osinska H., Hewett T. E., Boivin G., Gulick J., Ng W. A., Robbins J. (1996) J. Clin. Invest. 98, 1906–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim Y., Phan D., van Rooij E., Wang D. Z., McAnally J., Qi X., Richardson J. A., Hill J. A., Bassel-Duby R., Olson E. N. (2008) J. Clin. Invest. 118, 124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hervy M., Hoffman L., Beckerle M. C. (2006) Curr. Opin. Cell Biol. 18, 524–532 [DOI] [PubMed] [Google Scholar]
- 16. Heidkamp M. C., Bayer A. L., Kalina J. A., Eble D. M., Samarel A. M. (2002) Circ. Res. 90, 1282–1289 [DOI] [PubMed] [Google Scholar]
- 17. Michel J. B. (2003) Arterioscler. Thromb. Vasc. Biol. 23, 2146–2154 [DOI] [PubMed] [Google Scholar]
- 18. Reginato M. J., Mills K. R., Paulus J. K., Lynch D. K., Sgroi D. C., Debnath J., Muthuswamy S. K., Brugge J. S. (2003) Nat. Cell Biol. 5, 733–740 [DOI] [PubMed] [Google Scholar]
- 19. Akiyama T., Dass C. R., Choong P. F. (2009) Mol. Cancer Ther. 8, 3173–3180 [DOI] [PubMed] [Google Scholar]
- 20. Klamut H. J., Bosnoyan-Collins L. O., Worton R. G., Ray P. N. (1997) Nucleic Acids Res. 25, 1618–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Andrés V., Cervera M., Mahdavi V. (1995) J. Biol. Chem. 270, 23246–23249 [DOI] [PubMed] [Google Scholar]
- 22. Potthoff M. J., Arnold M. A., McAnally J., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) Mol. Cell. Biol. 27, 8143–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hinits Y., Hughes S. M. (2007) Development 134, 2511–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Potthoff M. J., Wu H., Arnold M. A., Shelton J. M., Backs J., McAnally J., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) J. Clin. Invest. 117, 2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Balza R. O., Jr., Misra R. P. (2006) J. Biol. Chem. 281, 6498–6510 [DOI] [PubMed] [Google Scholar]
- 26. Risebro C. A., Searles R. G., Melville A. A., Ehler E., Jina N., Shah S., Pallas J., Hubank M., Dillard M., Harvey N. L., Schwartz R. J., Chien K. R., Oliver G., Riley P. R. (2009) Development 136, 495–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quach N. L., Rando T. A. (2006) Dev. Biol. 293, 38–52 [DOI] [PubMed] [Google Scholar]
- 28. Greer P. L., Greenberg M. E. (2008) Neuron 59, 846–860 [DOI] [PubMed] [Google Scholar]
- 29. Shalizi A. K., Bonni A. (2005) Curr. Top Dev. Biol. 69, 239–266 [DOI] [PubMed] [Google Scholar]
- 30. Flavell S. W., Cowan C. W., Kim T. K., Greer P. L., Lin Y., Paradis S., Griffith E. C., Hu L. S., Chen C., Greenberg M. E. (2006) Science 311, 1008–1012 [DOI] [PubMed] [Google Scholar]
- 31. Shalizi A., Gaudillière B., Yuan Z., Stegmüller J., Shirogane T., Ge Q., Tan Y., Schulman B., Harper J. W., Bonni A. (2006) Science 311, 1012–1017 [DOI] [PubMed] [Google Scholar]
- 32. Flavell S. W., Kim T. K., Gray J. M., Harmin D. A., Hemberg M., Hong E. J., Markenscoff-Papadimitriou E., Bear D. M., Greenberg M. E. (2008) Neuron 60, 1022–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thiel G., Cibelli G. (2002) J. Cell. Physiol. 193, 287–292 [DOI] [PubMed] [Google Scholar]
- 34. Li L., Carter J., Gao X., Whitehead J., Tourtellotte W. G. (2005) Mol. Cell. Biol. 25, 10286–10300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O'Donovan K. J., Tourtellotte W. G., Millbrandt J., Baraban J. M. (1999) Trends Neurosci 22, 167–173 [DOI] [PubMed] [Google Scholar]
- 36. Khachigian L. M. (2006) Circ. Res. 98, 186–191 [DOI] [PubMed] [Google Scholar]
- 37. Grote K., Bavendiek U., Grothusen C., Flach I., Hilfiker-Kleiner D., Drexler H., Schieffer B. (2004) J. Biol. Chem. 279, 55675–55681 [DOI] [PubMed] [Google Scholar]
- 38. Nadruz W., Jr., Kobarg C. B., Constancio S. S., Corat P. D., Franchini K. G. (2003) Circ. Res. 92, 243–251 [DOI] [PubMed] [Google Scholar]
- 39. Nadruz W., Jr., Corat M. A., Marin T. M., Guimarães Pereira G. A., Franchini K. G. (2005) Cardiovasc. Res. 68, 87–97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.