Abstract
A class of anti-virulence compounds, the salicylidene acylhydrazides, has been widely reported to block the function of the type three secretion system of several Gram-negative pathogens by a previously unknown mechanism. In this work we provide the first identification of bacterial proteins that are targeted by this group of compounds. We provide evidence that their mode of action is likely to result from a synergistic effect arising from a perturbation of the function of several conserved proteins. We also examine the contribution of selected target proteins to the pathogenicity of Yersinia pseudotuberculosis and to expression of virulence genes in Escherichia coli O157.
Keywords: Bacteria, Bacterial Genetics, Gene Regulation, Protein Drug Interactions, Protein Secretion, Protein Targeting, Transcription
Introduction
As the prevalence of antibiotic-resistant strains increases, targeting virulence determinants of pathogenic bacteria has become an attractive alternative to the use of traditional bactericidal antibiotics (1–5). A key feature of this strategy is that the virulence-blocking compounds spare the endogenous microflora and thereby exert less selective pressure, which in turn should reduce development of resistance. One potential target is the bacterial type three secretion system (T3SS),7 a conserved protein injection organelle that is central to the virulence of many human, animal, and plant pathogens including Chlamydia sp., enteropathogenic and enterohemorrhagic Escherichia coli, Pseudomonas aeruginosa, Salmonella sp., Shigella sp., and Yersinia sp. (6, 7). With the T3SS, the pathogen translocates effector proteins into the cytosol of the host cell and thereby creates a niche that allows bacterial growth. A class of virulence-blocking compounds, the salicylidene acylhydrazides, was originally identified as putative T3SS inhibitors in Yersinia pseudotuberculosis (8). In a number of publications the compounds have been shown to be broadly effective in negatively affecting the function of the T3SS in a number of pathogenic bacteria including Chlamydia sp. (9–12), Salmonella typhimurium (13, 14), Y. pseudotuberculosis (15), Shigella sp. (16), and E. coli O157 (17). Two reports describe activity of the compounds in vivo (13, 18) and thus indicate that T3SS inhibitors have the potential to be developed into novel anti-bacterial agents (19, 20). In addition, small molecule inhibitors can be used as chemical probes to study the role of T3SS in bacterial pathogenesis (12, 21, 22). To date, the mechanism of inhibition for the salicylidene acylhydrazides has been unclear with several mechanisms being postulated, including direct effects on the T3SS machinery or on regulatory proteins that affect T3SS expression (15–17). The activity of salicylidene acylhydrazides on Chlamydia trachomatis can be reversed by the addition of iron, suggesting a possible link to iron availability in the cell (11).
Although target-based screening can identify compounds that perturb the function of the selected protein, the activity is often lost when the compounds are tested on bacterial cells due to a lack of bacterial cell permeability (23). Phenotypic screens circumvent this challenge and directly provide compounds that are active on the cellular level. The salicylidene acylhydrazides were identified using this strategy, and this is likely the underlying reason for the broad activity spectrum observed for these T3SS inhibitors. The drawback is, however, that the mode of action at the molecular level has to be studied at a later stage. Identification of the cellular targets for the salicylidene acylhydrazides constitutes a crucial step in understanding their true mode of action. Moreover, such data are essential for the structure-led design and improvement of any potential therapeutic compound. Affinity chromatography, expression-cloning technologies, and microarrays are among the many strategies that can be employed for target deconvolution (24). To help understand the molecular mechanism by which salicylidene acylhydrazides affect the T3SS, we aimed to identify the target proteins bound by this class of novel antibacterial compounds using an affinity reagent strategy (25). In this study we describe synthesis of a T3SS inhibitor affinity reagent and the isolation of putative target proteins from E. coli O157. By a series of in vitro experiments, we show that the compounds directly and selectively interact with the target proteins WrbA, Tpx, and FolX. The genes encoding the target proteins were deleted individually in both E. coli O157 and Y. pseudotuberculosis, and we show that the proteins are involved in regulation of T3SS gene expression. Our work provides the first identification of the cellular targets for this group of compounds and convincing evidence that their mode of action is likely to result from a synergistic effect arising from a perturbation of the function of several conserved proteins.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Construction of Mutants
Bacterial strains and plasmids used in this study are listed in supplemental Table S1. Mutations in tpx were made using QuikChange II (Stratagene) and were confirmed by sequencing. Oligonucleotides (Invitrogen) are listed in supplemental Table S2. E. coli O157 strains were cultured in LB, DMEM, or MEM-HEPES media (Sigma) supplemented with standard concentrations of antibiotics (26).
Affinity Chromatography Using Affi-Gel-labeled Salicylidene Acylhydrazides
The stepwise synthesis of ME0055-Aff is described in supplemental Protocol S1. E. coli O157 strain TUV93–0 was cultured in MEM-HEPES media at 37 °C to an A600 = 0.8. Cells were harvested and lysed using a French pressure cell. No subsequent separation steps were used to ensure that cell wall, cell membrane, and cytosolic proteins were represented in the lysate. The bacterial lysate was mixed with PBS-balanced ME0055-Aff beads overnight at 4 °C. The beads were separated by centrifugation at 500 × g for 5 min. An aliquot of the supernatant was saved for analysis, and the rest of the supernatant was discarded. The beads were washed 10 times using 10 volumes of PBS and separation by centrifugation at 500 × g for 5 min. The beads were mixed gently in the same volume of 20 μm ME0055 and incubated at room temperature for 30 min. Supernatant was collected by centrifugation at 500 × g for 5 min. The elution and centrifugation step was repeated with 200 μm ME0055 and 1% acetic acid, respectively. Equal volumes of washes and eluates were loaded onto the protein gels. Samples were visualized using Colloidal Blue Stain (Invitrogen), and bands were excised for subsequent in-gel digestion and analysis (27). Proteins analyzed by peptide mass fingerprinting were given a MOWSE score (28) to indicate the probability of the identification being correct. We set a threshold MOWSE value of 100 or greater, ensuring a significance of p < 10−8.
Screening for Protein Secretion and Western Blot Analyses
Eighteen bacterial strains (supplemental Table S1) were cultured overnight in LB and diluted to an A600 ≤ 0.05 in prewarmed MEM-HEPES medium. Cultures were grown at 37 °C to an A600 = 0.8, and then secreted proteins were extracted by TCA precipitation as described previously (29). Proteins were analyzed by SDS-PAGE, and Western blotting for Tir and EspD was carried out as described previously (29). Strain 430 displayed no reduction in secretion upon the addition of 20 μm ME0052, ME0053, ME0054, or ME0055 to the culture when assessed by Western blotting for Tir or EspD.
Re-sequencing of ZAP 430
Genomic DNA for the ZAP 430 strain was extracted using a QIAamp DNA extraction kit. The sequencing was performed in the Sir Henry Wellcome Functional Genomics Facility, University of Glasgow, using the Illumina Genome Analyzer II platform. The purified genomic DNA was randomly fragmented with a Bioruptor Sonicator (Diagenode Inc., NJ) followed by the ligation of adapters, and genomic library generation was performed according to the standard Illumina protocol. The genomic library at a concentration of 3 pm was then loaded onto a single lane of the Illumina flow cell, and the DNA clusters were generated using the Illumina Single Read Cluster Generation kit Version 4 (Illumina Inc.). Subsequently, 70 cycles of single end sequencing were performed. The raw data containing read sequences and associated qualities were generated with Genome Analyzer Pipeline v.1.5 (Illumina Inc.). Of 15,712,834 70-bp-long reads generated, 14,347,057 were successfully mapped to the E. coli O157:H7 strain EC4115 complete genome (GenBankTM accession NC_011353) using Maq software with the default parameter set, which resulted in 179× genome coverage. Subsequently, the SNPs were identified with Maq and filtered with the command “maq.pl SNPfilter -q 40 -w 5 -N 2 -d 3 -D 256 -n 20 -Q 40,” where parameters were chosen as suggested for the bacterial genomes (30). The latter identified 1348 SNPs (supplemental Table S3).
Far Western Blotting
The stepwise synthesis of ME0052-Bio and ME0055-Bio are described in supplemental Protocol S1. Target proteins were overexpressed in E. coli BL21 (λDE3). Bacterial pellets were lysed using 8 m urea, and the supernatant was collected by centrifugation at 14,000 × g for 1 min. The Far Western analysis was performed as described previously (31) using Novex NuPAGE 4–12% BisTris SDS-PAGE gels (Invitrogen), 20 μm ME0052-Bio as the probe, and HRP-conjugated streptavidin (Invitrogen) for the detection of biotin. Protein loading was checked by using an anti-His antibody that showed equivalent levels of overexpressed target protein in each lane (data not shown).
Analyses of Transcript Levels
Transcriptional profiling of E. coli O157 strains was carried out essentially as described previously (32). Overnight cultures of E. coli TUV-930 or defined mutants were grown in MEM-HEPES (Sigma) supplemented with 250 nm Fe(NO3)2 and glucose to a final concentration of 0.2% and then diluted to an A600 = 0.1 in the same media. At an A600 = 0.8, 15-ml cultures were stabilized in an equal volume of RNAprotect (Qiagen), and RNA was extracted using a Qiagen RNeasy Mini kit. DNA contamination was removed by DNase I treatment (Ambion). Total RNA was assessed for quality and quantified using an Agilent 2100 Bioanalyzer. Synthesis of cDNA and labeling of total RNA was performed using an Amersham Biosciences CyScribe post-labeling kit as per the manufacturer's instructions. The cDNA was hybridized to 70-mer-spotted oligonucleotide arrays containing ORFs from E. coli K-12, E. coli Sakai VT2, and E. coli EDL933 (University of Birmingham) using a Genomic Solutions GeneTac hybridization machine. Hybridized slides were scanned using a Genepix 4000A scanner and GenePix 7.0 software (Axon Instruments, Union City, CA). Data were analyzed using Genespring GX 7.3.1 (Agilent).
Quantitative PCR
Triplicate wild-type and mutant cultures were grown up in MEM-HEPES. RNA extractions were carried out using a Qiagen RNeasy Mini kit, and cDNA synthesis was carried out using a Qiagen QuantiTectTM Reverse Transcription kit. Duplicate quantitative PCRs were carried out using a Qiagen QuantifastTM SYBR® Green PCR kit and Stratagene MX3000, and oligonucleotide primers are listed in supplemental Table S2. All the experiments were performed according to the manufacturer's instructions.
Visualization of Flagella and EspA Filaments
Flagella and EspA filaments were visualized using immunofluorescence microscopy (17). Images were processed and quantified using Volocity suite software (PerkinElmer Life Sciences). Bacterial motility was assayed using soft agar plates as described previously (33).
Purification and Biophysical Studies of Tpx from Y. pseudotuberculosis (ypTpx)
ypTpx and ypTpxC61S were cloned, expressed, and purified as described previously (34). For NMR spectroscopy, ypTpx was expressed in M9 minimal media, with the nitrogen source substituted with 15NH4Cl. Experiments to identify chemically-shifted residues were performed using triple-labeled Tpx (13C,2H,15N), produced by culture in algal hydrolysate-based media (Cambridge Isotopes Ltd). For chemical shift perturbation studies, ypTpx NMR samples were prepared to contain 100 μm protein in 13 mm KPi, pH 7.4, 33 mm NaCl, and 5% D2O, and 15N FAST-HSQC (35) spectra were recorded at 298 K and 14.1 T using a Bruker AVANCE 600 spectrometer equipped with a 5-mm TCI cryoprobe. Compound was added from a concentrated stock solution in deuterated DMSO (dDMSO), and reference spectra recorded with an equal concentration (0.33% v/v) of dDMSO added. Backbone resonance assignment was achieved using triple resonance, and TROSY (transverse relaxation optimized spectroscopy) triple resonance experiments were recorded from 150 to 500 μm samples at 308 K. Data were processed using AZARA and analyzed using CCPNMR analysis (36).
Analytical Ultracentrifugation (AUC) of Tpx and ME0052
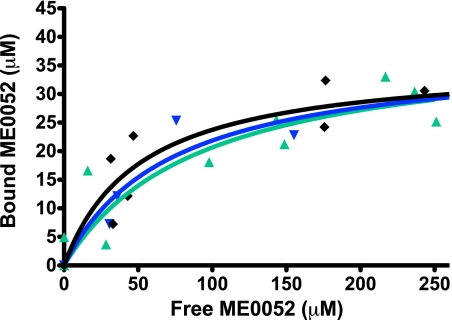
Before AUC analysis, ypTpx and ypTpx C61S were dialyzed against 20 mm Tris, pH 7.4, 50 mm NaCl. Sedimentation velocity experiments were performed using an Optima XL-I analytical ultracentrifuge (Beckman Coulter, Palo Alto, CA) and an An-50 Ti rotor. Data were collected at 49,000 rpm at 4 °C for ypTpx under oxidizing (with 10 mm H2O2) and reducing (with 5 mm DTT) conditions and for the C61S mutant. 360 μl of sample (100 μm protein solution with ME0052 at concentrations ranging from 0 to 250 μm) and reference solvent (20 mm Tris, pH 7.4, 50 mm NaCl, 1% v/v DMSO) were loaded into 3-mm path length charcoal-filled epon double-sector centerpieces. Rayleigh interference and absorbance (at 280 and 395 nm) data were collected every 15 min for 25 h (1 replicate of absorbance data in continuous mode with a radial step size of 0.005 cm). A wavelength of 395 nm was used to monitor the sedimentation of ME0052 only, as the protein does not contribute to the signal at this wavelength.
A two-stage analysis of sedimentation velocity data was used to determine the amount of ME0052 bound to oxidized and reduced ypTpx and the C61S mutant. First, the number of species in the system was evaluated using continuous distribution (c(s)) analysis, performed with SEDFIT software (37). The sedimentation coefficients (s20,0w) of oxidized and reduced ypTpx and the C61S mutant are 3.04, 2.62, and 2.80 s, respectively (data not shown); thus, a peak with this sedimentation coefficient in the c(s) distribution derived from sedimentation velocity data acquired at 395 nm could be attributed to ypTpx bound to ME0052. At ME0052 = 0 μm no peaks were resolved from the data; at higher concentrations two peaks were evident: one at very low s (corresponding to unbound ME0052) and the second close to 2.7 s (ME0052 bound to protein). The concentration of free and bound compound was determined by fitting the data with a (two species) non-interacting discrete species model in SEDFIT. An extinction coefficient of 11,100 m−1 cm2, determined from the linear dependence of absorbance at 395 nm with ME0052 concentration, was used to calculate the concentration of compound returned by the non-interacting discrete species model. The resultant reduced data were then fitted with a single-site-binding nonlinear regression model using the formula Y = Bmax × X/(Kd + X), where Y is the concentration of ME0052 bound to protein (determined by non-interacting discrete species), X is the total concentration of ME0052 (i.e. bound and unbound), Bmax is the concentration of compound required to saturate the protein (i.e. at maximal binding), and Kd is the equilibrium dissociation constant or concentration of compound required to reach half-maximal binding. The stoichiometry of interaction between ME0052 and ypTpx was determined by dividing Bmax by the total protein concentration.
RESULTS
Affinity Chromatography Identifies 16 Putative Salicylidene Acylhydrazide Target Proteins
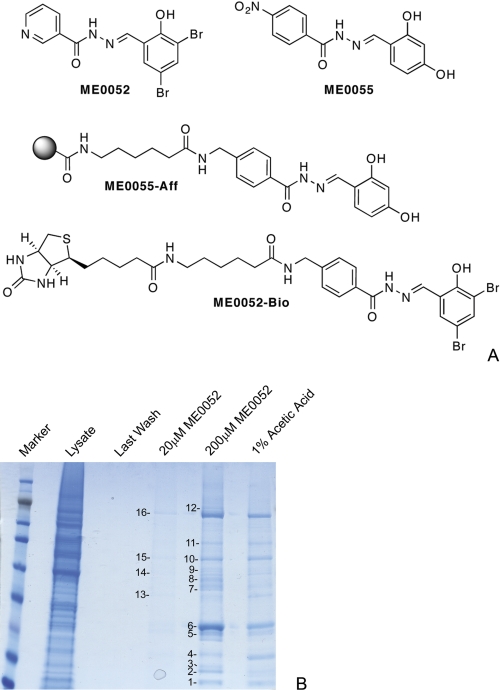
The two salicylidene acylhydrazides ME0052 and ME0055 were selected for chemical derivatization. ME0052 has been extensively studied and proved to display low toxicity against mammalian cells and activity against T3S in Y. pseudotuberculosis, C. trachomatis, Salmonella enterica, Shigella flexneri, and E. coli O157:H7 (10, 11, 14–17), and ME0055 was selected based on its low toxicity and high activity against T3S in E. coli O157 (17). The Affi-Gel- and biotin-labeled salicylidene acylhydrazide derivatives ME0055-Aff and ME0052-Bio (Fig. 1A) were synthesized by combined solid phase and solution phase chemistry. The position for attachment of the linker was based on previous structure-activity data (15, 38, 39). ME0055-Aff was used in affinity chromatography experiments to enrich for inhibitor binding proteins from E. coli O157 lysates prepared using a French pressure cell. After incubation, washing, and elution using stepwise increasing concentrations of unlabeled compound, proteins were resolved by SDS-PAGE and visualized with Colloidal Blue Stain (Fig. 1B). Bands were selected corresponding to the lanes derived from the fractions that had been eluted using unlabeled compound but that were less abundant in the lane associated with the acetic acid “stripping” of the beads. These were postulated to be true target proteins and not simply bound to the Affi-Gel bead itself. Bands from the gel were excised to permit in-gel trypsin digestion and analysis by liquid chromatography electrospray ionization tandem mass spectrometry (27). Using this approach, we identified 16 proteins (supplemental Table S4) that were bound by ME0055-Aff.
FIGURE 1.
A, shown are structures of compounds used and visualization of the proteins from the pulldown assay. Structures of the inhibitory compounds, ME0052 and ME0055, and derivatives, ME0055-Aff and ME0052-Bio, used for the identification of target proteins are shown. Supplemental Protocol S1 details the syntheses. ME0055-Aff was used in affinity chromatography experiments to enrich for putative target proteins; ME0055-Bio was used for Far Western experiments. B, visualization of the protein fractions from the affinity chromatography procedure is shown. Lysates from E. coli O157 were incubated with ME0055-Aff and washed, and then proteins were eluted using 20 and 200 μm unlabeled ME0055. The beads were stripped using 1% acetic acid. Equal volumes of samples from each fraction were separated by SDS-PAGE and then visualized using Colloidal Blue Stain. Proteins were excised, subjected to in-gel trypsin digestion, and analyzed by LC-electrospray ionization tandem MS. Each protein identified from the gel is numbered sequentially and is listed in supplemental Table S4.
Identification and Resequencing of an E. coli O157 Strain Insensitive to Salicylidene Acylhydrazides
As an independent screen, a panel of 18 clinical E. coli O157 isolates selected to represent a diverse range of phage types were tested for sensitivity against four salicylidene acylhydrazides (ME0052, ME0053, ME0054, and ME0055) that previously proved effective against E. coli O157 (17). Western blot analysis of Tir secretion in the presence of 20 μm concentrations of each of the compounds revealed a strain, ZAP 430, that showed no inhibition of Tir secretion upon the addition of any of the four compounds. Illumina re-sequencing of the genome (EMBL, ERP000335) and comparison against EC4115, a strain sensitive to the salicylidene acylhydrazides, demonstrated that ZAP 430 carried numerous mutations (supplemental Table S3). These mutations include the gene encoding WrbA, one of the target proteins identified from the affinity chromatography approach. Previous work has suggested that the salicylidene acylhydrazides may function by directly binding and perturbing the proteins associated with the T3SS basal apparatus (16). ZAP 430 had no mutations in the genes encoding the structural proteins of the T3SS; this insensitive strain contained T3SS proteins identical to both EDL933 and EC4115, reflecting the high level of conservation of these proteins.
ME0052-Bio Binds to Tpx, WrbA, and FolX from E. coli O157
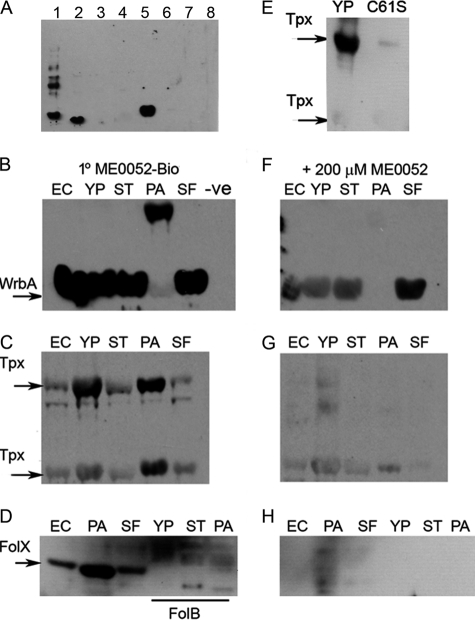
To facilitate studies of the putative binding proteins, we attempted to clone the corresponding genes. From the 16 amplified products, 12 genes were successfully cloned into a pET151 vector and verified by sequencing. Each of the 12 plasmids was tested for protein overexpression, with seven clones producing a protein of the expected size. These seven proteins were (or were encoded by): Tpx, FolX, z2714, z3974, WrbA, SurA, and StcE. Interactions with salicylidene acylhydrazides were tested in Far Western experiments using 20 μm concentrations of the biotin-labeled compound ME0052-Bio as a probe (Fig. 2A). From the seven overexpressed proteins, three gave clear signals after Far Western analysis: Tpx, a thiol peroxidase (40), WrbA a NAD(P)H quinine oxidoreductase (41), and FolX, a dihydroneopterin-tri-P-epimerase (42). No signal or a very weak signal was seen corresponding to the other four proteins or for E. coli lysates with no overexpressed target protein (Fig. 2A). We, therefore, decided to focus on Tpx, WrbA, and FolX for further studies.
FIGURE 2.
Far Western probing to examine binding of ME0052-Bio to putative target proteins. A, putative target proteins were cloned into pET151 and expressed in E. coli BL21 (λDE3) cells before lysis, separation, and transfer to a nitrocellulose membrane. ME0052-Bio was used as a probe, and interactions were detected using HRP-conjugated streptavidin. Proteins probed were (or encoded by) Tpx (1), FolX (2), z2714 (3), z3974 (4), WrbA (5), SurA (6), and StcE (7). Lane 8 contains bacterial lysate with no overexpressed protein as a negative control. The same protocol was used to subsequently probe proteins from E. coli O157 (EC), Y. pseudotuberculosis (YP), S. typhimurium (ST), P. aeruginosa (PA), and S. flexneri (SF) or empty vector control (−ve). Panels show overexpression of target proteins: WrbA (B), Tpx (C), FolX and FolB (D). Mutation of Tpx Cys-61 (C61S) affected binding of ME0052-Bio (E). The addition of 200 μm unlabeled ME0052 (F–H) strongly affected the binding of the biotinylated probe.
ME0052-Bio and ME0055-Bio Bind to WrbA, Tpx, and FolX from Multiple Pathogens
Previous work has demonstrated that salicylidene acylhydrazide compounds inhibit expression and function of the T3SS in numerous pathogens (19). We aimed to test if WrbA, Tpx, and FolX were conserved targets resulting in a common mechanism of inhibition. Cloning and overexpression of the genes encoding WrbA and Tpx were carried out from five different pathogens: E. coli O157, S. typhimurium, S. flexneri, P. aeruginosa, and Y. pseudotuberculosis. The gene encoding FolX, which is less conserved, was cloned from three pathogens: E. coli O157, P. aeruginosa, and S. flexneri. FolX is not present in Y. pseudotuberculosis, but this species does carry a structurally and functionally similar protein, FolB, as do P. aeruginosa and S. typhimurium. To examine the specificity of the inhibitor-target interaction further, we also cloned and overexpressed FolB from these three species. Using ME0052-Bio as a probe for Far Western experiments, we observed binding with overexpressed WrbA, Tpx, and FolX from all the species tested (Fig. 2, B–D). Protein loading was shown to be equivalent for each lane by probing the samples with an anti-His antibody (data not shown). After separation by SDS-PAGE, Tpx was detected as a monomer of 19 kDa as well as a dimer of 38 kDa, which was verified by Tandem MS identification of the excised bands. The presence of both monomer and dimer after SDS-PAGE indicated that Tpx had a stable structure and that heat and detergent were unable to completely denature the protein. The addition of ME0052-Bio resulted in a signal for both dimer and monomer of Tpx (Fig. 2C). Experiments were repeated using ME0055-Bio and gave the same results, albeit with a higher level of background signal. No interaction between ME0052-Bio and FolB was detected (Fig. 2D), indicating that the compound does not bind this protein when an equivalent amount of protein is used as bait. This is despite the fact that FolB and FolX from E. coli display 68% sequence similarity (as determined using ClustalW), and the structures superimpose with a root-mean-square-deviation of 1.7 Å (PDB codes 2O90 and 1B9L, respectively). The addition of 200 μm unlabeled ME0052 (Fig. 2, F–H), in competition with ME0052-Bio, resulted in a marked reduction in ME0052-Bio binding to WrbA, Tpx, and FolX irrespective of species.
To investigate the specificity of the interaction between target protein and inhibitor, we focused further on Tpx, a thiol peroxidase previously shown to be part of the oxidative stress defense system that uses reducing equivalents from thioredoxin (Trx1) and thioredoxin reductase (TrxR) to reduce alkyl hydroperoxides (40). Tpx is attractive to study as it is easy to purify, and conditions for protein crystallization have been determined (34, 43). The catalytically important residues of Tpx have been previously identified. Tpx is inactivated by mutation of cysteine 61 that is essential for intramolecular disulfide bond formation (40). The mutant protein species is present only in the structurally reduced form, as it cannot undergo the disulfide bond formation critical for the catalytic cycle of the protein. C61S Tpx formed a less stable dimer, and after SDS-PAGE, the majority of the protein was seen as a monomer (data not shown) rather than as a dimer for wild-type Tpx. The C61S mutation reduced ME0052-Bio binding (Fig. 2E).
NMR Chemical Shift Perturbation Shows That ME0052 Binds to Tpx from ypTpx
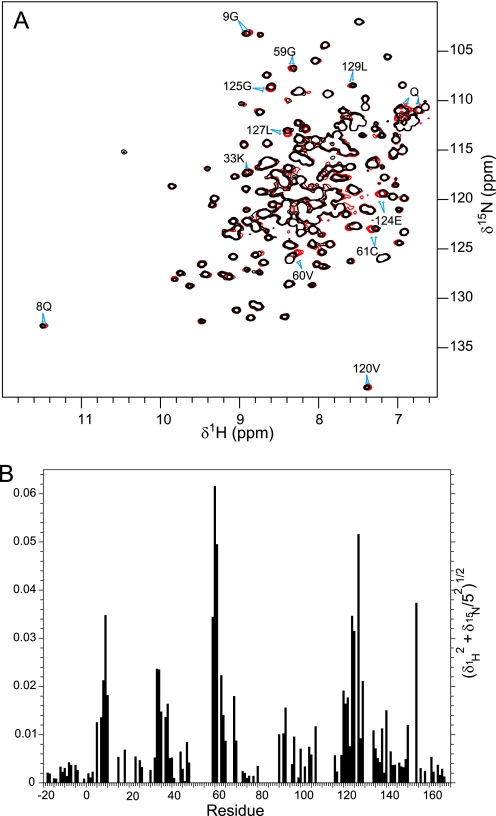
We validated the interaction between ME0052 and Tpx by NMR chemical shift perturbation. 15N heteronuclear Single Quantum Coherence (HSQC) spectra of 100 μm ypTpx recorded in the absence and presence of excess ME0052 revealed chemical shift changes specific to a small number of cross-peaks originating from particular backbone amides (Fig. 3). The shift changes increased gradually with increasing ligand concentration, indicating that the free and ligand-bound protein were in fast exchange and were saturated by 200 μm ME0052. No shifts were seen with the addition of DMSO alone. 81% of the backbone amide resonances were assigned, and the chemical shift changes mapped to the structure. These data indicate that the compound binds to a specific site on ypTpx, inducing only minor conformational changes.
FIGURE 3.
NMR chemical shift perturbation of ypTpx by ME0052. A, 15N HSQC spectra of 100 μm ypTpx in the absence (black) and presence (red) of 200 μm ME0052. A selection of the most perturbed cross peaks is indicated with their assignments, and a perturbed glutamine side chain is indicated (Q). B, a histogram of the weighted amide chemical shift perturbation by residue reveals clusters of residues affected by ligand binding.
Identification of Residues Involved in the Binding of ME0052 and Modeling of the Site
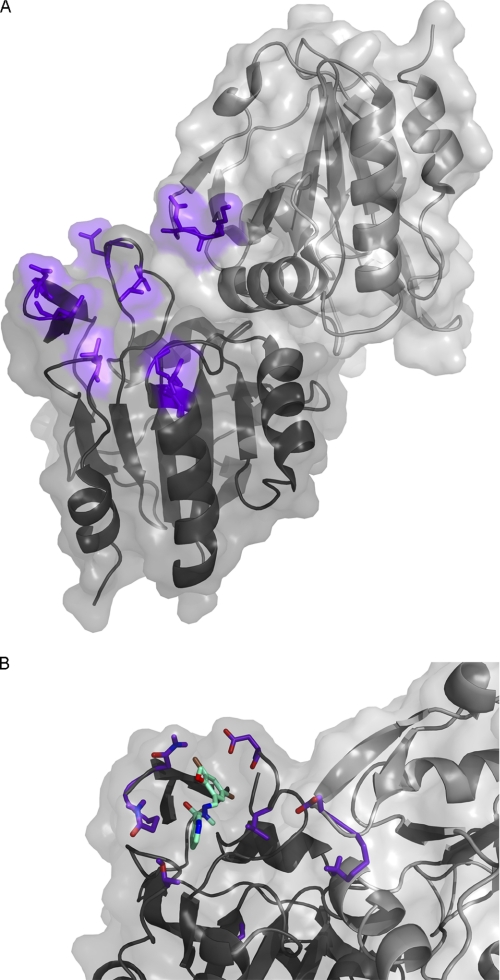
Based on the residues shown to be perturbed by NMR, ME0052 was docked into the published structure of E. coli O157 Tpx (ecTpx) (PDB code 3HVV (43)) using the program Autodock Vina in PyMOL (44, 45). There is a high sequence identity between Tpx from E. coli and Y. pseudotuberculosis, comprising 80% overall identity with 100% similarity in the residues affected by binding. As PDB code 3HVV is the structure of a C61S mutant, residue 61 was replaced with a cysteine to mimic the reduced structure. Residues that were shown to have the greatest NMR chemical shifts are colored purple for reference (Fig. 4). The binding site of ME0052 with Tpx forms near the dimer interface and comprises both subunits albeit with an unequal contribution, as can be seen on Fig. 4A. The residue with the largest shift is Val-60, adjacent to the catalytically active residue of Tpx.
FIGURE 4.
Mapping of residues involved in drug binding and ligand docking. A, shown is a schematic representation of the structure of ecTpx (PDB code 3HVV (43)) colored purple by the shifts observed when ME0052 is added to ypTpx. Subunit A is in dark gray, and subunit B is in lighter gray. B, shown is a surface representation of the ecTpx dimer, with the docked compound ME0052 (cyan) based on the residues with the highest shift (purple sticks) using Autodock Vina. The figures were created in PyMOL (56).
Determination of Binding Affinity and Stoichiometry
The salicylidene acylhydrazide compounds have a defined absorbance peak at 395 nm. This enabled binding of the compounds to ypTpx in solution to be examined using AUC. The concentration of ME0052 bound to ypTpx was determined for a range of total compound concentrations. These data were used to calculate the dissociation constant (Kd) and the stoichiometry of binding. Using this approach, the Kd for binding of ME0052 to oxidized ypTpx was determined to be 51 and 71 μm to reduced ypTpx (Fig. 5). The Kd for the C61S mutant revealed an approximate 2-fold reduction in binding (93 μm) (Fig. 5). The correlation coefficients (r = 0.8037, 0.9133, and 0.8440 for C61S, reduced, and oxidized ypTpx) of the nonlinear best fits confirmed the strength of dependence between the amount of bound drug and free drug. The stoichiometry was determined to be one molecule of ME0052 compound bound per ypTpx dimer.
FIGURE 5.
AUC analysis of ME0052 binding to ypTpx. A line graph shows the binding of ME0052 to purified ypTpx during AUC experiments. The lines represent different redox states for Tpx: oxidized (black diamonds), reduced (blue inverted triangles) and forced reduced mutant, C61S (green triangles). The amount of bound ME0052 was used to calculate the Kd values described in the text.
WrbA, Tpx, and FolX Contribute to Bacterial Gene Regulation
To assess any role of the three target proteins for the expression and function of the T3SS, the corresponding gene encoding each protein was deleted individually in both E. coli O157 and Y. pseudotuberculosis. In addition, mutagenesis was used to modify ecTpx and introduce a serine residue in place of the cysteine (Cys-61) that is essential for Tpx function. This was achieved by cloning the gene encoding ecTpx into a plasmid (pDW141), site-directed mutagenesis, and then allelic exchange to replace the wild-type copy of the tpx gene with the mutant allele. The final strain, E. coli Tpx_C61S, was used in subsequent assays to compare the phenotype of a deletion of Tpx with that of a non-functional mutant. The growth rate of all the E. coli O157 mutants tested was not affected during culture in MEM-HEPES medium used for the transcriptomic profiling. Using microarrays, the gene expression profile of each E. coli O157 mutant was compared with that of the wild type. Transcriptional changes for each mutant are listed in supplemental Table S5.
The most striking finding was that some 27 genes were significantly (p < 0.05) affected in all four mutants. These genes could be grouped into six Gene Ontology (GO) categories: cell motility, locomotory behavior, localization of the cell, behavior, locomotion, and flagellar motility. Nine genes required for flagellar synthesis were significantly (>2-fold change, p < 0.05) repressed in all four mutants. The genes encoding the T3SS were also affected, but in contrast to the flagellar genes, these were up-regulated, with the greatest changes seen for genes encoding the basal apparatus of the T3SS, such as escJ, escC, and escD (>2-fold change, p < 0.05). Previous work has demonstrated that the expression of flagella and the T3SS of E. coli O157 are cross-regulated to allow either motility or attachment as appropriate (44). The strong repression of flagella and stimulation of T3SS expression demonstrates that the three target proteins we have identified are required for normal expression and appropriate regulation of these key virulence factors.
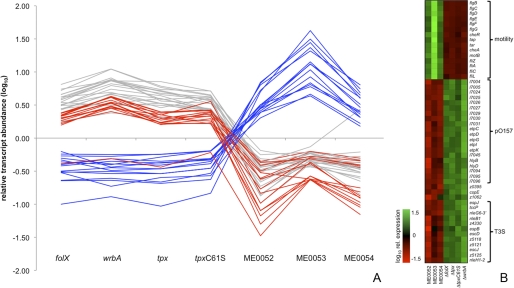
The transcriptomic data are of particular interest when compared with our previous work that determined the gene expression profile of E. coli O157 cultured in the presence or absence of salicylidene acylhydrazide compounds (17). In this earlier study we demonstrated that the addition of salicylidene acylhydrazides resulted in strong repression of the T3SS and up-regulation of flagella; that is, the reciprocal pattern of gene expression compared with the four mutants described here. Some 39 genes repressed by the addition of the inhibitors were up-regulated >1.5-fold in all mutants (15 of which were associated with the T3SS), and conversely, 15 genes stimulated by addition of the inhibitors were repressed in the mutants (13 of which were associated with motility). This pattern of gene expression can be most clearly seen by plotting on a single graph the -fold change of the genes that were significantly (p < 0.05) affected in both studies (Fig. 6, A and B). The C61S mutant displayed a transcriptional profile similar to that of the mutant with the defined deletion. The transcriptomic data were validated by quantitative PCR to examine transcript levels for selected genes; transcript levels for fliC confirmed the differences observed in flagellar expression with escJ tested to confirm the up-regulation of the T3SS genes. For example, the E. coli Tpx_C61S strain showed a 154-fold reduction of fliC expression and a 3-fold increase in escJ transcript level compared with the wild type.
FIGURE 6.
Comparison of gene expression between bacteria cultured with salicylidene acylhydrazides and defined mutants. Microarray data were filtered for transcripts that showed significant changes in gene expression (>1.5-fold, p < 0.05) when treated with salicylidene acylhydrazides (ME0052, ME0053, ME0054) and that also showed >5-fold changes in transcript abundance when compared with the defined mutants. A, shown is a graphic representation of gene expression. Red lines indicate genes associated with the T3SS, blue lines indicate transcripts associated with flagella and motility, and gray lines represent other genes. B, shown is a heatmap of gene expression. Gene names are indicated to the right of the heatmap, and experimental conditions (inhibitor or relevant gene deletion or mutation) are indicated below. Coloring of boxes indicates relative expression (green, increased; red, repressed) as indicated on the left. Genes have been grouped according to their function or genetic location in three major classes: motility, pO157, and T3SS.
Tpx Regulates Flagellar and T3SS Expression in E. coli O157
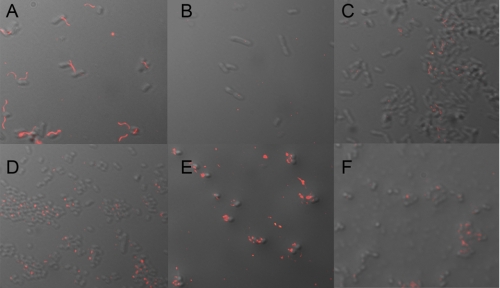
To determine whether the transcriptomic data translated into actual phenotypes, analyses of flagellar expression and effects on motility were tested using the E. coli O157 ΔTpx mutant. Immunofluorescence microscopy was used to visualize flagellar expression in the wild type, ΔTpx mutant, and in the complemented mutant. A culture of the bacteria was performed in LB medium at 28 °C as this is optimum for expression of flagella. More than 90% of wild-type bacteria expressed flagella in these conditions (Fig. 7A) with deletion of tpx reducing expression to <1% of bacteria (Fig. 7B). Restoration of flagellum expression could be achieved by introduction of tpx on a low copy number plasmid but not to wild-type levels (Fig. 7C). These data correlated well with the bacterial motility, which was markedly reduced for the ΔTpx mutant (supplemental Fig. S1). Treatment of wild-type E. coli O157 with ME0055 results in an increased proportion of bacteria expressing flagella (17). To examine effects on the expression of the T3SS, bacteria were cultured in DMEM. This medium provides an intermediate level of expression in wild-type strains, allowing any increases or decreases in the proportion of bacteria expressing the EspA filaments, the needle-like structures used to deliver effector proteins, to be measured. Some 45% of wild-type bacteria express EspA filaments in DMEM (Fig. 7D and supplemental Fig. S1), and deletion of tpx increased this proportion to 79% (Fig. 7E and supplemental Fig. S1), a result that is consistent with the transcriptomic data supporting the hypothesis that Tpx is involved in the regulation of both flagella and the T3SS, two key virulence factors in E. coli O157. Complementation resulted in a phenotype similar to that of wild type (Fig. 7F and supplemental Fig. S1).
FIGURE 7.
Role of Tpx for appropriate expression of flagella and EspA filaments. Bacteria were cultured in LB medium (A–C) or DMEM (D–F) to promote expression of flagella or EspA filaments. Wild-type bacteria (A) express flagella when visualized using immunofluorescence microscopy. The Tpx mutant (B) is non-motile and aflagellate, a phenotype that could be partially restored by introduction of plasmid-borne tpx (C). Culture in DMEM results in 45% of wild-type bacteria expressing EspA filaments (D). Deletion of tpx increased this proportion to 79% of the population (E), and complementation by plasmid-borne tpx returned this to wild-type levels (F). Images are composites created from differential interference contrast and immunofluorescence overlays.
Effect of Y. pseudotuberculosis Tpx and WrbA on Virulence in Vitro
To examine the contribution of the target proteins to pathogenesis, we created defined deletion strains of ΔTpx and ΔWrbA in Y. pseudotuberculosis and analyzed the effect on T3SS and virulence. Analysis of Yop secretion revealed that the secretion profiles of the ΔTpx and ΔWrbA mutants were elevated compared with that of wild-type bacteria (supplemental Fig. S2). This result correlates well with the transcriptomic data that also showed increased expression of genes associated with the T3SS. Secretion from all three strains could be reduced by the addition of 50 μm ME0052 (supplemental Fig. S2), indicating that the overall phenotype associated with the salicylidene acylhydrazide compounds results from affects on multiple target proteins. We also tested the ability of both ΔTpx and ΔWrbA mutants of Y. pseudotuberculosis to infect and kill a J774A macrophage-like cell line (10). Macrophage viability was measured by the addition of calcein AM and subsequent fluorescence analysis, with a ΔYopB translocation-deficient mutant as the negative control. Infection with wild-type bacteria resulted in 46 ± 16% viability compared with uninfected cells. The ΔTpx and ΔWrbA mutants were found to be as virulent as the wild-type strain with 42 ± 12 and 54 ± 4% viability, respectively. The addition of 25 μm of ME00052 rescued the macrophages infected with wild-type bacteria or with the ΔTpx and ΔWrbA mutants, restoring viability such that no macrophages were killed. Similarly, the ΔYopB mutant was avirulent, with no macrophages killed by this strain. These data demonstrate that no single target protein appears to be responsible for the phenotype associated with the addition of the compounds.
DISCUSSION
The identity, function, and three-dimensional structure of the target protein of a potential therapeutic compound constitute key information in a drug development process based on structure-based design (45, 46). The salicylidene acylhydrazide class of T3SS inhibitors was originally identified in a phenotypic bacterial reporter-gene screen (8). This strategy provides compounds that are active on the pathogenic organism and circumvents many of the drawbacks experienced in target-based screening based on proteins obtained by genomics (23). The salicylidene acylhydrazides have proved promising as T3SS inhibitors (19), and to further explore the potential of this compound class we have attempted to identify putative target proteins using an affinity reagent approach (47). In this work we have identified 16 proteins bound by the Affi-Gel-labeled compound, ME0055-Aff. These putative protein targets of the salicylidene acylhydrazides include WrbA, an NAD(P)H quinone oxidoreductase, and Tpx, a thiol peroxidase, both of which have been characterized in some detail (40, 41), and FolX, a dihydroneopterin-tri-P-epimerase. In contrast to WrbA and Tpx, the cellular function of FolX has been poorly characterized. How many of these proteins actually contribute to the phenotype associated with the addition of salicylidene acylhydrazide compounds is not clear. It has been established that metabolism is a key factor in the regulation of expression of type III secretion in P. aeruginosa (48). We would speculate that if the salicylidene acylhydrazides affect multiple proteins, then there could be a clear perturbation of normal metabolism, resulting in changes in gene expression and protein function. However, some interactions could have no consequence for virulence or result from weak and nonspecific binding. It is also feasible that the unlabeled compound binds additional, unidentified proteins, as the modification of ME0055 by the addition of the spacer and Affi-Gel bead could affect binding to target proteins by blocking critical functional groups. Given that we successfully confirmed binding for three of these target proteins using Far Western blotting and showed NMR chemical shift perturbations for Tpx using unlabeled ME0052, we would suggest that several of the putative target proteins are likely to be true interacting partners with the salicylidene acylhydrazides. Furthermore, we have demonstrated that WrbA, Tpx, and FolX all contribute to the normal regulation and expression of key virulence factors, specifically the T3SS and flagella, and would suggest that a combination of perturbations to normal protein activity for multiple target proteins is most likely to explain the phenotype observed upon the addition of the salicylidene acylhydrazides.
Quantitative structure-activity relationship (QSAR) models have been computed for focused libraries (38, 39, 49, 50) using a strategy based on statistical molecular design (51). The QSAR models for the salicylidene acylhydrazides were successfully validated with an external test set, and several compounds inhibited virulence in vitro (39). The QSAR models were, however, hard to interpret, which is likely the result of the compounds being evaluated in a cell-based assay and would be consistent with the notion that they target multiple proteins. The QSAR models reflect overall properties beneficial for interaction with several proteins and other processes including cell permeability. A mode of action that includes several targets is beneficial from an antibiotic resistance perspective as the pathogen must alter several proteins and pathways to escape the drug. However, further optimization of the salicylidene acylhydrazides against multiple targets is challenging (38, 39, 52), although polypharmacology is increasingly recognized as important in drug discovery and development (53).
Given that the compounds affect the function of the T3SS of several Gram-negative pathogens, the obvious implication is that the different species share common protein targets that result in similar phenotypes. At least three mechanisms of inhibition have been suggested; they are direct effects on the T3SS basal apparatus proteins (16), less-direct effects on proteins that then affect T3SS expression and function (17), and possible changes in iron availability (13). With reference to the latter mechanism, our previous work indicated that for E. coli O157:H7, no operons involved in iron metabolism were affected by the addition of the salicylidene acylhydrazides and, therefore, that iron availability was not sufficient to explain the inhibition observed with this strain (17). The three target proteins we have focused on are well conserved among Gram-negative pathogens; we have shown that both Tpx and WrbA from five different pathogens are bound by the salicylidene acylhydrazides and that the compounds bind FolX from three species. Additionally, our identification of the strain that was insensitive to the effects of the salicylidene acylhydrazides showed that this strain had no genetic changes that would affect the proteins that comprise the T3SS. This “insensitive” strain contained a mutant allele of wrbA, encoding one of the proposed target proteins. Given the large number of SNPs in this strain, we do not assume or conclude that this particular non-synonymous change results in the “insensitivity,” but it is an interesting observation that this target was identified by both approaches. The major conclusion from this aspect of the study was that the T3SS was highly conserved. Other genetic changes affecting cell wall structure, ABC transporters, or efflux pumps could all account for the phenotype observed with this strain. Overall, the weight of evidence in the present study favors a mechanism based on indirect effects that lead to changes in the regulation of the T3SS.
To understand the mechanism by which the salicylidene acylhydrazides affect individual target proteins, we focused on Tpx, a thiol peroxidase previously shown to be part of the oxidative stress defense system (40). One important result was the NMR chemical shift perturbation, as this showed that unlabeled compound binds to the target protein involving specific residues that cluster to a discrete area near the active site and the dimer interface. The binding site was modeled by docking the compound onto ecTpx (PDB code 3HVV), highlighting the residues identified by the chemical shift NMR experiments (Fig. 4).
Determination of both Kd and stoichiometry were achieved using an AUC-based method. This gave a Kd of ∼50 μm and a stoichiometry of 1 ME0052 molecule per ypTpx dimer. This result correlates well with our Far Western data that showed preferential binding of ME0052-Bio to the ypTpx dimer compared with the monomer. The C61S mutant forms a less stable dimer, resulting in the majority of the protein migrating as a monomer after SDS-PAGE and less binding of the ME0052-Bio probe. In contrast, when the C61S protein was used for AUC studies, the protein was present as a dimer and gave a Kd approximately twice that of the wild type but with the same stoichiometry as the wild-type protein. The lower Kd observed for the C61S mutant is consistent with the NMR result that identified Cys-61 as being a residue involved in binding of ME0052.
Previous work has shown that Tpx protects against exogenous hydrogen peroxide and is required for survival in macrophages (54). Our data also showed that a Y. pseudotuberculosis ΔTpx mutant was as virulent against J774A macrophages as wild-type bacteria and that the mutant remained sensitive to ME0052. This result is in agreement with the E. coli transcriptomic data and supports the hypothesis that the salicylidene acylhydrazides act on multiple targets. Although deletion of tpx in E. coli O157 resulted in clear effects on gene transcription, we were initially surprised to see an up-regulation of genes associated with the T3SS. Our working hypothesis was that the function of the T3SS proteins would be directly inhibited by the addition of the compounds. However, the data from the arrays showed that in E. coli O157, Tpx function leads to repression of the T3SS. This can be attributed to the peroxidase activity of Tpx rather than any direct regulatory effect of the protein itself, as the C61S mutant displayed a transcriptional profile similar to that of the mutant with the defined deletion. However, the mechanisms by which Tpx affects gene expression are not clear. As both Tpx and WrbA function to protect against oxidative stress, it does make biological sense for their activity to be linked to activation of motility, allowing the bacteria to evade such stresses. Previous work has shown a clear link between bacterial response to numerous stresses and control of motility (55).
Given that the phenotype associated with addition of the salicylidene acylhydrazide compounds is a repression of T3SS expression, the implication is that these compounds may actually increase activity of Tpx and WrbA, thereby enhancing their repressive effects on the T3SS. The addition of the salicylidene acylhydrazide compounds does not directly affect expression of tpx or wrbA (17), suggesting that the compounds might directly activate or stabilize the enzymes. The preferential binding to the Tpx dimer coupled with the stoichiometry and NMR chemical shift assignment show that the compounds bind very close to the active site of Tpx. It is possible that the binding of the compound near the active site results in a positive effect on protein activity. Future work will map the binding site of the salicylidene acylhydrazide to more target proteins, allowing any common binding site to be identified, a key step on the route to the development of this class of anti-virulence compounds.
Supplementary Material
Acknowledgments
We thank Dr. Roger Parton and Dr. Dan Walker for their critical reading of the manuscript. The sequencing of the ZAP 430 strain was carried out in the Sir Henry Wellcome Functional Genomics Facility at the University of Glasgow.
This work was generously supported by Medical Research Scotland Grant 223 ORG (to D. W., A. J. R. and R. B.) and Biotechnology and Biological Sciences Research Council (Swindon, United Kingdom) Grant BB/G011389/1 (to A. J. R. and M. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2, Tables S1–S5, and Protocol S1.
The Illumina data have been deposited on the EMBL database (ERP000335), and the MIAME compliant data are deposited on the Gene Expression Omnibus (GEO) at NBCI under the GEO accession number GSE23001.
- T3SS
- type three secretion system
- AUC
- analytical ultracentrifugation
- HSQC
- heteronuclear single quantum coherence
- ecTpx
- E. coli O157 Tpx
- QSAR
- quantitative structure-activity relationship
- ypTpx
- Y. pseudotuberculosis Tpx
- MEM
- minimum Eagle's medium
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SNP
- single-nucleotide polymorphism.
REFERENCES
- 1. Cegelski L., Marshall G. R., Eldridge G. R., Hultgren S. J. (2008) Nat. Rev. Microbiol. 6, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clatworthy A. E., Pierson E., Hung D. T. (2007) Nat. Chem. Biol. 3, 541–548 [DOI] [PubMed] [Google Scholar]
- 3. Escaich S. (2008) Curr. Opin. Chem. Biol. 12, 400–408 [DOI] [PubMed] [Google Scholar]
- 4. Lynch S. V., Wiener-Kronish J. P. (2008) Curr. Opin. Crit. Care 14, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rasko D. A., Sperandio V. (2010) Nat. Rev. Drug Discov. 9, 117–128 [DOI] [PubMed] [Google Scholar]
- 6. Hueck C. J. (1998) Microbiol. Mol. Biol. Rev. 62, 379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coburn B., Sekirov I., Finlay B. B. (2007) Clin. Microbiol. Rev. 20, 535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kauppi A. M., Nordfelth R., Uvell H., Wolf-Watz H., Elofsson M. (2003) Chem. Biol. 10, 241–249 [DOI] [PubMed] [Google Scholar]
- 9. Muschiol S., Bailey L., Gylfe A., Sundin C., Hultenby K., Bergström S., Elofsson M., Wolf-Watz H., Normark S., Henriques-Normark B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14566–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bailey L., Gylfe A., Sundin C., Muschiol S., Elofsson M., Nordström P., Henriques-Normark B., Lugert R., Waldenström A., Wolf-Watz H., Bergström S. (2007) FEBS Lett. 581, 587–595 [DOI] [PubMed] [Google Scholar]
- 11. Slepenkin A., Enquist P. A., Hägglund U., de la Maza L. M., Elofsson M., Peterson E. M. (2007) Infect. Immun. 75, 3478–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolf K., Betts H. J., Chellas-Géry B., Hower S., Linton C. N., Fields K. A. (2006) Mol. Microbiol. 61, 1543–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hudson D. L., Layton A. N., Field T. R., Bowen A. J., Wolf-Watz H., Elofsson M., Stevens M. P., Galyov E. E. (2007) Antimicrob. Agents Chemother. 51, 2631–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Negrea A., Bjur E., Ygberg S. E., Elofsson M., Wolf-Watz H., Rhen M. (2007) Antimicrob. Agents Chemother. 51, 2867–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nordfelth R., Kauppi A. M., Norberg H. A., Wolf-Watz H., Elofsson M. (2005) Infect. Immun. 73, 3104–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Veenendaal A. K., Sundin C., Blocker A. J. (2009) J. Bacteriol. 191, 563–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tree J. J., Wang D., McInally C., Mahajan A., Layton A., Houghton I., Elofsson M., Stevens M. P., Gally D. L., Roe A. J. (2009) Infect. Immun. 77, 4209–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chu H., Slepenkin A., Elofsson M., Keyser P., de la Maza L. M., Peterson E. M. (2010) Int. J. Antimicrob. Agents 36, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keyser P., Elofsson M., Rosell S., Wolf-Watz H. (2008) J. Intern. Med. 264, 17–29 [DOI] [PubMed] [Google Scholar]
- 20. Baron C. (2010) Curr. Opin. Microbiol. 13, 100–105 [DOI] [PubMed] [Google Scholar]
- 21. Puri A. W., Bogyo M. (2009) ACS Chem. Biol. 4, 603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hung D. T., Rubin E. J. (2006) Curr. Opin. Chem. Biol. 10, 321–326 [DOI] [PubMed] [Google Scholar]
- 23. Payne D. J., Gwynn M. N., Holmes D. J., Pompliano D. L. (2007) Nat. Rev. Drug Discov. 6, 29–40 [DOI] [PubMed] [Google Scholar]
- 24. Terstappen G. C., Schlüpen C., Raggiaschi R., Gaviraghi G. (2007) Nat. Rev. Drug Discov. 6, 891–903 [DOI] [PubMed] [Google Scholar]
- 25. Leslie B. J., Hergenrother P. J. (2008) Chem. Soc. Rev. 37, 1347–1360 [DOI] [PubMed] [Google Scholar]
- 26. Sambrook S., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbour Laboratory Press, New York [Google Scholar]
- 27. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 28. Pappin D. J., Hojrup P., Bleasby A. J. (1993) Curr. Biol. 3, 327–332 [DOI] [PubMed] [Google Scholar]
- 29. Roe A. J., Hoey D. E., Gally D. L. (2003) Biochem. Soc. Trans. 31, 98–103 [DOI] [PubMed] [Google Scholar]
- 30. Li H., Ruan J., Durbin R. (2008) Genome Res. 18, 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu Y., Li Q., Chen X. Z. (2007) Nat. Protoc. 2, 3278–3284 [DOI] [PubMed] [Google Scholar]
- 32. Roe A. J., Tysall L., Dransfield T., Wang D., Fraser-Pitt D., Mahajan A., Constandinou C., Inglis N., Downing A., Talbot R., Smith D. G., Gally D. L. (2007) Microbiology 153, 1350–1360 [DOI] [PubMed] [Google Scholar]
- 33. Lane M. C., Lockatell V., Monterosso G., Lamphier D., Weinert J., Hebel J. R., Johnson D. E., Mobley H. L. (2005) Infect. Immun. 73, 7644–7656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gabrielsen M., Zetterström C. E., Wang D., Beckham K. S., Elofsson M., Isaacs N. W., Roe A. J. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1606–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mori S., Abeygunawardana C., Johnson M. O., van Zijl P. C. (1995) J. Magn. Reson. B 108, 94–98 [DOI] [PubMed] [Google Scholar]
- 36. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 37. Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dahlgren M. K., Oberg C. T., Wallin E. A., Janson P. G., Elofsson M. (2010) Molecules 15, 4423–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dahlgren M. K., Zetterström C. E., Gylfe S., Linusson A., Elofsson M. (2010) Bioorg. Med. Chem. 18, 2686–2703 [DOI] [PubMed] [Google Scholar]
- 40. Baker L. M., Poole L. B. (2003) J. Biol. Chem. 278, 9203–9211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patridge E. V., Ferry J. G. (2006) J. Bacteriol. 188, 3498–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ploom T., Haussmann C., Hof P., Steinbacher S., Bacher A., Richardson J., Huber R. (1999) Structure 7, 509–516 [DOI] [PubMed] [Google Scholar]
- 43. Hall A., Sankaran B., Poole L. B., Karplus P. A. (2009) J. Mol. Biol. 393, 867–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Iyoda S., Koizumi N., Satou H., Lu Y., Saitoh T., Ohnishi M., Watanabe H. (2006) J. Bacteriol. 188, 5682–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simmons K. J., Chopra I., Fishwick C. W. (2010) Nat. Rev. Microbiol. 8, 501–510 [DOI] [PubMed] [Google Scholar]
- 46. Barker J. J. (2006) Drug Discov. Today 11, 391–404 [DOI] [PubMed] [Google Scholar]
- 47. Guiffant D., Tribouillard D., Gug F., Galons H., Meijer L., Blondel M., Bach S. (2007) Biotechnol. J. 2, 68–75 [DOI] [PubMed] [Google Scholar]
- 48. Rietsch A., Mekalanos J. J. (2006) Mol. Microbiol. 59, 807–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kauppi A. M., Andersson C. D., Norberg H. A., Sundin C., Linusson A., Elofsson M. (2007) Bioorg. Med. Chem. 15, 6994–7011 [DOI] [PubMed] [Google Scholar]
- 50. Dahlgren M. K., Kauppi A. M., Olsson I. M., Linusson A., Elofsson M. (2007) J. Med. Chem. 50, 6177–6188 [DOI] [PubMed] [Google Scholar]
- 51. Linusson A., Elofsson M., Andersson I. E., Dahlgren M. K. (2010) Curr. Med. Chem. 17, 2001–2016 [DOI] [PubMed] [Google Scholar]
- 52. Hillgren J. M., Dahlgren M. K., To T. M., Elofsson M. (2010) Molecules 15, 6019–6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Morphy R., Rankovic Z. (2009) Curr. Pharm. Des. 15, 587–600 [DOI] [PubMed] [Google Scholar]
- 54. Horst S. A., Jaeger T., Denkel L. A., Rouf S. F., Rhen M., Bange F. C. (2010) J. Bacteriol. 192, 2929–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nachin L., Nannmark U., Nyström T. (2005) J. Bacteriol. 187, 6265–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1, Schrodinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.