Abstract
Mycobacterium tuberculosis encodes five type VII secretion systems that are responsible for exporting a number of proteins, including members of the Esx family, which have been linked to tuberculosis pathogenesis and survival within host cells. The gene cluster encoding ESX-3 is regulated by the availability of iron and zinc, and secreted protein products such as the EsxG·EsxH complex have been associated with metal ion acquisition. EsxG and EsxH have previously been shown to form a stable 1:1 heterodimeric complex, and here we report the solution structure of the complex, which features a core four-helix bundle decorated at both ends by long, highly flexible, N- and C-terminal arms that contain a number of highly conserved residues. Despite clear similarities in the overall backbone fold to the EsxA·EsxB complex, the structure reveals some striking differences in surface features, including a potential protein interaction site on the surface of the EsxG·EsxH complex. EsxG·EsxH was also found to contain a specific Zn2+ binding site formed from a cluster of histidine residues on EsxH, which are conserved across obligate mycobacterial pathogens including M. tuberculosis and Mycobacterium leprae. This site may reflect an essential role in zinc ion acquisition or point to Zn2+-dependent regulation of its interaction with functional partner proteins. Overall, the surface features of both the EsxG·EsxH and the EsxA·EsxB complexes suggest functions mediated via interactions with one or more target protein partners.
Keywords: Bacteria, NMR, Protein Structure, Secretion, Zinc, EsxG/EsxH, Pathogenesis, Tuberculosis
Introduction
Mycobacterium tuberculosis is the primary causative agent of human tuberculosis and one of the oldest pathogens known to man, yet tuberculosis remains a major global health problem with an estimated 9.4 million new cases and over 1.3 million tuberculosis-related deaths annually (1). Analysis of the genome sequences for the closely related mycobacterial pathogens M. tuberculosis (2), Mycobacterium bovis (3), and Mycobacterium leprae (4) and comparative studies with attenuated M. bovis BCG strains identified a number of secreted proteins, including members of the Esx or CFP-10/ESAT-6 (10-kDa culture filtrate protein/6-kDa early secreted antigenic target) protein family, PE/PPE (proline-glutamic acid/proline-proline-glutamic acid) proteins, and MPT70/MPT83, which play essential, but as yet undefined, roles in mycobacterial pathogenesis.
M. tuberculosis encodes 23 Esx proteins, EsxA–W, which are generally characterized by their small size (∼100 residues), the presence of a central WXG motif, and their organization in pairs within the genome (5). The genes encoding the Esx pairs EsxA/EsxB (ESAT-6/CFP-10, Rv3875/Rv3874) and EsxG/EsxH (Rv0287/Rv0288) have been shown to be coordinately regulated forming small operons, and it is expected that all M. tuberculosis Esx genome pairs will behave similarly (6, 7). Studies have also shown that the protein products of several Esx pairs, including EsxA/EsxB, EsxG/EsxH, EsxR/EsxS (Rv3019c/Rv3020c), and EsxO/EsxP (Rv2346c/Rv2347c) form tight complexes, which are likely to be the functional form of these proteins (8–13).
Five of the 11 ESX loci (ESX-1 to ESX-5) within the M. tuberculosis genome appear to encode examples of the recently identified type VII secretion systems (T7SS), which have been shown to export a number of proteins, including Esx protein complexes and PE/PPE proteins. The best characterized of these systems is ESX-1 (rv3868/eccA to rv3883c/mycP), which is known to secrete the EsxA·EsxB protein complex, as well as at least seven other mycobacterial proteins, including EspA (Rv3616c), EspB (Rv3881c), EspC (Rv3615c), EspE (Rv3864), EspF (Rv3865), PE35 (Rv3872), and EspR (Rv3849) (14–19). Core components of type VII secretion systems include FtsK/SpoIIIE-like ATPases (Rv3870 and Rv3871 in ESX-1), transmembrane proteins (Rv3877 in ESX-1), and a subtilisin-like serine protease (MycP1 in ESX-1) (14, 20). The ESX systems are well conserved in M. tuberculosis and closely related mycobacteria, such as M. bovis, M. leprae, and Mycobacterium marinum, as well as more distantly related organisms such as Streptomyces coelicolor (20–22).
It appears that even within M. tuberculosis, the various ESX systems are differentially regulated and are likely to play different roles in infection. For example, ESX-1 and ESX-5 are thought to play a role in mycobacterial virulence and have been linked to granuloma formation, cell-to-cell spread of the mycobacteria, and escape from arrested phagosomes (14, 23–26), with ESX-1 reported to be under the control of multiple regulators, such as the DNA binding transcription factor EspR (Rv3849), the two-component system regulator PhoP, and the serine protease MycP1 (18, 27–29). In contrast, ESX-3, which encodes the proteins EsxG and EsxH, is required for optimal growth of M. tuberculosis and has been associated with essential processes such as iron and zinc acquisition (30–32). In M. tuberculosis, ESX-3 has been shown to be regulated by the iron-dependent transcriptional repressor IdeR, as well as the zinc-uptake regulator Zur (33, 34).
Despite growing evidence for diverse functional roles, secondary structure analysis by circular dichroism (CD), sequence comparisons, and helical wheel predictions suggest that M. tuberculosis Esx protein complexes are likely to adopt similar backbone topologies to the previously reported EsxA·EsxB complex, with distinct surface properties and features reflecting different functional roles (9, 12). Here we report the high resolution solution structure of the M. tuberculosis EsxG·EsxH protein complex, which confirms the expected similarity to the core structure of the EsxA·EsxB complex but reveals striking differences in surface features and properties, including the identification of a potential functional site and a specific Zn2+ binding site. In contrast to EsxA·EsxB, we obtained no evidence for a specific interaction between fluorescently labeled EsxG·EsxH complex and the surface of macrophage/monocyte-like cells. The surface features of both complexes point to roles mediated via interactions with target proteins or complexes. However, striking differences clearly suggest different binding partners, reflecting proposed roles for EsxA·EsxB in pathogen-host cell signaling and for EsxG·EsxH in iron and zinc acquisition by infecting mycobacteria.
EXPERIMENTAL PROCEDURES
Protein Expression Vectors
The full-length coding regions for EsxG (Rv0287) and EsxH (Rv0288) were amplified by PCR from pET28a expression vectors containing EsxG and EsxH, respectively (10). EsxG was ligated into the pET23a Escherichia coli expression vector and expressed as a full-length protein without the N-terminal His tag. EsxH was cloned into the pLeic01 E. coli expression vector by ligation-independent cloning using the In-Fusion dry down PCR cloning kit (Clontech). EsxH was expressed as a full-length protein with an N-terminal His tag and a tobacco etch virus cleavage site (ENLYFQSM).
Protein Expression, Refolding, and Purification
Unlabeled and uniformly 15N-, 13C-, and 15N/13C-labeled EsxG and EsxH were expressed individually from pET23a- and pLeic01-based vectors in E. coli BL21(DE3) as described previously (9, 10, 35). The two proteins were obtained as inclusion bodies, which were solubilized in buffer containing guanidine hydrochloride and co-refolded to produce soluble EsxG·EsxH complex, essentially as reported previously (9, 10, 35). The refolded EsxG·EsxH complex was purified by nickel affinity chromatography followed by gel filtration. The N-terminal His tag attached to EsxH, expressed from the pLeic01 vector, was removed by cleavage with tobacco etch virus protease.
NMR Spectroscopy
NMR spectra were acquired from 0.35-ml samples of 0.7–1.0 mm EsxG·EsxH complex in 25 mm NaH2PO4, 100 mm NaCl, 0.02% (w/v) NaN3, 0.1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, pH 6.5, containing either 10% D2O, 90% H2O or 100% D2O as appropriate. All NMR data were acquired and processed as described by Ilghari et al. (35). 15N/1H HSQC6 spectra of EsxG·EsxH were acquired in the presence and absence of equimolar Zn2+ and Fe3+ to determine whether the complex contained a specific metal ion binding site. In these experiments, equimolar amounts of ZnCl2 or FeCl3 were added to either 100 μm 15N-labeled EsxG·unlabeled EsxH or 100 μm unlabeled EsxG·15N-labeled EsxH in a 20 mm Bis-Tris, 100 mm NaCl, 0.02% (w/v) NaN3, 0.1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, pH 6.5, buffer. The spectra were acquired at 35 °C on 600-MHz Bruker Avance or DRX systems. Typical acquisition times for the HSQC spectra were 60 ms in F2 (1H), and 30 ms in F1 (15N), with the spectra collected over ∼1.5 h. The spectra were processed using Topspin (Bruker Biospin Ltd.) with linear prediction used to extend the effective acquisition times by up to 2-fold in F1.
Structural Calculations
The family of converged EsxG·EsxH structures was determined in a two-stage process using the program CYANA (36). Initially, the combined automated NOE assignment and structure determination protocol (CANDID) was used to automatically assign the intra- and intermolecular NOE cross-peaks identified in three-dimensional 15N- and 13C-edited NOESY spectra. Subsequently, several cycles of simulated annealing combined with redundant dihedral angle constraints to increase convergence were used to produce the final converged EsxG·EsxH structures (37, 38).
The input for the CANDID stage consisted of essentially complete 15N, 13C, and 1H resonance assignments for the non-exchangeable groups in the EsxG·EsxH complex (35), four manually picked three-dimensional NOE peak lists corresponding to all NOEs involving amide protons (1538) and all NOEs between aliphatic protons (2222), and two manually picked two-dimensional NOE peak lists corresponding to all NOEs involving aromatic side chain protons (127). The CANDID stage also included 264 backbone torsion angle constraints (Φ/Ψ) for the EsxG·EsxH complex determined by the protein backbone dihedral angle prediction program TALOS (39). CANDID calculations were carried out using the default parameter settings in CYANA, with chemical shift tolerances set to 0.04 ppm (direct and indirect 1H) and 0.4 ppm (15N and 13C). The final converged EsxG·EsxH structures were produced from 100 random starting coordinates using a standard torsion angle-based simulated annealing protocol combined with five cycles of redundant dihedral angle constraints (37, 38). The calculations were based upon 2323 non-redundant NOE-derived upper distance limits (maximum value 6.0 Å), assigned to unique pairs of protons using CANDID, 264 Φ/Ψ torsion angle constraints derived from TALOS, and 24 hydrogen bond constraints involving slowly exchanging backbone amides in regions of regular helical structure (four per hydrogen bond). Analysis of the family of structures obtained was carried out using the programs CYANA and MOLMOL (36, 40).
Mapping of the Zn2+ Binding Site
The minimal shift approach was used to analyze the changes in the positions of EsxG·EsxH backbone amide NMR signals resulting from Zn2+ binding, as described previously (41).
Cell Culture
Monocyte-like U937 cells (85011440, European Collection of Cell Cultures (ECACC)) were maintained in RPMI 1640 medium (Invitrogen) containing 10% fetal calf serum (FCS) (Invitrogen) and 2 mm glutamine (Invitrogen) at 37 °C and 5% CO2. For fluorescence microscopy experiments, cells were attached to glass coverslips precoated with either 160 μg/ml poly-l-lysine (Sigma) or 5 μg/ml fibronectin (Sigma) by incubation at 37 °C and 5% CO2 for ∼10 min.
Fluorescence Microscopy
Alexa Fluor 546-labeled full-length EsxG·EsxH complex was prepared as described for EsxA·EsxB, and potential binding of the full-length complex to the surface of U937 cells was investigated as reported previously for the EsxA·EsxB complex (12).
RESULTS AND DISCUSSION
Structure of the EsxG·EsxH Complex
The combined automated NOE assignment and structure determination module CANDID (36) was used to determine unique assignments for the NOEs identified in two- and three-dimensional NOE-based spectra. Assignments were obtained for 87% of the total NOE peaks identified, producing 2323 non-redundant 1H to 1H upper distance limits. The final family of EsxG·EsxH structures was determined using a total of 2611 NMR-derived structural constraints (an average of 13.5 per residue), which are summarized in Table 1. Following the final round of CYANA calculations, 30 satisfactorily converged structures were obtained from 100 random starting structures. The residual constraint violations and the structural statistics for the family of converged EsxG·EsxH structures are shown in Table 1.
TABLE 1.
NMR constraints and structural statistics for the EsxG·EsxH complex
| NMR constraints | ||
| No. of constraints used in final structural calculation | ||
| Intraresidue NOEs | 544 | |
| Sequential (short range) NOEs (i, i + 1) | 598 | |
| Medium range NOEs (i, i ≤ 4) | 635 | |
| Long range NOEs (i, i ≥ 5) | 546 | |
| Intermolecular | 339 | |
| Intramolecular | 207 | |
| Torsion angles | 264 (132Φ and 132Ψ) | |
| Hydrogen bonding | 24 | |
| Maximum (left column) and total (right column) constraint violations in 30 converged EsxG·EsxH structures | ||
| Upper distance limits (Å) | 0.32 ± 0.00 | 6.95 ± 0.05 |
| Lower distance limits (Å) | 0.10 ± 0.00 | 0.90 ± 0.04 |
| van der Waals contacts (Å) | 0.35 ± 0.00 | 6.04 ± 0.04 |
| Torsion angle ranges (°) | 3.09 ± 0.32 | 52.8 ± 0.74 |
| Average CYANA target function (Å2) | 0.62 ± 0.15 | |
| Structural statistics | ||
| Structural statistics for the family of converged EsxG·EsxH structures | ||
| Residues within regions of the Ramachandran plot | ||
| Allowed regions | 96.6% | |
| Generously allowed | 3.4% | |
| Disallowed | 0 | |
| r.m.s.d. for structured region (residues 15–79 of EsxG and residues 17–75 of EsxH) | ||
| Backbone | 1.11 ± 0.21 Å | |
| Heavy atoms | 1.48 ± 0.25 Å | |
| r.m.s.d. for α-helical regions (residues 18–42 and 49–76 of EsxG and 19–38 and 51–73 of EsxH) | ||
| Backbone | 0.85 ± 0.21 Å | |
| Heavy atoms | 1.16 ± 0.23 Å | |
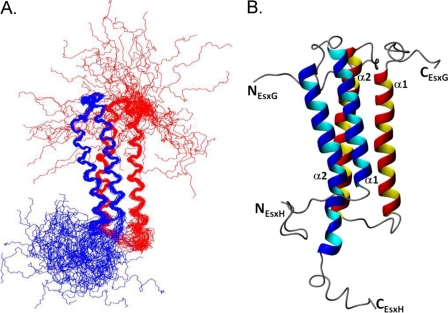
The overlays of the protein backbones for the 30 converged structures obtained are shown in Fig. 1, and together with relatively low root mean square deviation (r.m.s.d.) values to the mean structure for both the backbone and the heavy atoms (Table 1), indicate that the solution structure of the EsxG·EsxH complex has been determined to fairly high resolution. Within the complex, both EsxG and EsxH adopt helix-turn-helix hairpin structures, which are arranged antiparallel to each other, forming a four-helix bundle. The core of the complex is well defined (residues 15–79 in EsxG and 17–75 in EsxH), although there appears to be significant flexibility in the hairpin loop of EsxG (residues 40–47) as well as the N and C termini of EsxG and EsxH, which form flexible arms at both ends of the complex (Fig. 1). The two long helices in the hairpin structures are formed from residues Phe-18–Phe-42 (α1) and Ala-49–Leu-76 (α2) in EsxG and Ala-19–Ala-38 (α1) and Tyr-51–Ser-73 (α2) in EsxH. The helices in EsxG are completely α-helical, whereas in EsxH, helix α2 terminates with a single turn of 310 helix (Ser-74–His-76) followed by another single turn of α-helix (Glu-77–Met-81). The exposed C-terminal region of EsxH shows some propensity to adopt a helical conformation (reflected in both patterns of NOEs and backbone chemical shifts reported by Ilghari et al. (35)), and the conservation of aromatic and hydrophobic residues located in the C-terminal regions of both EsxG and EsxH implies some functional significance for these flexible regions. In particular, Tyr-94 and Phe-97 of EsxG are well conserved in EsxG orthologues from other mycobacteria (Fig. 2C) as well as in the closely related M. tuberculosis EsxS protein; however, these residues are not conserved throughout the M. tuberculosis Esx protein family, implying a functional role specific to the EsxG·EsxH complex.
FIGURE 1.
Solution structure of the EsxG·EsxH complex. A, a best-fit superposition of the protein backbone for the family of 30 converged structures obtained for EsxG·EsxH, with EsxH shown in blue and EsxG shown in red. B, ribbon representations of the backbone topology of the EsxG·EsxH complex based on the converged structure closest to the mean, with EsxG shown in red and EsxH shown in blue. Panels A and B clearly show the two helix-turn-helix hairpin structures formed by the individual proteins and the flexible N and C termini of both proteins within the complex.
FIGURE 2.
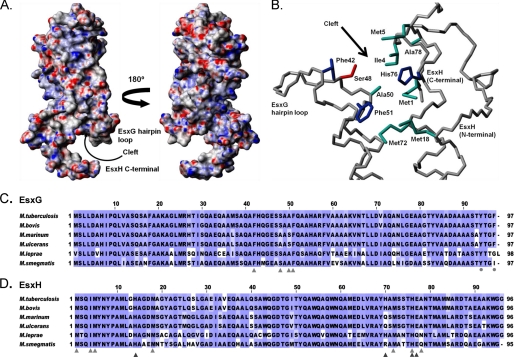
Surface view of the EsxG·EsxH complex, including identification and conservation of amino acids found within the EsxG·EsxH cleft. A, the surface of the complex is colored according to electrostatic potential (40), with areas of significant negative charge shown in red, areas of significant positive charge are shown in blue and neutral areas are shown in white. An interesting feature of the EsxG·EsxH complex is the cleft that is formed between the loop linking the helices of EsxG and the C-terminal arm of EsxH. The structures shown here are rotated by approximately −45°/135° about the y axis in comparison with the ribbon representation shown in Fig. 1. B, orientation of key amino acid side chains that contribute to the formation of the cleft on the surface of the EsxG·EsxH complex, where aliphatic residues with hydrophobic side chains (Ala, Ile, and Met) are shown in cyan, aromatic residues (His and Phe) are colored blue, and the polar residue (Ser) is shown in red. C and D show multiple sequence alignments highlighting the conservation of amino acids in EsxG (C) and EsxH (D) from M. tuberculosis, M. bovis, M. marinum, M. ulcerans, M. leprae, and Mycobacterium smegmatis. Residues forming the cleft are highlighted by light gray triangles, and conserved aromatic residues at the C-terminal of EsxG are highlighted by light gray circles. Dark gray triangles indicate residues from EsxH that form the Zn2+ binding site.
As predicted previously (9), the contact surface between EsxG and EsxH is essentially hydrophobic in nature and accounts for ∼15% (∼1340 Å2) of the total surface area of both proteins. The residues found at the intermolecular interface include 19 residues from EsxG (Phe-18, Lys-21, Met-25, Thr-28, Ala-32, Ala-35, Ala-50, Phe-51, Ala-54, Arg-57, Phe-58, Ala-61, Lys-64, Val-65, Leu-68, Val-71, Ala-72, Asn-75, and Leu-76) and 21 residues from EsxH (Met-18, Tyr-21, Leu-25, Leu-28, Glu-31, Ile-32, Glu-35, Leu-39, Ala-42, Trp-43, Thr-47, Ile-49, Trp-54, Gln-57, Trp-58, Ala-61, Leu-65, Ala-68, Tyr-69, Ala-71, and Met-72). The stabilizing interactions between EsxG and EsxH appear to rely almost entirely on favorable van der Waals contacts; however, an intermolecular salt bridge (Lys-21–Glu-31) appears to stabilize the interaction between the N-terminal region of helix α1 in EsxG and the C-terminal region of the equivalent helix in EsxH. The interactions within the helical hairpins are also primarily based on van der Waals interactions; however, close inspection of the structure reveals the potential for the formation of two salt bridges within the EsxG hairpin (Arg-26–Asp-70 and Glu-33–His-55).
Analysis of the electrostatic surface of the complex reveals a fairly even distribution of positive and negative charge (Fig. 2A) with no significant hydrophobic patches on the surface of the complex, which together with solubility to over 1 mm in aqueous solution argues against a membrane-spanning role. In addition to the long flexible N- and C-terminal arms of the proteins, another notable feature of the EsxG·EsxH complex is the presence of a cleft on the surface of the structure, which could indicate a potential binding site for an interaction partner (Fig. 2, A and B). The cleft is formed by elements corresponding to the flexible N-terminal region of EsxH (specifically residues Met-1, Ile-4, Met-5, and Met-18), the hairpin turn region of EsxG (residues Phe-42, Ser-48, Ala-50, and Phe-51), and the C-terminal region of the α2 helix in EsxH (residues Met-72, His-76, and Ala-78) (Fig. 2, B–D). Met-18 and Met-72 from EsxH form the base of the cleft, with the other residues remaining partially exposed to the solvent and therefore also accessible to any potential binding partner. The cleft is predominantly hydrophobic in nature, and although relatively narrow, the precise shape is variable across the family of EsxG·EsxH structures, reflecting the flexibility in the N-terminal region of EsxH. The hydrophobic and aromatic residues that form the cleft are conserved in EsxG·EsxH orthologues from M. bovis, M. marinum, Mycobacterium ulcerans, and M. leprae (Fig. 2, C and D), as well as the closely related M. tuberculosis EsxR and EsxS molecules (Fig. 3). However, they are not conserved throughout other members of the M. tuberculosis ESX protein family (9, 12), which suggests a functional site specific to EsxG·EsxH and EsxR·EsxS complexes. Overall, the surface features of the EsxG·EsxH complex suggest a function most probably mediated via interactions with one or more target proteins, involving either the cleft and/or the flexible N- or C-terminal arms at either end of the complex.
FIGURE 3.
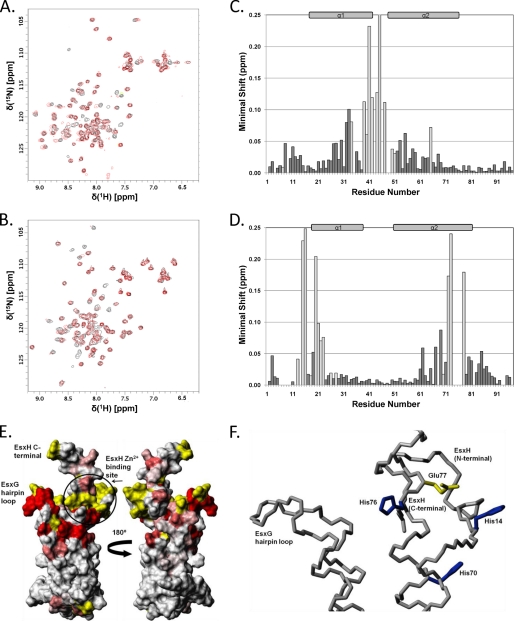
Mapping of the Zn2+ binding site on EsxG·EsxH. A, an overlay of two 15N/1H HSQC spectra of uniformly 15N-labeled EsxG bound to unlabeled EsxH (100 μm) acquired in the presence (red) and absence (black) of equimolar amounts of Zn2+. B, an overlay of 15N-labeled EsxH bound to unlabeled EsxG in the presence (red) and absence (black) of Zn2+. For clarity, the region containing the four tryptophan indolic signals from EsxH is not shown. C and D, histograms of the minimal chemical shift changes observed for backbone amide groups of EsxG (C) and EsxH (D) on binding of Zn2+ to the EsxG·EsxH complex. The positions of the helices in the EsxG·EsxH complex are highlighted above the histograms. Residues that showed significant line broadening of backbone amide signals upon the addition of Zn2+ are shown as light gray bars. For clarity, the minimal shift shown for Gly-45 of EsxG has been truncated (minimal shift 0.8 ppm). E, surface views of the EsxG·EsxH complex where residues are colored according to the perturbation of the backbone amide signals induced by Zn2+ binding. Residues that showed a minimal shift of less than 0.02 ppm are shown in white, residues with a shift over 0.1 ppm are in red, and residues with a shift between 0.02 and 0.1 ppm are colored according to the magnitude of the shift on a linear gradient between white and red. Residues for which no data were obtained are shown in yellow. The structures in E are rotated by 180° about the x axis and −45°/135° about the y axis in comparison with the ribbon representation shown in Fig. 1. F, positions of key amino acid side chains, in the structure closest to the mean, which contribute to the formation of the Zn2+ binding site on the EsxG·EsxH complex. The three histidine residues (His-14, His-70, and His-76) are colored blue, and the glutamic acid residue (Glu-77) is shown in yellow. The family of NMR structures indicates that the side chains of residues His-14, His-76, and Glu-77 are fairly flexible; however, upon binding Zn2+, these side chains are likely to adopt a single orientation and become less flexible.
Previous studies have shown the importance of the flexible C-terminal region of EsxB in binding the EsxA·EsxB complex to the surface of host monocyte/macrophage cells (12). Therefore, to determine whether this is also the case for the EsxG·EsxH complex, similar fluorescence microscopy studies to those described for EsxA·EsxB (12) were carried out using Alexa Fluor 546-labeled EsxG·EsxH. Interestingly, the experiments provided no convincing evidence of a specific interaction of labeled EsxG·EsxH with the surface of host cells as the low level of cell-associated fluorescence seen (500-ms exposure times as compared with 100–200 ms for EsxA·EsxB) showed no significant reduction in the presence of a 15-fold molar excess of non-labeled EsxG·EsxH. These results, combined with the reported up-regulation of all ESX-3 genes under conditions of low iron/zinc (31, 33, 34), suggest that potential host cell binding partners for the EsxG·EsxH complex are more likely to be found within the host cell rather than on the cell surface.
Specific Zn2+ Binding by the EsxG·EsxH Complex
The role of ESX-3 and its secreted soluble factors, such as EsxG·EsxH, in iron and zinc acquisition is not yet fully understood, although it has been reported that ESX-3 has been linked to iron acquisition via the mycobactin pathway (31). It is possible that EsxG·EsxH could play a role in iron acquisition; however, it is unlikely that the complex interacts with the M. tuberculosis siderophore mycobactin. The chemical structure of mycobactin T from M. tuberculosis (42), combined with examination of the crystal structure of CD1a protein with a mycobactin-like molecule bound in a hydrophobic cleft (43; PDB 1XZO), suggests that the width and depth of the cleft on the surface of EsxG·EsxH would not be large enough to accommodate mycobactin.
To assess potential Zn2+ or Fe3+ binding by EsxG·EsxH, samples of the complex in which either EsxG or EsxH was uniformly 15N-labeled were prepared containing equimolar amounts of the individual metal ions. The addition of Zn2+ resulted in dramatic changes in the 15N/1H HSQC spectra of both proteins in the complex, with substantial shifts and/or line broadening observed for a significant number of backbone amide signals (Fig. 3, A and B), which clearly indicates relatively tight binding of Zn2+. In marked contrast, the addition of Fe3+ had no effect on the 15N/1H HSQC of either protein in the complex. The location of the specific Zn2+ binding site on EsxG·EsxH was mapped by minimal shift analysis of the changes seen in the backbone amide signals (41), which is summarized for both proteins in the histograms shown in Fig. 3, C and D. In the case of EsxG, this reveals that the majority of the substantially perturbed signals arise from residues located toward the closed end of the hairpin structure, in particular, from residues near the C terminus of helix 1 and in the flexible loop between the two helices. In contrast, signals from residues positioned close to the open ends of the EsxH hairpin are most affected by Zn2+ binding, which is consistent with the antiparallel arrangement of the two hairpin structures within the EsxG·EsxH complex and localizes the effects of Zn2+ coordination to one end of the complex, as illustrated in Fig. 3E.
Examination of the surface regions of the EsxG·EsxH complex perturbed by Zn2+ binding revealed a cluster of three histidine residues from EsxH (His-14, His-70, and His-76) with side chains able to accommodate tetrahedral coordination of a single zinc ion. This region of EsxH also contains an appropriately positioned glutamic acid residue (Glu-77), which is likely to provide the fourth Zn2+-coordinating group (Fig. 3F). The identification of this cluster of residues as the probable zinc ion binding site was further supported by analysis of the family of EsxG·EsxH structures using the program FEATURE (44), which recognized this region as a very likely Zn2+ coordination site. The three histidine residues involved in Zn2+ binding are conserved across mycobacterial species with an obligate pathogenic lifestyle, such as M. tuberculosis and M. leprae, but His-70 is replaced by glutamine or arginine in environmental mycobacteria and opportunistic pathogens (Fig. 2D). Similarly, His-70 of EsxH is substituted by a glutamine residue in the very closely related EsxR (Fig. 4E). This suggests that the functional importance of zinc ion binding by the EsxG·EsxH complex may be restricted to survival or growth within infected host cells and implies that the very closely related EsxR·EsxS complex may not be fully functionally equivalent.
FIGURE 4.
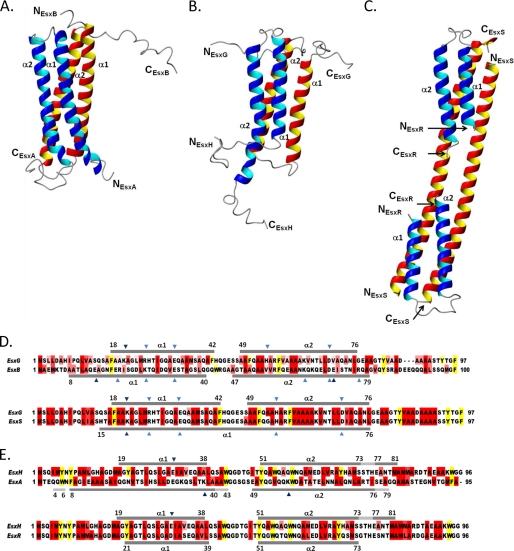
Comparison of EsxG·EsxH with family members EsxA·EsxB and EsxR·EsxS. A–C, equivalent views of ribbon representations of the solution structures of the heterodimeric EsxA·EsxB (A) and EsxG·EsxH (B) complexes, alongside the crystal structure of the heterotetrameric form of the EsxR·EsxS (C) complex. In EsxA·EsxB and EsxG·EsxH, the individual proteins form helix-turn-helix hairpin motifs, which are arranged antiparallel to each other, forming a four-helix bundle. In comparison, the domain-swapped heterotetramer of EsxR·EsxS is composed of two molecules of EsxR, which form helix-turn-helix hairpin motifs, and two molecules of EsxS, which form long antiparallel α-helices (8). The helices within the EsxG·EsxH complex, particularly the N-terminal helix of EsxH, are significantly shorter than the equivalent helices in the EsxA·EsxB complex. Among the most striking features of the EsxA·EsxB and EsxG·EsxH complexes are the flexible N and C termini of both proteins, which are not present in the EsxR·EsxS crystal structure. D and E show optimized sequence alignments of EsxG and EsxH with EsxA/B and EsxR/S, respectively. D, EsxG aligned with EsxB (32% identity) and EsxS (95% identity). E, EsxH aligned with EsxA (19% identity) and EsxR (85% identity). The α-helical regions observed in the solved complex structures are indicated by dark gray bars, and regions of 310 helix are indicated by light gray bars. Dark blue triangles indicate residues involved in intermolecular salt bridges, and light blue triangles show residues involved in intramolecular salt bridges. Residues are highlighted as follows: aliphatic residues with hydrophobic side chains (Ala, Ile, Leu, Met, and Val) are in red, and aromatic residues (Phe, Trp, and Tyr) are in yellow.
All the residues of EsxH with backbone amide resonances substantially perturbed by zinc ion binding lie either within or adjacent to the identified coordination site, and the spectral changes seen almost certainly reflect direct involvement of this region in Zn2+ chelation. In contrast, the affected but fairly distant residues within the hairpin loop of EsxG appear to have no direct role in zinc ion binding (Fig. 3, E and F), and the significant line broadening observed for backbone amide signals here suggests that Zn2+ coordination leads to a significant change in the mobility of this region, which may affect the interaction of potential functional partners with the cleft discussed previously. The specific Zn2+ binding site present on EsxG·EsxH may reflect a direct role in zinc ion acquisition but could also point to Zn2+-dependent regulation of its interaction with one or more functional partner proteins.
Comparative Analysis between EsxG·EsxH and Other M. tuberculosis Esx Complexes
Close analysis of the structure of the EsxA·EsxB complex, combined with optimized sequence alignments for members of the M. tuberculosis Esx protein family and helical wheel predictions, indicate that all M. tuberculosis Esx complexes are likely to form 1:1 heterodimers with core structures similar to the four-helix bundle of EsxA·EsxB (9, 12). Key hydrophobic residues are well conserved throughout the M. tuberculosis Esx family, allowing the complexes to adopt similar backbone structures. Residues located on the external surfaces and at the N and C termini of the proteins are somewhat less well conserved, which suggests significant differences in surface features and distinct functional sites, potentially reflecting diverse roles for Esx family complexes (14, 23–26, 30, 31).
Comparative analysis of the solution structure of EsxG·EsxH with EsxA·EsxB reveals that the overall backbone folds of the two complexes are highly similar. In both cases, the individual proteins adopt helix-turn-helix structures, which are arranged antiparallel to each other, resulting in the formation of stable four-helix bundles (Fig. 4). The similarity is reflected in comparisons of the backbone atom coordinates, which yield an r.m.s.d. of 1.8 Å for the superposition of residues Phe-18–Gln-40 and Ala-50–Leu-76 from EsxG and Ala-19–Ala-38 and Tyr-51–Ser-73 from EsxH with residues Phe-18–Gln-40 and Ala-50–Ile-76 of EsxB and Gln-19–Lys-38 and Tyr-51–Ala-73 of EsxA. However, despite obvious similarities, such as the overall folds of the protein complexes, as well as disordered N and C termini, there are some striking differences between these two related complexes. Firstly, helices of the proteins forming the EsxG·EsxH complex, in particular the N-terminal helices of EsxG and EsxH, are significantly shorter (10–11 residues) than those in the EsxA·EsxB complex (Fig. 4). Secondly, the C-terminal region of EsxB in the EsxA·EsxB complex shows a distinct tendency to adopt a helical conformation; however, in the EsxG·EsxH complex, it is the C-terminal region of EsxH (EsxA-related) that has a propensity to adopt a helical conformation. Finally, other than the flexible arms of EsxA and EsxB, no obvious functional sites were apparent on the surface of the EsxA·EsxB complex (12), whereas the EsxG·EsxH structure reveals a noticeable cleft (Fig. 2, A and B), suggesting the presence of a functional binding site, and contains a specific Zn2+ binding site not present on EsxA·EsxB.
The EsxG protein contains a modified WXG motif, where the tryptophan residue has been substituted with a histidine residue. Comparison of the WXG loops from EsxH, EsxA, and EsxB reveals significant variability in conformation, which seems to be influenced by long range contacts with their partner protein, as illustrated by EsxH (Figs. 1 and 4). The most notable difference with the modified HXG loop of EsxG is an apparent increase in flexibility as compared with the WXG loops of EsxH (Fig. 1A), EsxA, and EsxB (12). The HXG motif is conserved in EsxG orthologues from other mycobacterial species (Fig. 2C), as well as the closely related M. tuberculosis EsxS protein (Fig. 3D), suggesting that the additional flexibility of the WXG loop may be important to the function of the EsxG·EsxH complex.
Interestingly, recently reported studies of the M. tuberculosis EsxR and EsxS proteins, which share 85 and 95% sequence identity with EsxH and EsxG, respectively, have shown the formation of a major heterodimeric complex and a minor heterotetrameric complex (15:1 ratio) (8). The structure of the predominant EsxR·EsxS heterodimer has not been determined. However, due to the high level of amino acid conservation with EsxG and EsxH, it is very likely that this complex will form a structure that is essentially identical to the one reported here for the EsxG·EsxH complex. Arbing et al. (8) solved the crystal structure of the minor heterotetrameric EsxR·EsxS complex, which revealed that the two EsxS molecules each formed single, long α-helices arranged antiparallel to each other, whereas the two copies of EsxR both formed helix-turn-helix hairpin structures, which were closely associated with the ends of the EsxS pair (Fig. 4C). Overall, the positions of helical regions, as well as intra- and intermolecular salt bridges reported for the heterotetrameric complex, fit well with the helical regions and salt bridges observed for the EsxG·EsxH heterodimeric complex (Fig. 4). Comparison of the backbone coordinates show that the EsxH and EsxR molecules are highly similar as the superposition of residues 20–74 from both molecules yields an r.m.s.d. of 1.06 Å. This superposition also yields backbone r.m.s.d. values of 2.60 and 1.71 Å for residues 18–41 (α1) and residues 49–76 (α2), respectively, of EsxG as compared with EsxS. These r.m.s.d. values indicate significant similarities between the structures of the EsxG·EsxH heterodimer and the EsxR·EsxS heterotetramer, as expected for a higher order complex produced by domain swapping.
Despite clear evidence from reported gel filtration experiments for the formation of EsxR·EsxS heterotetrameric complexes (8), our studies with M. tuberculosis EsxA·EsxB and EsxG·EsxH complexes have so far found no indication of higher order complex formation. For example, during gel filtration purification (pH 6.5), we see only a single EsxA·EsxB or EsxG·EsxH peak (supplemental Fig. 1), which corresponds to the expected size of the heterodimeric complex. However, under different solution conditions, the proteins may have the potential to form domain-swapped heterotetramers like EsxR·EsxS.
Conclusions
The findings reported here, together with previous work (9, 12), clearly show that it is possible to predict with confidence a core structure for the Esx family proteins. However, significant differences seen in surface features mean that it will be necessary to solve the high resolution structures of individual complexes to identify the functionally important, complex-specific surface features. The surface features and properties of both the EsxA·EsxB and the EsxG·EsxH complexes suggest roles mediated via binding to one or more functional partner proteins, which remain to be identified and could be of either host cell or mycobacterial origin. The distinct surface features and expression profiles for the two Esx family complexes clearly point to distinct functional roles, with EsxA·EsxB strongly implicated in pathogen-host cell signaling (12, 14, 24–26) and EsxG·EsxH potentially involved in metal ion scavenging, either directly through the zinc ion binding site identified or by Zn2+-dependent modulation of its interaction with functional partners (30, 31, 33). Clearly, multiple gene duplication events in M. tuberculosis have allowed the evolution of diverse functions for Esx family complexes, which presumably exploit the flexibility offered by functional complexes.
Supplementary Material
This work was supported by program and project grants from the Wellcome Trust (Grants 080085 and 083629, respectively).
The atomic coordinates and structure factors (code 2KG7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- HSQC
- heteronuclear single quantum correlation
- r.m.s.d.
- root mean square deviation
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. World Health Organization (2009) Global Tuberculosis Control: Epidemiology, Strategy, Financing Who Report 2009, Publication WHO/HTM/TB/2009.411, Geneva, Switzerland [Google Scholar]
- 2. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 3. Garnier T., Eiglmeier K., Camus J. C., Medina N., Mansoor H., Pryor M., Duthoy S., Grondin S., Lacroix C., Monsempe C., Simon S., Harris B., Atkin R., Doggett J., Mayes R., Keating L., Wheeler P. R., Parkhill J., Barrell B. G., Cole S. T., Gordon S. V., Hewinson R. G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cole S. T., Eiglmeier K., Parkhill J., James K. D., Thomson N. R., Wheeler P. R., Honoré N., Garnier T., Churcher C., Harris D., Mungall K., Basham D., Brown D., Chillingworth T., Connor R., Davies R. M., Devlin K., Duthoy S., Feltwell T., Fraser A., Hamlin N., Holroyd S., Hornsby T., Jagels K., Lacroix C., Maclean J., Moule S., Murphy L., Oliver K., Quail M. A., Rajandream M. A., Rutherford K. M., Rutter S., Seeger K., Simon S., Simmonds M., Skelton J., Squares R., Squares S., Stevens K., Taylor K., Whitehead S., Woodward J. R., Barrell B. G. (2001) Nature 409, 1007–1011 [DOI] [PubMed] [Google Scholar]
- 5. Pallen M. J. (2002) Trends Microbiol. 10, 209–212 [DOI] [PubMed] [Google Scholar]
- 6. Berthet F. X., Rasmussen P. B., Rosenkrands I., Andersen P., Gicquel B. (1998) Microbiology 144, 3195–3203 [DOI] [PubMed] [Google Scholar]
- 7. Okkels L. M., Andersen P. (2004) J. Bacteriol. 186, 2487–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arbing M. A., Kaufmann M., Phan T., Chan S., Cascio D., Eisenberg D. (2010) Protein Sci. 19, 1692–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lightbody K. L., Ilghari D., Waters L. C., Carey G., Bailey M. A., Williamson R. A., Renshaw P. S., Carr M. D. (2008) J. Biol. Chem. 283, 17681–17690 [DOI] [PubMed] [Google Scholar]
- 10. Lightbody K. L., Renshaw P. S., Collins M. L., Wright R. L., Hunt D. M., Gordon S. V., Hewinson R. G., Buxton R. S., Williamson R. A., Carr M. D. (2004) FEMS Microbiol. Lett. 238, 255–262 [DOI] [PubMed] [Google Scholar]
- 11. Meher A. K., Bal N. C., Chary K. V., Arora A. (2006) FEBS J. 273, 1445–1462 [DOI] [PubMed] [Google Scholar]
- 12. Renshaw P. S., Lightbody K. L., Veverka V., Muskett F. W., Kelly G., Frenkiel T. A., Gordon S. V., Hewinson R. G., Burke B., Norman J., Williamson R. A., Carr M. D. (2005) EMBO J. 24, 2491–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Renshaw P. S., Panagiotidou P., Whelan A., Gordon S. V., Hewinson R. G., Williamson R. A., Carr M. D. (2002) J. Biol. Chem. 277, 21598–21603 [DOI] [PubMed] [Google Scholar]
- 14. Abdallah A. M., Gey van Pittius N. C., Champion P. A., Cox J., Luirink J., Vandenbroucke-Grauls C. M., Appelmelk B. J., Bitter W. (2007) Nat. Rev. Microbiol. 5, 883–891 [DOI] [PubMed] [Google Scholar]
- 15. Bitter W., Houben E. N., Bottai D., Brodin P., Brown E. J., Cox J. S., Derbyshire K., Fortune S. M., Gao L. Y., Liu J., Gey van Pittius N. C., Pym A. S., Rubin E. J., Sherman D. R., Cole S. T., Brosch R. (2009) PLoS Pathog. 5, e1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fortune S. M., Jaeger A., Sarracino D. A., Chase M. R., Sassetti C. M., Sherman D. R., Bloom B. R., Rubin E. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10676–10681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McLaughlin B., Chon J. S., MacGurn J. A., Carlsson F., Cheng T. L., Cox J. S., Brown E. J. (2007) PLoS Pathog. 3, e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raghavan S., Manzanillo P., Chan K., Dovey C., Cox J. S. (2008) Nature 454, 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu J., Laine O., Masciocchi M., Manoranjan J., Smith J., Du S. J., Edwards N., Zhu X., Fenselau C., Gao L. Y. (2007) Mol. Microbiol. 66, 787–800 [DOI] [PubMed] [Google Scholar]
- 20. Gey Van Pittius N. C., Gamieldien J., Hide W., Brown G. D., Siezen R. J., Beyers A. D. (2001) Genome Biol. 2, RESEARCH0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akpe San Roman S., Facey P. D., Fernandez-Martinez L., Rodriguez C., Vallin C., Del Sol R., Dyson P. (2010) Microbiology 156, 1719–1729 [DOI] [PubMed] [Google Scholar]
- 22. Stinear T. P., Seemann T., Harrison P. F., Jenkin G. A., Davies J. K., Johnson P. D., Abdellah Z., Arrowsmith C., Chillingworth T., Churcher C., Clarke K., Cronin A., Davis P., Goodhead I., Holroyd N., Jagels K., Lord A., Moule S., Mungall K., Norbertczak H., Quail M. A., Rabbinowitsch E., Walker D., White B., Whitehead S., Small P. L., Brosch R., Ramakrishnan L., Fischbach M. A., Parkhill J., Cole S. T. (2008) Genome Res. 18, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abdallah A. M., Savage N. D., van Zon M., Wilson L., Vandenbroucke-Grauls C. M., van der Wel N. N., Ottenhoff T. H., Bitter W. (2008) J. Immunol. 181, 7166–7175 [DOI] [PubMed] [Google Scholar]
- 24. Davis J. M., Ramakrishnan L. (2009) Cell 136, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van der Wel N., Hava D., Houben D., Fluitsma D., van Zon M., Pierson J., Brenner M., Peters P. J. (2007) Cell 129, 1287–1298 [DOI] [PubMed] [Google Scholar]
- 26. Volkman H. E., Clay H., Beery D., Chang J. C., Sherman D. R., Ramakrishnan L. (2004) PLoS Biol. 2, e367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frigui W., Bottai D., Majlessi L., Monot M., Josselin E., Brodin P., Garnier T., Gicquel B., Martin C., Leclerc C., Cole S. T., Brosch R. (2008) PLoS Pathog. 4, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gonzalo-Asensio J., Mostowy S., Harders-Westerveen J., Huygen K., Hernández-Pando R., Thole J., Behr M., Gicquel B., Martín C. (2008) PLoS ONE 3, e3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohol Y. M., Goetz D. H., Chan K., Shiloh M. U., Craik C. S., Cox J. S. (2010) Cell Host Microbe 7, 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Serafini A., Boldrin F., Palù G., Manganelli R. (2009) J. Bacteriol. 191, 6340–6344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Siegrist M. S., Unnikrishnan M., McConnell M. J., Borowsky M., Cheng T. Y., Siddiqi N., Fortune S. M., Moody D. B., Rubin E. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18792–18797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sassetti C. M., Boyd D. H., Rubin E. J. (2003) Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 33. Maciag A., Dainese E., Rodriguez G. M., Milano A., Provvedi R., Pasca M. R., Smith I., Palù G., Riccardi G., Manganelli R. (2007) J. Bacteriol. 189, 730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodriguez G. M., Voskuil M. I., Gold B., Schoolnik G. K., Smith I. (2002) Infect. Immun. 70, 3371–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ilghari D., Waters L. C., Veverka V., Muskett F. W., Carr M. D. (2009) Biomol. NMR Assign. 3, 171–174 [DOI] [PubMed] [Google Scholar]
- 36. Herrmann T., Güntert P., Wüthrich K. (2002) J. Biomol. NMR 24, 171–189 [DOI] [PubMed] [Google Scholar]
- 37. Güntert P., Wüthrich K. (1991) J. Biomol. NMR 1, 447–456 [DOI] [PubMed] [Google Scholar]
- 38. Güntert P., Mumenthaler C., Wüthrich K. (1997) J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 39. Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 40. Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 41. Waters L. C., Veverka V., Böhm M., Schmedt T., Choong P. T., Muskett F. W., Klempnauer K. H., Carr M. D. (2007) Oncogene 26, 4941–4950 [DOI] [PubMed] [Google Scholar]
- 42. Snow G. A. (1965) Biochem. J. 97, 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zajonc D. M., Crispin M. D., Bowden T. A., Young D. C., Cheng T. Y., Hu J., Costello C. E., Rudd P. M., Dwek R. A., Miller M. J., Brenner M. B., Moody D. B., Wilson I. A. (2005) Immunity 22, 209–219 [DOI] [PubMed] [Google Scholar]
- 44. Ebert J. C., Altman R. B. (2008) Protein Sci. 17, 54–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.