Abstract
Apicomplexa are protist parasites that include Plasmodium spp., the causative agents of malaria, and Toxoplasma gondii, responsible for toxoplasmosis. Most Apicomplexa possess a relict plastid, the apicoplast, which was acquired by secondary endosymbiosis of a red alga. Despite being nonphotosynthetic, the apicoplast is otherwise metabolically similar to algal and plant plastids and is essential for parasite survival. Previous studies of Toxoplasma gondii identified membrane lipids with some structural features of plastid galactolipids, the major plastid lipid class. However, direct evidence for the plant-like enzymes responsible for galactolipid synthesis in Apicomplexan parasites has not been obtained. Chromera velia is an Apicomplexan relative recently discovered in Australian corals. C. velia retains a photosynthetic plastid, providing a unique model to study the evolution of the apicoplast. Here, we report the unambiguous presence of plant-like monogalactosyldiacylglycerol and digalactosyldiacylglycerol in C. velia and localize digalactosyldiacylglycerol to the plastid. We also provide evidence for a plant-like biosynthesis pathway and identify candidate galactosyltranferases responsible for galactolipid synthesis. Our study provides new insights in the evolution of these important enzymes in plastid-containing eukaryotes and will help reconstruct the evolution of glycerolipid metabolism in important parasites such as Plasmodium and Toxoplasma.
Keywords: Chloroplast, Evolution, Glycerolipid, Mass Spectrometry (MS), Membrane Lipids, Apicomplexa, Chromera velia, Apicoplast, Galactolipid/Galactosyltransferase, Plastid Evolution
Introduction
Galactolipids, i.e. monogalactosyldiacylglycerol (MGDG)5 and digalactosyldiacylglycerol (DGDG), are the most abundant lipid classes in the membranes of plant and algal plastids, where they represent up to 85% of the total lipid composition (1). This unique lipid composition is only otherwise found in cyanobacteria (2), the lineage from which the plastid was derived by endosymbiosis (3). In plants and algae, MGDG and DGDG syntheses are catalyzed by two galactosyltransferases localized in the plastid envelope (1, 4). In these organisms, MGDG is synthesized in a single step by MGDG synthases that transfer a β-galactosyl moiety from a UDP-galactose (UDP-Gal) donor to the sn-3 position of diacylglycerol (DAG). In cyanobacteria, MGDG synthesis occurs via a two-step process in which a glucosyltransferase, referred to as monoglucosyldiacylglycerol (MGlcDG) synthase, transfers a glucosyl from UDP-glucose (UDP-Glc) donor onto DAG to form MGlcDG, which is subsequently epimerized into MGDG (5). In both plants and cyanobacteria, a DGDG synthase catalyzes the transfer of a second α-galactose from UDP-Gal to MGDG. As the major lipids of thylakoid and plastid membranes, MGDG and DGDG are thought to be essential for the biogenesis and function of these membranes (6). Molecular disruption of the MGDG synthetic pathway in Arabidopsis thaliana showed that galactolipids play an essential role in early embryo development and biogenesis of a functional photosynthetic apparatus (6). Furthermore, both MGDG and DGDG were found tightly associated with crystallized photosystems and light harvesting complexes (7), and DGDG might be required for their stability and proper activity (8–12). Galactolipids have long been thought to be restricted to plastid localization and functions. However, several important studies showed that DGDG could also be exported and become a critical component of extraplastidic compartments. Under specific environmental conditions, such as phosphate shortage, DGDG can substitute for phospholipids in extraplastidic membranes such as the plasma membrane, mitochondria, and tonoplast (1). Galactolipids are therefore involved in critical functions of membrane biogenesis and homeostasis, as well as developmental and physiological processes in autotrophic organisms.
The Apicomplexa include a large group of protozoa that are the causative agents of major human diseases such as malaria (Plasmodium spp.) and toxoplasmosis (Toxoplasma gondii). Apicomplexa acquired a plastid, termed the apicoplast, by the secondary endosymbiosis of a red alga (13, 14). The apicoplast no longer retains photosynthetic enzymes but is essential for the viability of these organisms, probably due to the retention of other key metabolic functions (15, 16). The discovery of the apicoplast immediately raised questions regarding the possible occurrence of plastidial galactolipids, their synthesis, and putative role in these parasites. None of the genes encoding for galactolipid synthesis have been found in Apicomplexa genomes to date (17). However, the synthesis of galactolipids co-migrating with spinach MGDG and DGDG could be detected in Plasmodium falciparum and T. gondii lysates by metabolic labeling with radioactive UDP-Gal (18). Hydrolysis with α- and β-galactosidases combined with alkaline hydrolysis confirmed that these galactolipids contained α- and β-galactoses as polar heads and esterified fatty acids as their hydrophobic tail, making them very similar to plant galactolipids (18). Furthermore, a lipid with chromatographic properties similar to plant DGDG was detected in total lipid extract from P. falciparum and T. gondii (19). A digalactolipid reacting against an anti-DGDG antibody was localized within the pellicle membranes of T. gondii, but not in the apicoplast (20). This plant-like digalactolipid was proposed to be the same as those detected in previous labeling experiments. Lipidomic analysis performed in T. gondii showed the presence of two types of hexosyl(galacto) lipid classes as minor membrane components as follows: (i) plant-like hexosylglycerolipids and (ii) hexosylceramides (20, 21). Although these data are suggestive of a pathway of galactolipid biosynthesis in some Apicomplexan parasites, many questions remain. Are Apicomplexan galactolipids generated via a plant-like or cyanobacterium-like pathway using highly divergent enzymes? Is the galactolipid biosynthetic machinery located in the plastid or other compartments?
Very recently, Chromera velia, a photosynthetic protist closely related to Apicomplexa, was discovered in corals in Sydney Harbor (22). C. velia contains a four membrane-bound plastid that is still photosynthetic and is therefore an interesting model to study the conversion of secondary plastids from autotrophic to heterotrophic state, as happened to the apicoplast. Here, we report the unambiguous characterization of plant-like galactoglycerolipids in C. velia via HPTLC, GC-MS, and LC/MS-MS approaches and determine their localization by immunocytochemistry. Metabolic labeling is used to characterize a plant-like galactolipid synthesis pathway in C. velia. Finally, we identify the candidate genes encoding galactosyltransferases responsible for galactolipid synthesis in C. velia and discuss their importance for the evolution of this pathway in photosynthetic eukaryotes.
EXPERIMENTAL PROCEDURES
C. velia Culture and Lipid Extraction
C. velia was maintained as described previously (14) and harvested by centrifugation at 700 × g, and total lipids were extracted in chloroform/methanol (2:1, v/v) and chloroform/methanol/water (1:2:0.8, v/v). After removal of insoluble material by centrifugation (15,000 × g, 10 min), extracts were dried under nitrogen and subjected to biphasic partitioning in 1-butanol and water (2:1, v/v). The organic phase was dried, and lipids were resuspended in 1-butanol. Total lipid was analyzed by HPTLC using aluminum-backed silica gel sheets (Merck). One-dimensional HPTLCs were developed in chloroform/methanol/water (65:35:2, v/v). Glycolipids were stained and visualized with orcinol/H2SO4.
Sugar Head Analysis by Gas Chromatography-Mass Spectrometry
Individual glycolipid species were extracted from silica scrapings from one-dimensional HPTLCs using chloroform/methanol/water (10:10:3, v/v), and the supernatant was dried under nitrogen and desalted by 1-butanol/water biphasic partitioning (2:1, v/v). The sugar head of each glycolipid was analyzed by GC-MS after methanolysis and trimethylsilyl derivatization (40).
Mass Spectrometric Analysis of MGDG and DGDG
The MGDG or DGDG fraction was diluted 10-fold in 10 mm ammonium formate in methanol and then the lipid was flow-injected to Agilent 6410 triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA) at the rate of 0.2 ml/min in the solvent of H2O/MeOH/tetrahydrofuran = 14:20:66 (v/v). The capillary voltage was set at 4000 V, and the gas temperature and flow rate were set to 250 °C and 7 liters/min, respectively. Nebulizer was set at 40 p.s.i. MS/MS experiments were conducted in positive mode with a fragmentation voltage of 135 V and collision energy of 15 eV.
Immunofluorescence Assay
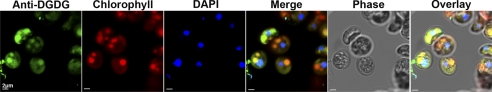
All media and antibody dilutions used for immunofluorescence assays were prepared in PHEM buffer, pH 6.9 (PIPES 60 mm, HEPES 25 mm, EGTA 10 mm, MgCl2 2 mm). C. velia cells were fixed in 4% paraformaldehyde for 45 min on ice prior to permeabilization in 0.1% Triton X-100 for 10 min on ice. After being blocked with 3% BSA in PHEM for 45 min at 4 °C, cells were then incubated with polyclonal rabbit serum anti-DGDG (1:25) for 1 h at 4 °C with agitation. Labeled cells were incubated with secondary anti-rabbit Alexa488 (1:1000) for 1 h at 4 °C, followed by incubation with DAPI (1:15,000) for 15 min at 4 °C. Cells were mounted on slides and coverslips and observed with a Leica confocal microscope.
Activity Assay
C. velia was harvested by centrifugation at 700 × g and resuspended in 5 volumes (v/v) of lysis buffer (MOPS-NaOH, pH 7.9, 50 mm, 20% glycerol (v/v), sodium acetate 0.8 m, DTT 10 mm) followed by sonication for three intervals of 15 s on ice with 2 min resting on ice between each pulse (Branson sonifier 250, output 40%). 2.5 g of pre-frozen (in liquid N2) spinach leaves were crushed with a mortar and a pestle in the presence of 2.5 ml of lysis buffer. Spinach lysate was resuspended in 5 volumes (w/v) of lysis buffer. DTT was added (10 mm) prior to centrifugation at 500 × g, 4 °C for 20 min. Then a 90-μl aliquot was preincubated at 30 °C for 5 min with either UDP-[14C]galactose or UDP-[14C]glucose (PerkinElmer Life Sciences, 6.3 Ci/mmol, final concentration, 1 μCi/reaction) added to start the enzyme reaction. After 2 h, reactions were stopped by addition of 375 μl of chloroform/methanol (1:2, v/v) and vortexing. After centrifugation at 4000 × g for 10 min, the supernatant was pooled, and another chloroform/methanol/water (1:2:0.8, v/v) was added for further extraction of lipids. The supernatant was recovered and mixed with the pooled supernatant and dried up under nitrogen gas. The dried material was biphasically partitioned by 1-butanol/water (2:1, v/v). The butanol phase was recovered and dried. Again the lipid was dissolved in the 10 μl of water-saturated 1-butanol. Total lipid was analyzed by HPTLC using aluminum-backed silica gel sheets (Merck). HPTLC was migrated in the solvent system of chloroform/methanol/water (65:35:2, v/v). The HPTLC was exposed to high sensitivity x-ray film, Kodak BiomaxMR film (Eastman Kodak Co.), to detect 14C-labeled lipids.
RNA Isolation, RT-PCR, RACE, and Sequencing
Short fragments of MGDG and DGDG synthase genes were identified in the 454-pyrosequencing of C. velia genomic DNA (14). Total RNA was isolated using TRIzol® reagent (Invitrogen) and purified using RNeasy MinElute cleanup kit (Qiagen). For each gene, fragments determined in the 454-data survey were linked using RT-PCR with specific oligonucleotides. 3′-RACE was used to obtain a complete 3′ sequence of the MGDG synthase transcript. In DGDG synthase, 3′-RACE repeatedly produced a single amplicon containing an incomplete 3′ portion of the transcript, probably due to the oligo(dT) adaptor misannealing to an A-rich region upstream of the stop codon. All PCR amplicons were cloned and several clones from each of these amplicons sequenced. All sequences for each gene were unambiguously assembled into a single contig. Our attempts to obtain 5′ ends of the transcripts were unsuccessful and lea only to nonspecific amplification products. MGDG and DGDG synthase sequences of C. velia were deposited under GenBankTM accessions numbers JF912518 and JF912519.
Phylogenetic Analyses
For genomic sequence survey of C. velia, GenBankTM protein and EST databases were searched using multiple rounds of blastp and tblastn similarity searches using different queries (MGDG synthase from A. thaliana, Cyanidioschyzon merolae, and Phaeodactylum tricornutum; MGlcDG synthase from Synechocystis PCC6803; DGDG synthase from A. thaliana and Synechocystis PCC6803; and Escherichia coli MURG). Nearly identical paralogs likely derived from different gene models on identical genomic loci (e.g. in Oryza and Populus) were limited to the most canonical model. Highly divergent sequences of some bacteria were excluded from these sequence sets. Two principal datasets from each gene were created as follows: (i) global dataset for assessing the monophyly of eukaryotic homologs and their position within bacterial genes, and (ii) eukaryotic dataset for exploring the branching patterns with eukaryotes and the position of C. velia. In global phylogenies of DGDG synthase, the relationships of both the plant-like and the cyanobacterial form (also present in Cyanophora and cyanidiales) were explored. All dataset were aligned in MAFFT using the local pair (LINSI) algorithm and ambiguously aligned positions were trimmed in Gblocks using following parameters: −b1 = 50% +1; −b2 = 50% +1; −b3 = 12 −b4 = 4 −b5 = h. This procedure resulted in following matrix sizes: MGDG phylogenies in supplemental Fig. S4, A and B, and Fig. 8A = 160, 183, and 326 sites, respectively; DGDG phylogenies in supplemental Fig. S5, and Fig. 8B = 112, and 364 sites (alignments available on request). Global phylogenies were computed in PhyML 3.0 using LG model (parameters, γ 8, I, SPR) with aLRT branch supports. Phylogenies from eukaryotic datasets were computed under identical conditions in PhyML 3.0 and supported by PhyML and RaxML bootstrap analyses.
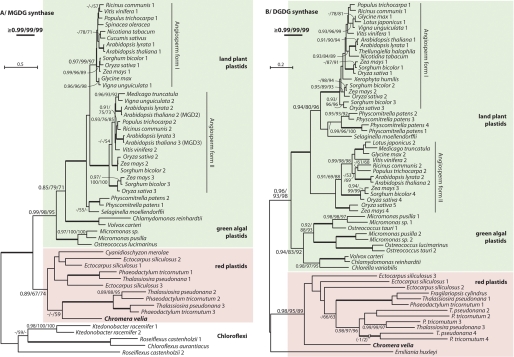
FIGURE 8.
Eukaryotic phylogeny of C. velia MGDG and DGDG synthases. Maximum likelihood phylogenies of 52 eukaryotic MGDG synthase (A) and 61 DGDG synthase (B) genes. The trees display PhyML aLRT and PhyML bootstrap and RaxML bootstrap branch supports (for details see “Experimental Procedures”). Highly supported branches are thickened as indicated.
RESULTS
C. velia Contains MGDG and DGDG Enriched in Long Polyunsaturated Fatty Acid Chains
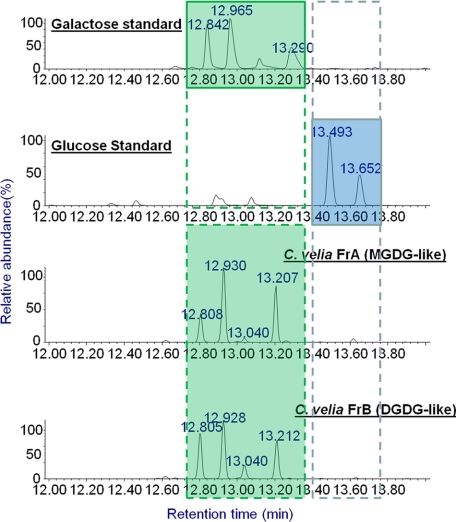
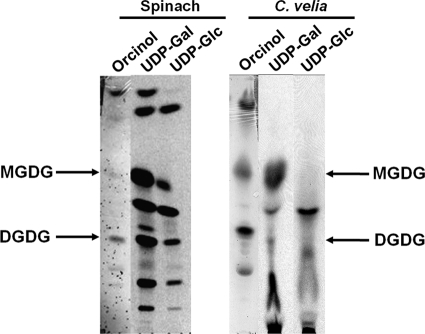
To assess the presence of plant/plastid-like galactolipids in C. velia, total lipids were analyzed by HPTLC. As shown in Fig. 1, C. velia synthesized a number of abundant orcinol-positive glycolipids that had a similar HPTLC migration to the plastid MGDG and DGDG galactolipids and sulfoquinovosyldiacylglycerol (SQDG) from spinach (Spinacia oleracea) leaf extracts. MGDG and DGDG fractions from S. oleracea and the corresponding C. velia fractions (fraction A and fraction B, respectively) were extracted from the HPTLC sheets and reanalyzed by HPTLC in a different solvent system. Again the extracted C. velia MGDG-like and DGDG-like fractions showed similar mobility to the S. oleracea galactolipids. Fractions A and B were subjected to mild alkaline treatment to hydrolyze ester-linked fatty acids and release the galacto-glycerol backbone. The glycan headgroups of the C. velia glycolipids (fractions A and B) displayed similar chromatographic behavior on HPTLC to those obtained from hydrolyzed S. oleracea MGDG and DGDG (supplemental Fig. S1). The presence of galactolipids was confirmed by GC-MS analysis of monosaccharides released by solvolysis and trimethylsilylation. These analyses showed that the major sugar in fractions A and B was galactose (Fig. 2).
FIGURE 1.
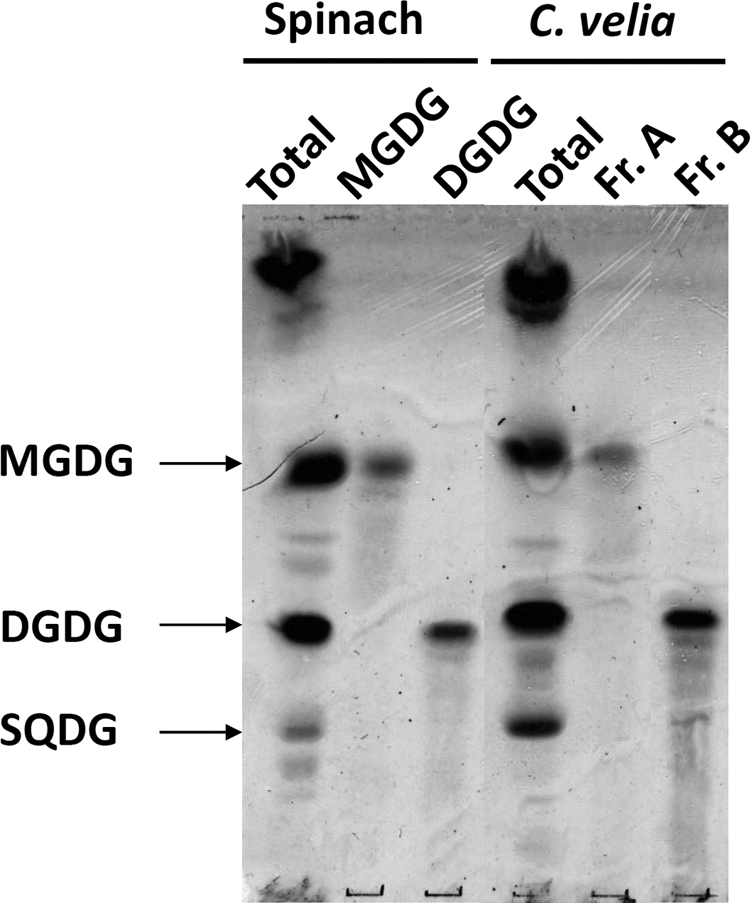
C. velia contains plant-like glycolipids. Total lipids from spinach and C. velia were extracted from whole leaves and whole cells, respectively. Extracted lipids were migrated by HPTLC in chloroform/methanol/water (65:25:25, v/v) and sprayed with orcinol/sulfuric acid to allow glycolipid detection. Plastidial MGDG, DGDG, and SQDG are indicated. Fr., fraction.
FIGURE 2.
Identification of MGDG and DGDG in C. velia using GC-MS analysis. C. velia glycolipid fractions (Fr. A and Fr. B in Fig. 1) were subjected to GC-MS after methanolysis and tetramethylsilyl derivatization. The corresponding chromatograms were compared with those of galactose and glucose standards.
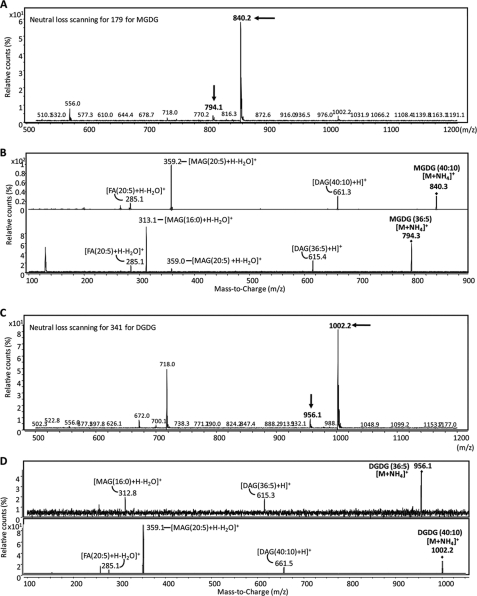
To further define the structures of the C. velia galactolipids, the HPTLC-purified glycolipids were subjected to LC/MS-MS analysis (Fig. 3). Neutral loss scanning for m/z 179 ion and m/z 341 ion in positive mode gave the characteristic ions for an ammonium-adducted and dehydrated monohexose from MGDG and an ammonium-adducted and dehydrated dihexose from DGDG, respectively (Fig. 3, A and C). The parent ions that were detected by neutral loss scanning of m/z 179 from the S. oleracea galactolipids had masses of m/z 792.3 and 764.2, corresponding to molecular species with 36:6 (36 carbon and 6 double bonds) and 34:6 fatty acyl chains, respectively (supplemental Fig. S2). Similarly, neutral loss scan for m/z 341 in S. oleracea lipids detected DGDG species with masses of m/z 954.2 and 932.3, corresponding to molecular species with the same lipid composition (36:6 and 34:3). Similar analyses of the C. velia galactolipid fractions A and B allowed detection of a different set of parent ions indicating a distinct fatty acyl composition. Neutral loss scanning for m/z 179 and 341 revealed a series of parent ion peaks in the m/z 500–1100 mass ranges. Parent ion masses of m/z 840.3 and 794.3 corresponded to MGDG [M + NH4]+ ion containing 36:5 and 40:10 acyl chains, respectively (Fig. 3A). Similarly, DGDG parent ions at m/z 956.1 and 1002.2 corresponded to [M + NH4]+ ion with 36:5 and 40:10 acyl chains, respectively (Fig. 3C). When molecular species at m/z 840.3 and 794.3 were fragmented, ions derived from loss of galactose (m/z 179), diacylglycerol ([DAG (40:10) + H]+ (m/z 661.3) or [DAG (36:5) + H]+ (m/z 615.4)), and monoacylglycerol ([MAG (20:5) + H − H2O]+ (m/z 359) and/or [MAG (16:0) + H − H2O]+ (m/z 313)) were detected (Fig. 3B). These fragmentation patterns are the same as those for S. oleracea MGDG except for the difference of fatty acid species confirming the galactolipid fraction as MGDG. Similarly, MS/MS fragmentation of m/z 956.1 and 1002.2 generated characteristic ions corresponding to loss of digalactose (m/z 341), a peak of [DAG + H]+, and peaks of MAG (20:5) and/or MAG (16:0) (Fig. 3D). These fragmentation patterns also corresponded to those of DGDG from S. oleracea confirming galactolipid fraction B as DGDG. These data demonstrate that the C. velia galactolipids are equivalent to the plant galactoglycerolipids. Furthermore, the presence of SQDG sulfolipid was confirmed by the precursor ion scan for the characteristic ion of SQDG, m/z 225 in negative mode. The major SQDG were detected as [M − H]− ions at m/z 817 and 843, SQDG (34:2) and SQDG (36:3), respectively (supplemental Fig. S3).
FIGURE 3.
LC-MS/MS analysis of C, velia MGDG and DGDG molecular species. A, total lipid extracts of C. velia were analyzed by positive ion LC-MS/MS in neutral loss scanning mode. Neutral loss scanning for m/z 179 (loss of hexose) identified parent ions at m/z 840.3 and 794.3 corresponding to MGDG [M + NH4]+ molecular species with 36:5 and 40:10 (total carbon length/degree of unsaturation) fatty acyl chains, respectively (arrows). B, MS/MS of these parent ions gave fragment ions containing the diacylglycerol ([DAG (40:10) + H]+ at m/z 661.3 or [DAG(36:5) + H]+ at m/z 615.4) and the monoacylglycerol ([MAG (20:5) + H − H2O]+ at m/z 359 and/or 16:0 MAG [MAG (16:0) + H − H2O]+ at m/z 313) moieties. C, DGDG species were detected by neutral loss scanning for m/z 341 in positive ion mode. Parent ions at m/z 956.1 and 1002.2 corresponded to [M + NH4]+ ion of DGDG molecular species with 36:5 and 40:10 acyl chains, respectively (arrow). D, MS/MS of the DGDG parent ions gave fragment ions containing the same diacylglycerol and monoacyl species as in MGDG.
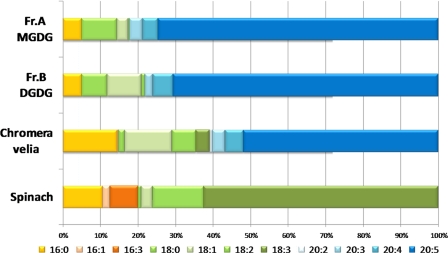
The fatty acyl chain composition of the galactolipids was further supported by GC-MS fatty acid analysis of C. velia MGDG, DGDG, and total lipids (Fig. 4). Total lipids from both S. oleracea and C. velia are highly enriched in unsaturated fatty acids (89 and 84%, respectively). MGDG and DGDG fractions from C. velia followed a similar pattern in terms of their unsaturated fatty acid composition (i.e. 86 and 88%, respectively). By contrast with spinach total lipids, which are highly enriched in linolenic acid (C18:3; 62%), C. velia total lipid fraction contained a large majority of eicosapentenoic acid (C20:5, 52%). A similar composition was found in both MGDG- and DGDG-like fractions, whereas eicosapentenoic acid contributed to 74 and 70% of the fractions, respectively. C. velia lipids predominantly contained very long fatty acid chains (>18 carbons) as follows: 61% in the total lipid fraction and 77% for both MGDG and DGDG fractions, and S. oleracea only contained shorter fatty acid chains.
FIGURE 4.
C. velia fatty acid composition analyzed via GC-MS. Fatty acid compositions of C. velia MGDG, DGDG, and total lipid fractions were analyzed by GC-MS analysis. Each fatty acid class is as indicated. C. velia MGDG (Fr. A) and DGDG (Fr. B) fractions contained unsaturated fatty acid (i.e. 86 and 88%, respectively). Spinach total lipids contained linolenic acid (C18:3; 62%), C. velia total lipid fraction contained eicosapentenoic acid (C20:5, 52%).
DGDG Is Detected in the C. velia Plastid
In plants and algae, galactolipids are predominantly found within the plastid membranes. In contrast, preliminary analyses of the T. gondii DGDG-like lipid indicated localization to the pellicle membranes (20). To assess the subcellular localization of C. velia galactolipids, anti-DGDG serum was used to detect this glycolipid on whole cells (18, 20, 23). Cells were fixed and permeabilized prior to their incubation with the anti-DGDG serum, and fluorescence was detected by confocal microscopy (Fig. 5). Because the plastid of C. velia is photosynthetically active and contains chlorophyll, it can be observed directly courtesy of the chlorophyll autofluorescence. The chlorophyll signal showed that the single plastid of C. velia occupies a large part of the cell and overlaps with the signal detected by anti-DGDG fluorescence suggesting that the galactolipid is localized within the plastid membranes, as observed in plants and algae.
FIGURE 5.
C. velia DGDG localizes at the plastid. C. velia whole cells were fixed and permeabilized prior to labeling with rabbit anti-DGDG as the primary antibody followed by AlexaFluor anti-rabbit 488 as the secondary antibody. Nuclei were visualized by Hoechst staining and plastids detected by autofluorescence of chlorophyll.
C. velia Contains an Active MGDG Synthase
Plant MGDG is generated by the transfer of a galactosyl polar head onto a DAG, a reaction catalyzed by a multigenic family of galactosyltransferases called MGDG synthases. By contrast, cyanobacteria synthesize MGDG by glucosylation of DAG and epimerization of the glucosyl polar head to synthesize MGDG, which is believed to be the ancestral pathway. To determine whether C. velia uses the plant-like or cyanobacterium-like pathway to initiate the galactolipid synthesis, we reconstituted this pathway in cell-free lysates. Whole C. velia and S. oleracea cells were lysed, and an organellar fraction was incubated with either UDP-[14C]galactose or UDP-[14C]glucose prior to total lipid extraction. Total lipids were separated by HPTLC and visualized by autoradiography. Synthesis of both MGDG and DGDG was detected in S. oleracea lysate after incubation with UDP-[14C]galactose (Fig. 6). Markedly less synthesis of MGDG and DGDG was observed with UDP-[14C]glucose; we interpret this as epimerase activity converting UDP-glucose to UDP-galactose. In Chromera, however, only the lysate incubated with UDP-[14C]galactose resulted in labeling of radioactive MGDG and DGDG (Fig. 6). No galactolipid was detected in the lysate incubated with UDP-[14C]glucose (Fig. 6). We conclude that MGDG synthesis operates in a higher plant-like fashion in C. velia.
FIGURE 6.
Activity assay shows that C. velia synthesizes its galactolipids in plant-like fashion. Spinach leaves and C. velia whole cell lysate were incubated with UDP-[14C]glucose (UDP-Glc) or UDP-[14C]galactose (UDP-Gal). Total lipids were then extracted and subjected to HPTLC, which was exposed to an x-ray film to detect possible radioactive lipids. MGDG and DGDG are indicated by arrows.
Identification of MGDG Synthase and DGDG Synthase Genes in C. velia
The C. velia genome has not yet been sequenced. However, we made use of a C. velia genome sequence survey (14) to search for potential galactolipid synthesis genes using MGDG, MGlcDG, and DGDG synthase sequences as queries (see under “Experimental Procedures”). We identified two candidate fragments corresponding to a MGDG synthase gene, three candidate hits for the DGDG synthase, but no candidate hit for the cyanobacterial MGlcDG synthase. We tried to obtain full-length versions of the putative C. velia MGDG and DGDG synthases by RT-PCR and 5′- and 3′-RACE approaches. An almost complete sequence of MGDG synthase (likely missing only a portion of its 5′ end) and an incomplete sequence from DGDG synthase were obtained (Fig. 7).
FIGURE 7.
Identification of MGDG and DGDG synthase genes in C. velia. Candidate sequences for MGDG and DGDG synthases were initially identified by mining a C. velia genomic survey using sequence queries from A. thaliana, P. tricornutum, C. merolae, and Synechocystis. An almost complete sequence of the MGDG synthase (missing a part of its 5′ end) and an incomplete sequence of the DGDG synthase were obtained by 5′- and 3′-RACE approaches using specific primers designed from candidate hits. Corresponding protein sequence from C. velia MGDG synthase (A) and DGDG synthase (B) were aligned with homologues from Arabidopsis (AtMGD1), spinach (SoMGD1), and diatom (PtMGD and PtDGD).
Phylogenic History of MGDG and DGDG Synthases in Eukaryotes
Using sequences from bacterial glycosyltransferases and eukaryotic homologs from both MGDG and DGDG synthases, we conducted a two-step phylogenic analysis for each of the two genes: (i) global phylogeny evaluating the monophyly of eukaryotic forms and their relationships to cyanobacterial and bacterial sequences (supplemental Figs. S4 and S5), and (ii) phylogeny restricted to eukaryotic homologs to assess the evolution of eukaryotic forms in general and C. velia in particular (Fig. 8). Similarity searches with MGDG synthase queries (see under “Experimental Procedures”) confirmed that eukaryotic and cyanobacterial forms are distantly related, as suggested previously (17). In global MGDG synthase phylogenies, all eukaryotes formed a monophyletic clade nested within diverse bacterial glycosyltransferases (supplemental Fig. S4, A and B), with the closest affiliation to those from chloroflexi, firmicutes, deinococci, and spirochetes. This affiliation received absolute support when the divergent bacterial MURG sequences were excluded (data not shown). In contrast, cyanobacterial MGlcDG synthase was related to a distinct group of bacterial glycosyltransferases (data not shown). The eukaryotic phylogeny of MGDG synthase in Fig. 8A is largely consistent with vertical inheritance within the major lineages; chlorophytes and streptophytes form a group to the exclusion of the red algal lineage (red alga C. merolae and secondary red plastids). The MGDG synthase sequence from C. velia was consistently associated with the red algal lineage, as expected if it was vertically inherited from a secondary red algal endosymbiont (Fig. 8A).
Homology searches with DGDG synthase also revealed that the plant-like form that is present in the majority of eukaryotes that possess DGDG synthase does not closely resemble the cyanobacterial form (see “Experimental Procedures”). However, in two eukaryotic lineages, the glaucophyte Cyanophora and cyanidiales red algae (represented by C. merolae and Cyanidium caldarium), only the cyanobacterial form was detected (supplemental Fig. S5). Importantly, DGDG synthase is encoded in the plastid genome of the cyanidiales, providing direct evidence that cyanobacterial DGDG synthase was ancestral to all eukaryotes, but has been substituted in some lineages by an unrelated galactosyltransferase. The plant-like DGDG synthase formed a clade with three α-proteobacterial sequences. Unfortunately, these were too divergent to serve as a reliable outgroup (data not shown), so the phylogeny of eukaryotic DGDG synthase genes lacks an outgroup (Fig. 8B). Nevertheless, the phylogeny of eukaryotic DGDG synthase is consistent with vertical inheritance and suggests acquisition of this novel enzyme took place prior to divergence of green and red primary plastids. The C. velia DGDG synthase branched with high support among other red plastids, as a sister to heterokont algae, once again consistent with the conclusion that it was vertically acquired from a red algal endosymbiont.
DISCUSSION
C. velia is a close relative of parasites like malaria and T. gondii and has a photosynthetic plastid homologous to the nonphotosynthetic plastid (apicoplast) typical of these parasites. Here, we have shown that C. velia contains abundant galactolipids with two acyl chains esterified to a glycerol backbone, namely MGDG and DGDG. Immunolocalization of DGDG revealed it to be concentrated within the C. velia plastid membranes. C. velia incorporates galactose into MGDG and DGDG from UDP-galactose similar to plant chloroplasts, probably courtesy of plant-like MGDG and DGDG synthase enzymes encoded by nuclear C. velia genes.
Galactolipids are the principal components of the membranes of plastids from plants, red and green algae, and are also abundant in cyanobacteria (1). Galactolipids are believed to play a key role in photosynthesis (6, 24). Synthesis of galactolipids is reasonably well understood in cyanobacteria (5) and in primary plastids of plants and some red and green algae. Less is known about the galactolipid synthetic pathway in the primary plastids of glaucophytes (17) and very little in organisms bearing secondary plastids like C. velia. LC-MS/MS analysis confirmed that the chemical structures of C. velia galactolipids are similar to higher plant chloroplast MGDG and DGDG (Fig. 3). Indeed, fraction A (MGDG-like) and fraction B (DGDG-like) contained a monohexosyl and dihexosyl corresponding to their glycosyl polar head, as detected by the typical neutral loss of m/z 179 and 341 ions, respectively. Together with the results obtained by GC-MS and HPTLC (Figs. 1 and 2), this confirms that fractions A and B are monogalactolipid and digalactolipid, respectively. Further analysis of the LC-MS/MS-produced fragments of both fractions A and B showed that they contain a DAG backbone, confirming a structure similar to that of chloroplast MGDG and DGDG, respectively. We also found that C. velia galactolipids are enriched in very long polyunsaturated fatty acid chains, especially C20:5 (Figs. 3 and 4). Overall levels of unsaturated fatty acids are quite similar between S. oleracea and C. velia (i.e. 89 and 84%, respectively). However, both MGDG and DGDG of C. velia contained a fatty acid combination of C20:5 and C16:0, as opposed to the canonical C18:3 and C16:3 combination typical of most plant galactolipids (25, 26). Whether the C20 chain derives solely from de novo synthesis by a plastidial FASII pathway or through a cooperative synthesis involving both FASII and elongase(s) remains to be established.
MGDG and DGDG have also been identified in dinoflagellates; alveolate protists that also possess a secondary plastid (27). Electrospray ionization tandem mass spectrometry approaches showed that dinoflagellate galactolipids mainly contain C20:5, C18:5, and C18:4 fatty acids (28). Abundance of C20:5 fatty acid in both C. velia and dinoflagellate galactolipids is consistent with them both having obtained their plastids through a common ancestral secondary endosymbiosis of a red alga (14). Intriguingly, growth temperature modulates the level of unsaturation in dinoflagellate DGDG; low temperature favors C18:5 over C18:4, and C20:5 remained constant (28). We did not explore the effect of growth temperature on saturation levels in C. velia galactolipids, but this may be of interest in the general context of coral reef response to temperature variations.
We detected an abundance of galactose-containing lipids in whole cells of C. velia by HPTLC and GC-MS (Figs. 1 and 2). In plant chloroplasts, MGDG and DGDG typically represent 75% of the total lipids (1), and high levels of galactolipids in C. velia, a photosynthetic alveolate, are consistent with the importance of these lipids for photosynthesis. By contrast, previous lipidomic analysis of the Apicomplexan parasite T. gondii concluded that hexosyl lipids were only a minor lipid class (20, 21). Because most Apicomplexa are obligate intracellular parasites and depend on their host to provide most nutrients and precursors for membrane biogenesis, the apparent difference (on the basis of HPTLC) in relative abundance of galactolipids between C. velia and T. gondii likely reflects life style difference. Galactolipids are definitely present in Apicomplexa, as demonstrated by metabolic labeling and immunodetection approaches, but they are a minor component and apparently do not occur in the relict plastid of these parasites (18, 20). C. velia is thus like other autotrophic organisms but is different from related Apicomplexan parasites in that it contains a considerable amount of galactolipids that are mainly localized within its photosynthetic plastid (Fig. 5). We do not know which of the C. velia plastid membranes contain galactolipids. In plant chloroplasts, galactolipids occur in the thylakoids and the two bounding membranes. C. velia has an additional set of two bounding membranes (four in all), acquired during the secondary endosymbiotic process (29). Whether these extra membranes contain galactolipids will require subfractionation of C. velia plastids.
In the parasite T. gondii, a digalactolipid-like epitope was immunolocalized to the plasma membrane and the underlying alveoli (inner membrane complex), which together form the pellicle structure (20). Here, we show digalactolipids of the related C. velia mainly localize to the plastid. How is this difference rationalized? Plants are known to relocalize DGDG from the plastid to extraplastidial membranes during phosphate starvation (23, 30–32), allowing them to remobilize vital phosphates from phospholipids replaced by DGDG (24). One possible hypothesis is that Apicomplexa also delocalize DGDG from the apicoplast to extra-plastidial membranes. We tested this hypothesis by characterizing genes likely responsible for C. velia galactolipid synthesis and using these to search for similar synthesis pathways in Apicomplexa.
We identified two C. velia genes encoding putative galactosyltransferases sharing high sequence identity with plant chloroplast MGDG synthase and DGDG synthase (Fig. 7). Based on the glycosyltransferase classifications (CAZy is available on line (33)), the putative C. velia MGDG and DGDG synthases (CvMGD and CvDGD, respectively) belong to the typical GT28 and GT4 families, respectively (20, 34). In Arabidopsis, there are three isoforms of MGDG synthases as follows: AtMGD1, AtMGD2, and AtMGD3. MGD1 synthesizes the bulk of MGDG required for expansion of photosynthetic membranes, and MGD2 and -3 are up-regulated under stress conditions such as phosphate deprivation (26). AtMGD1 is nucleus-encoded but localizes to the inner membrane of the plant chloroplasts in plants courtesy of an N-terminal transit peptide (35). The C. velia cDNA we recovered is incomplete preventing us from predicting if CvMGD is plastid-targeted. A structural model of the plant MGD1, established using E. coli MURG as a template and validated by site-directed mutagenesis, identified residues critical for activity (17). UDP-Gal binding residues are conserved between CvMGD and plant MGD1 (Fig. 7), which is congruent with our substrate incorporation analyses. Critical DAG-binding site residues (17, 36) are also present in CvMGD, again consistent with our structural analyses showing DAG is the backbone. Plant MGDG synthase is activated by phosphatidic acid and phosphatidylglycerol (36), and two residues proposed to be involved in activation of AtMGD1 (Arg-260 and Trp-287) occur in C. velia (Fig. 7), perhaps suggesting the algal enzyme also undergoes such activation. Interestingly, CvMGD has an unusual insertion, from residues Phe-98 to Glu-115, not present in plants or unicellular algae. The insertion localizes in the vicinity of the hinge separating the two subdomains on the predicted MGDG synthase structural model (17). Two additional shorter insertions flank the β′2-α′2 region of the complete fold model in C. velia MGDG and could have roles in substrate discrimination.
Higher plants possess two DGDG synthase isoforms, DGD1 and DGD2. DGD2 forms a functionally related group with most algae. A structural model for A. thaliana DGDG synthase 2 (AtDGD2) identified residues critical for activity (37), and CvDGD has essential residue Val-32 (Fig. 7). AtDGD2 lacks transmembrane domains but associates with the outer plastid membrane via hydrophobic interactions, and tryptophan residues oriented toward the surface of AtDGD2 are important for the enzyme activity (37). CvDGD apparently also lacks transmembrane domains, and a hydrophobic region, particularly the critical Trp-26 and Trp-57, may mediate membrane interaction as in AtDGD2 (Fig. 7) (37). Moreover, Trp-139 and Trp-177, which retained partial activity in AtDGD2 when mutated from tryptophan to phenylalanine in AtDGD2 (37), occur as phenylalanine in CvDGD (Fig. 7), further supporting the authenticity of C. velia DGDG synthase.
Phylogenetic trees of MGDG and DGDG synthases (Fig. 8) show that both sequences from C. velia group with those of heterokont algae (diatoms and phaeophytes) to the exclusion of green algae and plants. These data are consistent with other evidence that the alveolate plastid is derived from a red algal endosymbiont rather than from a green lineage (14).
Considering the importance of galactolipids for oxygenic photosynthesis, it is somewhat surprising that the phylogenetic histories of enzymes involved had not been investigated in much detail. We therefore sought to provide a comprehensive picture of the galactolipid synthesis origin and evolution in eukaryotes. Our analyses revealed that enzymes of plant-like galactolipid biosynthesis are not closely related to cyanobacterial enzymes. This is surprising because galactolipids, although abundant in cyanobacteria and plastids, are rarely found in other bacteria (9), and it might therefore be anticipated that eukaryotic plastid enzymes were acquired from the cyanobacterial ancestor of the plastid. However, the phylogenies show that plant-like MGDG synthase is derived from a bacterial glycosyltransferase distinct from cyanobacterial MGDG (supplemental Fig. S4), probably by horizontal gene transfer. Remarkably, however, several bacterial species from this group (e.g. Chloroflexus auranticus and Treponema sp.) have also been shown to contain MGDG galactolipids (38, 39). None of these bacterial glycosyltransferases have yet been characterized, but this coincidence leads us to propose that they constitute a previously overlooked type of β-galactosyltransferase that catalyzes MGDG synthesis in a fashion similar to land plants. If confirmed, this would also explain why eukaryotes could replace a two-step cyanobacterium-type MGDG synthesis with a single enzyme while retaining the galactolipid product essential for their thylakoid membranes.
Plant-like DGDG synthase was also apparently acquired by a horizontal gene transfer event, although the gene donor is unclear in this case. However, because a glaucophyte and two red algae still retain the cyanobacterial DGDG synthase (encoded by the plastid genome), two evolutionary scenarios are possible. On one hand, the plant-like DGDG synthase may have been acquired before the divergence of the green and red plastid lineage (or all primary plastids), which co-existed in parallel with the plastid genome-encoded DGDG synthase of cyanobacterial origin for a certain amount of time, and eventually the two forms were differentially lost resulting in the current pattern. On the other hand, the plant-like DGDG synthase may have been acquired in the ancestor of the green algal lineage and horizontally transferred to the red plastid lineage after the divergence of cyanidiales. Both scenarios are complex considering the timing and course of plastid evolution, but it is possible they can be distinguished by presence/absence patterns of the two enzyme forms in an expanded data base of red algae and glaucophytes.
The eukaryotic phylogenies of MGDG and DGDG synthases show that multimeric enzyme forms found in some land plants originated by an ancestral duplication preceding the angiosperm divergence. This division is also in agreement with functional divergence between MGD1 and MGD2/MGD3 enzymes in A. thaliana, which have acquired distinctive roles in galactolipid biosynthesis depending on cellular conditions (26). Interestingly, our analyses also revealed the existence of many previously unrecognized paralogs of both MGDG and DGDG synthases in several other eukaryotic species. For example, the diatoms and the phaeophyte Ectocarpus contain three to four paralogs of the enzymes (Fig. 8, A and B). Although the particular functions of these paralogs remain unknown, it is noteworthy that a majority of them retains conserved essential residues involved in galactolipid synthesis in land plants. It is therefore likely that presence of several MGDG and DGDG synthase isoforms in these species reflects segregation of functions within the same biosynthetic pathway, perhaps in a fashion similar to those of A. thaliana.
CONCLUSION
Galactolipids are essential components of photosynthetic plastids and the most abundant polar lipids on earth. Here, we provide the first comprehensive study of galactolipids in a secondary plastid containing alga C. velia, a close relative of plastid containing Apicomplexan parasites of medical importance. The identification of galactolipids in photosynthetic plastids of C. velia, and their apparent absence in apicoplasts of Apicomplexan parasites, supports the notion that an abundance of MGDG and DGDG correlates with photosynthesis. Galactolipid metabolism in C. velia provides an extant model for a process that was either lost or remodeled after photosynthetic loss in Apicomplexa. Because the C. velia MGDG and DGDG synthase genes characterized here have no identifiable homologs in Apicomplexan parasite genomes, we must assume that synthesis of galactolipids in Apicomplexa is achieved by an alternative means. This is fascinating given that eukaryotes apparently invented an alternative galactolipid synthesis mechanism and abandoned the cyanobacterium-like endosymbiont mechanism during integration of the symbiont. Our data show that this pathway too was subsequently lost in preference for yet another system when photosynthesis was relinquished by Apicomplexa in favor of parasitism. In any case, galactolipid synthesis is absent in animal cells and constitutes an attractive target against which to develop drugs to combat malaria or toxoplasmosis.
Supplementary Material
Acknowledgments
We thank Dr. Falconet for help with the immunofluorescence analysis. We also thank W. A. Webster for technical assistance.
This work was supported in part by European Research Council FP7 Marie Curie Actions via an International Outgoing Fellowship Marie Curie fellowship (to C. Y. B.), National Health and Medical Research Council of Australia, CNRS, and Agence Nationale de la Recherche ReGal grant (to E. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- MGDG
- monogalactosyldiacylglycerol (1,2-diacyl-3-O-(β-d-galactopyranosyl)-sn-glycerol)
- DGDG
- digalactosyldiacylglycerol (1,2-diacyl-3-O-(α-d-galactopyranosyl-(1→6)-O-β-d-galactopyranosyl)-sn-glycerol)
- DAG
- 1,2-diacylglycerol-sn-glycerol
- HPTLC
- high precision thin layer chromatography
- GC-MS
- gas chromatography-mass spectrometry
- SQDG
- sulfoquinovosyl-diacylglycerol
- MGlcDG
- monoglucosyl-diacylglycerol 91,2-diacyl-3-O-(α-d-glucopyranosyl)-sn-glycerol
- MURG
- UDP-α-d-(N-acetyl)-glucosamine:N-acetylmuramyl-(pentapeptide)pyrophosphoryl-undecaprenol-4β-d-9N-acetyl)-glucosamyltransferase
- RACE
- rapid amplification of cDNA ends
- MGD
- monogalactosyldiacylglycerol
- DGD
- digalactosyldiacylglycerol.
REFERENCES
- 1. Jouhet J., Maréchal E., Block M. A. (2007) Prog. Lipid Res. 46, 37–55 [DOI] [PubMed] [Google Scholar]
- 2. Joyard J., Teyssier E., Miege C., Berny-Seigneurin D., Marechal E., Block M. A., Dorne A. J., Rolland N., Ajlani G., Douce R. (1998) Plant Physiol. 118, 715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keeling P. J. (2004) Am. J. Bot. 91, 1481–1493 [DOI] [PubMed] [Google Scholar]
- 4. Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2010) Prog. Lipid Res. 49, 128–158 [DOI] [PubMed] [Google Scholar]
- 5. Awai K., Kakimoto T., Awai C., Kaneko T., Nakamura Y., Takamiya K., Wada H., Ohta H. (2006) Plant Physiol. 141, 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kobayashi K., Kondo M., Fukuda H., Nishimura M., Ohta H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17216–17221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dörmann P., Benning C. (2002) Trends Plant Sci. 7, 112–118 [DOI] [PubMed] [Google Scholar]
- 8. Dörmann P., Hoffmann-Benning S., Balbo I., Benning C. (1995) Plant Cell 7, 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hölzl G., Zähringer U., Warnecke D., Heinz E. (2005) Plant Cell Physiol. 46, 1766–1778 [DOI] [PubMed] [Google Scholar]
- 10. Guo J., Zhang Z., Bi Y., Yang W., Xu Y., Zhang L. (2005) FEBS Lett. 579, 3619–3624 [DOI] [PubMed] [Google Scholar]
- 11. Ivanov A. G., Hendrickson L., Krol M., Selstam E., Oquist G., Hurry V., Huner N. P. (2006) Plant Cell Physiol. 47, 1146–1157 [DOI] [PubMed] [Google Scholar]
- 12. Steffen R., Eckert H. J., Kelly A. A., Dörmann P., Renger G. (2005) Biochemistry 44, 3123–3133 [DOI] [PubMed] [Google Scholar]
- 13. McFadden G. I., Reith M. E., Munholland J., Lang-Unnasch N. (1996) Nature 381, 482. [DOI] [PubMed] [Google Scholar]
- 14. Janouskovec J., Horák A., Oborník M., Lukes J., Keeling P. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 10949–10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ralph S. A., van Dooren G. G., Waller R. F., Crawford M. J., Fraunholz M. J., Foth B. J., Tonkin C. J., Roos D. S., McFadden G. I. (2004) Nat. Rev. Microbiol. 2, 203–216 [DOI] [PubMed] [Google Scholar]
- 16. Fichera M. E., Roos D. S. (1997) Nature 390, 407–409 [DOI] [PubMed] [Google Scholar]
- 17. Botté C., Jeanneau C., Snajdrova L., Bastien O., Imberty A., Breton C., Maréchal E. (2005) J. Biol. Chem. 280, 34691–34701 [DOI] [PubMed] [Google Scholar]
- 18. Maréchal E., Azzouz N., de Macedo C. S., Block M. A., Feagin J. E., Schwarz R. T., Joyard J. (2002) Eukaryot. Cell 1, 653–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bisanz C., Bastien O., Grando D., Jouhet J., Maréchal E., Cesbron-Delauw M. F. (2006) Biochem. J. 394, 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Botté C., Saïdani N., Mondragon R., Mondragón M., Isaac G., Mui E., McLeod R., Dubremetz J. F., Vial H., Welti R., Cesbron-Delauw M. F., Mercier C., Maréchal E. (2008) J. Lipid Res. 49, 746–762 [DOI] [PubMed] [Google Scholar]
- 21. Welti R., Mui E., Sparks A., Wernimont S., Isaac G., Kirisits M., Roth M., Roberts C. W., Botté C., Maréchal E., McLeod R. (2007) Biochemistry 46, 13882–13890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moore R. B., Oborník M., Janouskovec J., Chrudimský T., Vancová M., Green D. H., Wright S. W., Davies N. W., Bolch C. J., Heimann K., Slapeta J., Hoegh-Guldberg O., Logsdon J. M., Carter D. A. (2008) Nature 451, 959–963 [DOI] [PubMed] [Google Scholar]
- 23. Jouhet J., Maréchal E., Baldan B., Bligny R., Joyard J., Block M. A. (2004) J. Cell Biol. 167, 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benning C. (2009) Annu. Rev. Cell Dev. Biol. 25, 71–91 [DOI] [PubMed] [Google Scholar]
- 25. Moreau P., Bessoule J. J., Mongrand S., Testet E., Vincent P., Cassagne C. (1998) Prog. Lipid Res. 37, 371–391 [DOI] [PubMed] [Google Scholar]
- 26. Awai K., Maréchal E., Block M. A., Brun D., Masuda T., Shimada H., Takamiya K., Ohta H., Joyard J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10960–10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leblond J. D., Chapman P. J. (2000) J. Phycol. 36, 1103–1108 [Google Scholar]
- 28. Leblond J. D., Dahmen J. L., Evens T. J. (2010) Eur. J. Phycol. 45, 13–18 [Google Scholar]
- 29. Kalanon M., McFadden G. I. (2010) Biochem. Soc. Trans. 38, 775–782 [DOI] [PubMed] [Google Scholar]
- 30. Härtel H., Dormann P., Benning C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andersson M. X., Stridh M. H., Larsson K. E., Liljenberg C., Sandelius A. S. (2003) FEBS Lett. 537, 128–132 [DOI] [PubMed] [Google Scholar]
- 32. Andersson M. X., Larsson K. E., Tjellström H., Liljenberg C., Sandelius A. S. (2005) J. Biol. Chem. 280, 27578–27586 [DOI] [PubMed] [Google Scholar]
- 33. Coutinho P. M., Henrissat B. (1999) J. Mol. Microbiol. Biotechnol. 1, 307–308 [PubMed] [Google Scholar]
- 34. Kelly A. A., Dörmann P. (2002) J. Biol. Chem. 277, 1166–1173 [DOI] [PubMed] [Google Scholar]
- 35. Miège C., Maréchal E., Shimojima M., Awai K., Block M. A., Ohta H., Takamiya K., Douce R., Joyard J. (1999) Eur. J. Biochem. 265, 990–1001 [DOI] [PubMed] [Google Scholar]
- 36. Dubots E., Audry M., Yamaryo Y., Bastien O., Ohta H., Breton C., Maréchal E., Block M. A. (2010) J. Biol. Chem. 285, 6003–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ge C., Georgiev A., Öhman A., Wieslander Å., Kelly A. A. (2011) J. Biol. Chem. 286, 6669–6684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Livermore B. P., Johnson R. C. (1974) J. Bacteriol. 120, 1268–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Knudsen E. J., Bryn K., Ormerod J. G., Sirevag R. (1982) Arch. Microbiol. 132, 149–154 [Google Scholar]
- 40. McConville M. J., Homans S. W., Thomas-Oates J. E., Dell A., Bacic A. (1990) J. Biol. Chem. 265, 7385–7394 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.